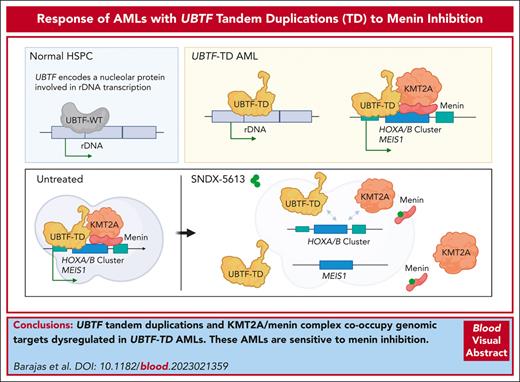
UBTF tandem duplications and KMT2A/menin complex co-occupy genomic targets dysregulated in UBTF-TD acute myeloid leukemias.
UBTF-TD acute myeloid leukemias are sensitive to menin inhibition.
Visual Abstract
UBTF tandem duplications (UBTF-TDs) have recently emerged as a recurrent alteration in pediatric and adult acute myeloid leukemia (AML). UBTF-TD leukemias are characterized by a poor response to conventional chemotherapy and a transcriptional signature that mirrors NUP98-rearranged and NPM1-mutant AMLs, including HOX-gene dysregulation. However, the mechanism by which UBTF-TD drives leukemogenesis remains unknown. In this study, we investigated the genomic occupancy of UBTF-TD in transformed cord blood CD34+ cells and patient-derived xenograft models. We found that UBTF-TD protein maintained genomic occupancy at ribosomal DNA loci while also occupying genomic targets commonly dysregulated in UBTF-TD myeloid malignancies, such as the HOXA/HOXB gene clusters and MEIS1. These data suggest that UBTF-TD is a gain-of-function alteration that results in mislocalization to genomic loci dysregulated in UBTF-TD leukemias. UBTF-TD also co-occupies key genomic loci with KMT2A and menin, which are known to be key partners involved in HOX-dysregulated leukemias. Using a protein degradation system, we showed that stemness, proliferation, and transcriptional signatures are dependent on sustained UBTF-TD localization to chromatin. Finally, we demonstrate that primary cells from UBTF-TD leukemias are sensitive to the menin inhibitor SNDX-5613, resulting in markedly reduced in vitro and in vivo tumor growth, myeloid differentiation, and abrogation of the UBTF-TD leukemic expression signature. These findings provide a viable therapeutic strategy for patients with this high-risk AML subtype.
Introduction
The upstream binding transcription factor (UBTF) gene encodes for a nucleolar protein (UBTF/UBF) involved in nucleolar formation that is primarily associated with active transcription of ribosomal DNA (rDNA).1,2UBTF is necessary for embryonic development past the morula stage, and UBTF knockout cells displayed nucleolar disassembly, abnormal heterochromatin distribution on active rDNA, and loss of rRNA synthesis.3
Recently, UBTF has gained increased interest in hematological malignancies, such as acute myeloid leukemia (AML) and B-cell acute lymphoblastic leukemia.4-11 Recurrent exon 13 tandem duplications (TD) in UBTF have emerged as a major subtype-defining genomic alteration in pediatric AML that is associated with poor prognosis4,5,11,12 and has also recently been reported in ∼3% of adults aged 18 to 60 years9 and 1.2% of all individuals over 18 years of age.10UBTF-TDs have a high variant allele frequency and are preserved during disease progression. Patients with UBTF-TD AML are associated with high measurable residual disease positivity and poor outcomes, suggesting an overall poor response to conventional chemotherapy. Although expression of UBTF-TD in cord-blood CD34+ (cbCD34+) cells leads to increased myeloid proliferation and clonogenicity,4 the molecular mechanisms by which UBTF-TD promotes leukemogenesis remain poorly understood.
Here, we use transcriptomic and epigenomic profiling to provide mechanistic insights into UBTF-TD leukemias and ultimately propose a therapeutic strategy for individuals with this high-risk AML subtype. We show that UBTF-TD/KMT2A/menin colocalize and interact with chromatin at the homeobox gene clusters and other target loci to maintain a leukemic molecular signature that is dependent on UBTF-TD genomic localization to these regions. Based on these mechanistic findings, and recent work demonstrating that HOX-dysregulated AMLs are sensitive to menin inhibitors,13-16 we hypothesized that menin inhibition is a viable therapeutic option for UBTF-TD AMLs. Our data demonstrate that UBTF-TD AMLs are sensitive to SNDX-5613 (revumenib) in vitro and in vivo. Collectively, we provide a mechanism of how UBTF-TD alterations drive myeloid leukemias and nominate menin inhibition as a therapeutic option for patients with UBTF-TD AMLs.
Methods
Cell culture and analysis of cbCD34+ cell models
UBTF-TD cbCD34+ models were generated as previously described.4 Briefly, commercially available cbCD34+ cells were obtained from Lonza (catalog no. 2C-101, Lot#1ITL248959) or isolated (cat#130-1-453, Miltenyi Biotec) from cord blood obtained from Carolinas Cord Blood Bank/ Duke University. Cells were cultured in StemSpan SFEMII media (#09655, STEMCELL Technologies) supplemented with penicillin/streptomycin, l-glutamine, and recombinant human SCF, FLT-3, TPO, and IL-6 (all 50 ng/mL PeproTech), UM729 (#72332, STEMCELL Technologies), and SR-1 (1 μmol/L, #72344, STEMCELL Technologies). Cells were transduced with lentiviral particles for MND-PGK-mCherry (MPC) vector control or MPC constructs expressing N-terminus human infuenza hemagglutinin (HA)-tagged UBTF–wild-type (WT), N-terminus HA-tagged UBTF-TD, and N-terminus FKBP12F36V-3XHA-UBTF-TD. Transduced mCherry+ cells were sorted and expanded. Molecular experiments were performed at 40 to 60 days after sorting, unless otherwise noted.
UBTF-TD patient derived xenograft model for SNDX-5613 treatment
UBTF-TD luciferase-labeled patient derived xenograft (PDX) model was generated at St Jude Children’s Research Hospital and is available through the PROPEL repository at St Jude Children’s Research Hospital (https://propel.stjude.cloud/). PDX cells were slowly thawed and resuspended in phosphate-buffered saline (PBS) + 20% fetal bovine serum (FBS). Cells were then subjected to mouse cell depletion (STEMCELL Technologies, cat# 19849), and 0.5 million live human blast cells were injected via tail vein into 20 NSG-SGM3 mice (NOD.Cg-Prkdcscid, Il2rgtm1Wjl, Tg(CMV-IL3, CSF2, KITLG)1Eav/MloySzJ, Jackson Laboratory). Mice were then monitored weekly by in vivo imaging system and biweekly by flow cytometry. At 2 weeks after transplant, we assessed human CD45% (human chimerism > 0.1%) and in vivo imaging system bioluminesence (BL) total flux p/s > 1.2E6) in 20 mice. Two mice did not show engraftment and were excluded from the study. The remaining 18 mice were then randomly split into a vehicle and SNDX group (n = 9 per group). The SNDX group was treated with SNDX-5613 (cat# HY-136175, MedChemExpress) (dissolved in 0.5% methylcellulose (type 400 cPs) in ultrapure water acidified with fumaric acid, at 10 mg/mL) at 75mg/kg by oral gavage twice daily (5 days on, 2 days off). The vehicle group was treated with vehicle control (0.5% methylcellulose (type 400 cPs) in ultrapure water acidified with fumaric acid). Treatment was continued for 5 weeks for 3 mice per group; these mice were sacrificed at week 6 of the experiment to assess molecular and pathological phenotypes. For the colony-forming unit assay, 1000 UBTF-PDX cells were plated in methylcellulose (#H4435, STEMCELL Technologies). For serial transplant experiments, cells from each group were pooled and transplanted via tail vein injection into 8-week-old NSG-SGM3 mice. Treatment for the remaining 6 mice from each group continued for 6 weeks. These mice were monitored and sacrificed at the first sign of morbidity or leukemic cell burden above thresholds. Leukemic cell burden was assessed by flow cytometry in the peripheral blood (PB), and mice were sacrificed when human CD45+ reached > 75% chimerism in the PB or when mice displayed hunched posture, lethargy, loss of appetite and weight, labored breath, and a rough coat. All animal studies, including husbandry, breeding, and experimental procedures, were performed in accordance with protocols approved by the St Jude Children’s Research Hospital Institutional Animal Care and Use Committee.
CUT&RUN and data analysis
Cleavage under targets and release using nuclease (CUT&RUN)17 was performed to assess genomic occupancy and histone modifications with antibodies for H3K4me3 (pAb cat# 13-0041, EpiCypher, 1:50), H3K27ac (mAb #8173, Cell Signaling Technology, clone: D5E4, 1:50), HA-tagged UBTF-WT or UBTF-TD (mAb #3724, Cell Signaling Technology, clone: C29F4, 1:25), UBTF (mAb Cat# sc-13125, Santa Cruz Biotechnology, clone F-9, 1:10), menin (pAb Cat# A300-105A, Bethyl Laboratories, 1:50), KMT2A (pAB A300-086A, Bethyl Laboratories, 1:50), and RNA Pol II (mAb cat# 05-623, Millipore, clone: CTD4H8, 1:50) in cbCD34+ cell models and PDXs according to the manufacturer’s protocol (cat#14-1048, EpiCypher, kit v3.3). After DNA purification, ∼5ng of DNA from each sample was subjected to DNA library prep (cat#E7103, NEB, NEBNext Ultra II DNA Library Prep Kit for Illumina) per manufacturer’s instructions for enrichment of short fragments (1.8x AMPure Beads) and sequenced on NovaSeq (PE, 100 bp, 10 million reads). To obtain quality reads, raw reads were processed and trimmed with Trim_galore (v0.4.4) from cutadapt18 and FASTQC analysis.19 A default quality score cutoff of Q20 is used. Quality-trimmed reads were then mapped to hg38+rDNA genome build20 by bwa (v0.7.12-r1039)21 and converted to a bam file by samtools (v1.2).22 Duplicate reads were marked with biobambam2 (v2.0.87).23 Uniquely mapped and properly paired reads were then extracted with samtools (v1.2) and biobambam2. Fragments with a size shorter than 2000 bp were extracted, and the center 80 bp of each fragment was used to generate bigwig track files by UCSC tools (v4)24 and visualized using IGV (v2.16.0).25 Narrow peaks were called with MACS2 (v2.1.1.20160309),26 and broad peaks were called by SICER (v1.1).27 For UBTF peak calls, artifacts at LINC02408, ST8SIA3, APBB1IP, OR2K2, PUM2, CDH11, BIK, FAM183BP, TMEM86B, and MAP3K1 were removed for all samples.
RNA sequencing
RNA sequencing (RNA-Seq) was performed in cbCD34+ expressing MPC-[FKBP12F36V-UBTF-TD] like previously done.4 Briefly, cells were treated with dimethyl sulfoxide or dTAG-13 (1μM) for 3 days. Total RNA was extracted from 1×106 cells using the Quick-RNA Miniprep Kit (cat# R1054, Zymo Research). A total of 250 ng RNA was subjected to library preparation using the TruSeq Stranded Total RNA library kit (Illumina, CA) and sequenced on NovaSeq (100 million reads, PE, 100bps).
Primary AML cells
The primary AML samples obtained with written informed consent were provided by the Princess Maxima Center for Pediatric Oncology or St. Jude Children’s Research Hospital. Five UBTF-TD samples with co-occurring FLT3-ITD and WT1 mutations and 1 sample with RUNX1::RUNX1T1 fusion were used for in vitro coculture experiments. Patient-derived AML cells were cultured in a serum-free condition on healthy bone marrow–derived mesenchymal stem cells (MSCs). MSCs were seeded at a density of 7500 cells/cm2 in Dulbecco modified Eagle medium, low glucose, GlutaMAX(TM), pyruvate (Gibco BRL, #21885) medium supplemented with 20% FBS, 8 ng/mL fibroblast growth factor-2 (PeproTech, London, United Kingdom) and 100 U/ml penicillin/streptomycin (Gibco BRL, Life Technologies, Breda, The Netherlands), and cultivated in a 37°C, 5% CO2 incubator until reaching 70% confluence. Primary AML cells were thawed and seeded over an MSC layer at a density of 5×105 cells/mL in SFEMII medium (STEMCELL Technologies, Cologne, Germany) supplemented with 100 U/mL penicillin/streptomycin (Gibco BRL), 10 ng/mL FLT3 ligand, 10 ng/mL GM-CSF, 10 ng/mL IL-3, 150 ng/mL SCF, 100 ng/mL TPO (all from PeproTech), 750 nM SR1 (Biogeme, Lausanne, Switzerland), and 1.35 μM UM729 (STEMCELL Technologies). Cocultures were maintained at 37°C with 5% CO2 and expanded by adjusting AML cell numbers to 5 × 105 cells/mL every 4 days.
Proximity ligation assay (PLA)
Duolink in situ detection reagent red (DUO92008; Sigma) was used to perform PLA in accordance with the manufacturer’s protocol. Briefly, cells were washed twice with 1X PBS, cytospun on a glass slide, fixed, permeabilized, and then incubated with blocking buffer (all reagents provided in the kit) for 1 hour. Subsequently, the cells were probed with primary antibodies against MLL (14197, Cell Signaling Technology, 1:250), UBTF (F-9, Santa Cruz #SC-13125; 1:50), and menin (AbCam ab2605, 1:250). For negative reaction controls, cells were incubated with only 1 of each antibody. The slides were washed and incubated (1 hour, 37°C), with specific plus and minus Duolink PLA probes (1:5). After washing, the slides were further incubated with ligation-ligase solution (30 min, 37°C) followed by incubation with amplification polymerase solution (2 h, 37°C). The slides were finally incubated with DAPI (300 nM) for 5 minutes in the dark and washed twice. Images were acquired using a confocal microscope (Leica, SP8) and processed by Leica LAS AF Lite software (Leica). For flow cytometry analysis of PLA, all the steps were performed in 96-well plates, and data were acquired by a Beckman Coulter CytoFLEX LX with subsequent data analysis using FlowJo software (V10.0.7; TreeStar, Ashland, OR).
Statistics
Details about statistical comparisons are provided in each figure legend. For survival data, Kaplan-Meier estimates were computed by the treatment group. The difference in survival distribution between genotypes was examined using an exact log-rank test. All the computations were done using R or GraphPad Prism, and all P values are 2-sided.
Results
UBTF-TD localizes to genomic loci that are dysregulated in UBTF-TD AMLs
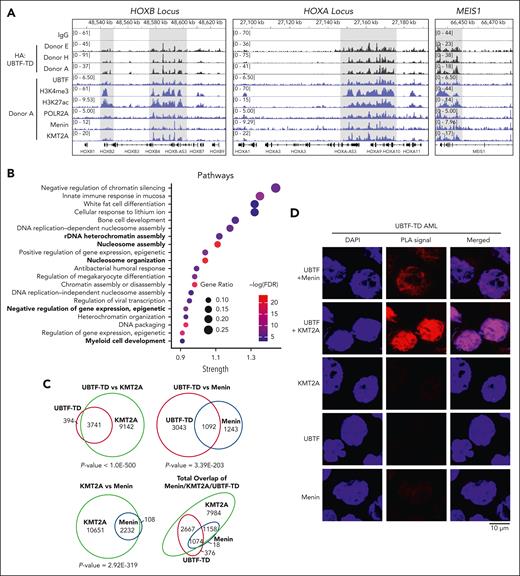
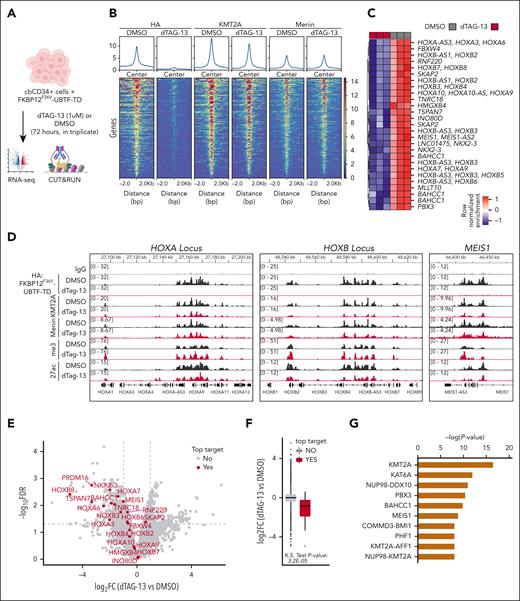
We hypothesized that UBTF-TD may interact with genomic loci that define the molecular signature of UBTD-TD AML, such as the HOXA and HOXB clusters. We, therefore, assessed the genomic occupancy of UBTF-TD using CUT&RUN.17 We first assessed the genomic distribution of UBTF-TD in cbCD34+ from 3 independent donors transduced with an N-terminus HA-tagged UBTF-TD expressing lentiviral construct (Figure 1A).4 Genome-wide annotation of UBTF-TD occupancy peaks showed that UBTF-TD binding is preferentially associated with promoters (supplemental Figure 1A-B). We then found that UBTF-TD consistently localized to regions dysregulated in UBTF-TD leukemias like HOXB, HOXA, and MEIS1 in all 3 biological replicates (supplemental Figure 1C). The peaks at these regions colocalized with H3K4me3, H3K27ac, and RNA Pol II (POLR2A) peaks, indicating active transcriptional status (Figure 1A). GO Enrichment analysis of UBTF-TD targets common in 3 biological replicates enriched in molecular pathways associated with myeloid cell development and nucleosome assembly, in addition to canonical functions of UBTF involved in rDNA activity (Figure 1B). We also assessed KMT2A and menin binding, considering their known role in HOX gene activation in certain leukemia subtypes.13,28-30 We found significant overlaps of occupied genomic targets between UBTF-TD and KMT2A (P value = 1.2E-142), UBTF-TD and menin (P value = 3.2E-95), and KMT2A and menin (P value = 2.0E-268) (Figure 1C). These nuclear colocalizations were validated in a primary UBTF-TD patient sample by PLA (Figure 1D).
UBTF-TD/menin/KMT2A co-occupy genomic loci of genes dysregulated in UBTF-TD AML. (A) IGV tracks of HA:UBTF-TD (black), demonstrating features from 3 cbCD34+ donors transduced with UBTF-TD–expressing lentiviral vectors and maintained in culture for 40 days. CUT&RUN for UBTF, H3K4me3, H3K27ac, POLR2A, menin, and KMT2A in blue were performed in donor A. (B) GO-enrichment pathway analysis of significant targets occupied by UBTF-TD in all 3 donors (n = 226). (C) Overlap of genomic regions occupied by UBTF-TD (HA), KMT2A, and menin. Significance of overlap was calculated using hypergeometric distribution. (D) In situ PLA of endogenous UBTF/menin and UBTF/KMT2A in a UBTF-TD patient sample. Single targets (menin, KMT2A, and UBTF) are shown as controls.
UBTF-TD/menin/KMT2A co-occupy genomic loci of genes dysregulated in UBTF-TD AML. (A) IGV tracks of HA:UBTF-TD (black), demonstrating features from 3 cbCD34+ donors transduced with UBTF-TD–expressing lentiviral vectors and maintained in culture for 40 days. CUT&RUN for UBTF, H3K4me3, H3K27ac, POLR2A, menin, and KMT2A in blue were performed in donor A. (B) GO-enrichment pathway analysis of significant targets occupied by UBTF-TD in all 3 donors (n = 226). (C) Overlap of genomic regions occupied by UBTF-TD (HA), KMT2A, and menin. Significance of overlap was calculated using hypergeometric distribution. (D) In situ PLA of endogenous UBTF/menin and UBTF/KMT2A in a UBTF-TD patient sample. Single targets (menin, KMT2A, and UBTF) are shown as controls.
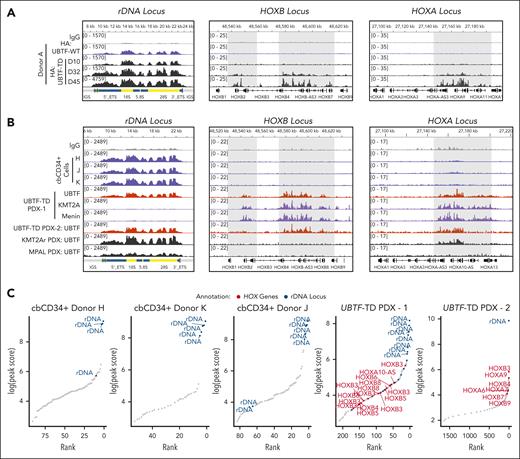
To assess whether localization to HOXA or HOXB loci and other regions that characterize UBTF-TD AMLs was unique to UBTF-TD and not UBTF-WT, we first compared the genomic binding of HA-tagged UBTF-WT or HA-tagged UBTF-TD using an HA antibody in a cbCD34+ lentiviral expression model (Figure 2A). We could clearly detect both UBTF-WT and UBTF-TD at rDNA loci at early time points when introduced into primary cbCD34+ cells (day 10 after transduction and sorting). Although we did not observe strong UBTF-TD occupancy at the HOXA or HOXB loci until day 32, some signal could be detected at these loci on day 10 for UBTF-TD but not for UBTF-WT. UBTF-WT expression in cbCD34+ does not transform cells, and cells begin to differentiate in vitro beyond day 20, and thus not enough material can be collected for a similar analysis.4 Alternatively, we tested endogenous UBTF occupancy in 3 independent biological replicates of normal cbCD34+ cells, 2 aggressive UBTF-TD patient-derived xenograft (PDX) models, a KMT2A-rearranged (KMT2A-r) PDX (KMT2A::MLLT3 fusion with UBTF-WT), and a mixed phenotype leukemia (MPAL with UBTF-WT) using an antibody that detects both UBTF-WT and UBTF-TD (Figure 2B). These data show that UBTF is bound at rDNA in normal hematopoietic progenitor cells as well as in UBTF-TD-PDX, KMT2A-r-PDX, and MPAL-PDX. However, UBTF was detected at HOXB and HOXA regions only in the UBTF-TD PDX cells and at lower levels at the HOXA locus in the KMT2A-r-PDX but not in normal cbCD34+ cells or MPAL PDX. Furthermore, like in the cbCD34+ UBTF-TD model, we also observed that KMT2A and menin colocalize at HOXB and HOXA loci with UBTF-TD (Figure 2B). We then assessed the top UBTF peaks localized to promoters or rDNA in these samples and found that UBTF was enriched at rDNA, but not HOXA/HOXB regions, in normal cbCD34+ cells (Figure 2C). In contrast, both UBTF-TD PDX included top peaks for both rDNA and the HOXA/HOXB loci. These collective data demonstrate that the localization of UBTF to rDNA regions is a shared feature of both UBTF-WT and UBTF-TD, but that localization to targets dysregulated in UBTF-TD AMLs, such as the HOXA and HOXB clusters, is enhanced by UBTF-TD.
UBTF is localized to HOXA/HOXB regions in UBTF-TD AML but not in normal hematopoietic progenitors. (A) Genomic occupancy of UBTF-WT and UBTF-TD in cbCD34+ cells. UBTF occupancy was assessed via HA:CUT&RUN in cbCD34+ cells expressing HA-UBTF-WT (blue) or HA:UBTF-TD at days 10 (D10) for UBTF-WT and UBTF-TD, as well as day 32 (D32) and day 45 (D45) for UBTF-TD. (B) IGV tracks of UBTF at the rDNA, HOXB, and HOXA genomic loci of normal cbCD34+ cells from 3 biological replicates (blue), 2 different UBTF-TD PDX (colored in red), a KMT2A-r PDX (KMT2A::MLLT3, colored in black), and an MPAL PDX (black). (C) Rank ordered peak scores of UBTF peaks subset to promoters or rDNA in normal cbCD34+ cells, UBTF-TD PDX from B (UBTF-TD PDX–1), and the second UBTF-TD PDX (UBTF-TD PDX–2). Peaks at HOXA/HOXB (red) or rDNA (blue) are annotated.
UBTF is localized to HOXA/HOXB regions in UBTF-TD AML but not in normal hematopoietic progenitors. (A) Genomic occupancy of UBTF-WT and UBTF-TD in cbCD34+ cells. UBTF occupancy was assessed via HA:CUT&RUN in cbCD34+ cells expressing HA-UBTF-WT (blue) or HA:UBTF-TD at days 10 (D10) for UBTF-WT and UBTF-TD, as well as day 32 (D32) and day 45 (D45) for UBTF-TD. (B) IGV tracks of UBTF at the rDNA, HOXB, and HOXA genomic loci of normal cbCD34+ cells from 3 biological replicates (blue), 2 different UBTF-TD PDX (colored in red), a KMT2A-r PDX (KMT2A::MLLT3, colored in black), and an MPAL PDX (black). (C) Rank ordered peak scores of UBTF peaks subset to promoters or rDNA in normal cbCD34+ cells, UBTF-TD PDX from B (UBTF-TD PDX–1), and the second UBTF-TD PDX (UBTF-TD PDX–2). Peaks at HOXA/HOXB (red) or rDNA (blue) are annotated.
UBTF-TD expression and genomic localization is required for myeloid cell expansion
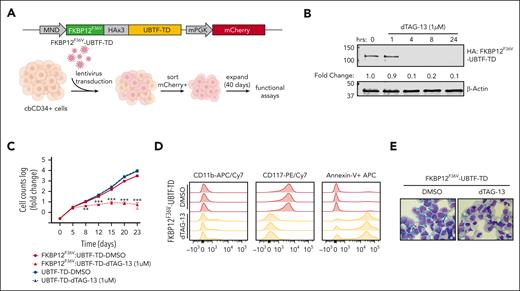
We next sought to determine if UBTF-TD expression and aberrant genomic localization are driving and maintaining a leukemic phenotype. To test this, we first designed a FKBP12F36V::HA-UBTF-TD fusion construct, which allows for rapid degradation of UBTF-TD protein upon dTAG-13 treatment,31 and expressed it in cbCD34+ cells (Figure 3A). We found that the FKBP12F36V tag did not impair the ability of UBTF-TD to promote transformation in cbCD34+ cells, as measured by cell proliferation and colony-forming unit assays, and that levels of expression of this FKBP12F36V::HA-UBTF-TD were comparable to endogenous UBTF (supplemental Figure 2A-C). Treatment with dTAG-13 resulted in rapid degradation (< 4 hours) of UBTF-TD protein (Figure 3B) and induced a decline in proliferation in vitro (Figure 3C). This change was accompanied by morphologic and immunophenotypic features of myeloid differentiation, including an increase in CD11b expression and a loss of stem cell marker CD117, along with an increase in apoptotic cells as measured by annexin V staining (Figure 3D-E). This contrasts with cells that express untagged UBTF-TD, which were unaffected by dTAG-13 treatment (supplemental Figure 2D).
Leukemic transformation is dependent on UBTF-TD expression. (A) Schematic of FKBP12F36V-HA-UBTF-TD lentiviral construct and generation of cbCD34+ UBTF-TD degradation model. (B) Time-course degradation of FKBP12F36V-HA-UBTF-TD after treatment with dTAG-13 (1 μM) in cbCD34+ cells. Immunoblot with HA- and β-actin–specific antibodies. (C) Growth rate after dTAG-13 treatment. cbCD34+ cells expressing FKBP12F36V-HA-UBTF-TD or HA-UBTF-TD were counted over time while treated with DMSO or 1μM dTAG-13. Log of fold change was calculated from starting cell number of 200K cells. (∗∗∗ indicates P value < .001; ∗∗ indicates P value < .01, after adjustment by 2-stage step-up method (Benjamini, Krieger, and Yekutieli). (D) Immunophenotyping of cells from panel C after 10 days of treatment. (E) Wright-Giemsa staining to assess cellular morphology of cells from panels A and B after 10 days of treatment. DMSO, dimethyl sulfoxide.
Leukemic transformation is dependent on UBTF-TD expression. (A) Schematic of FKBP12F36V-HA-UBTF-TD lentiviral construct and generation of cbCD34+ UBTF-TD degradation model. (B) Time-course degradation of FKBP12F36V-HA-UBTF-TD after treatment with dTAG-13 (1 μM) in cbCD34+ cells. Immunoblot with HA- and β-actin–specific antibodies. (C) Growth rate after dTAG-13 treatment. cbCD34+ cells expressing FKBP12F36V-HA-UBTF-TD or HA-UBTF-TD were counted over time while treated with DMSO or 1μM dTAG-13. Log of fold change was calculated from starting cell number of 200K cells. (∗∗∗ indicates P value < .001; ∗∗ indicates P value < .01, after adjustment by 2-stage step-up method (Benjamini, Krieger, and Yekutieli). (D) Immunophenotyping of cells from panel C after 10 days of treatment. (E) Wright-Giemsa staining to assess cellular morphology of cells from panels A and B after 10 days of treatment. DMSO, dimethyl sulfoxide.
To further validate if genomic occupancy of UBTF-TD to dysregulate in UBTF-TD AML was driving the leukemic phenotype, we performed CUT&RUN and RNA-seq after a 3-day treatment with 1 μM dTAG-13 (Figure 4A). This treatment was previously determined to be sufficient to degrade UBTF-TD but not long enough for phenotypic changes in proliferation, cell death, or differentiation to take place (supplemental Figure 3A-E). This treatment also had no detectable effect on normal cbCD34+ cells (supplemental Figure 3F-G). After the 3-day treatment with dTAG-13, UBTF-TD occupancy was significantly reduced in 266 regions (false discovery rate < 0.05) (Figure 4B). Importantly, the top UBTF-TD–depleted regions (n = 29) were loci dysregulated in UBTF-TD leukemias, such as the HOXA/HOXB and MEIS1 loci (Figure 4C-D). However, KMT2A binding was only slightly reduced, and menin displayed little to no change in occupancy, suggesting that loss of UBTF-TD in these regions does not immediately disrupt KMT2A/menin localization. Yet, loss of UBTF-TD occupancy was associated with decreased mRNA levels for these top targets (Figure 4E-F), suggesting a dependency on UBTF-TD for the maintenance of active transcription but not for KMT2A and menin localization. Furthermore, upstream network analysis revealed that significantly depleted UBTF-TD target genes are those predicted to be regulated by KMT2A (Figure 4G). Despite these findings, no interaction motif present in most of the regions could be identified for UBTF-TD (supplemental Figure 4A-B). This is similar to previous reports showing that DNA binding of UBTF-WT is not driven by a specific motif.32 Collectively, we show that UBTF-TD localization to DNA loci dysregulated in UBTF-TD AML is necessary for leukemic cell expansion and expression of the UBTF-TD molecular signature.
UBTF-TD genomic localization is required for gene activation. (A) Experimental design of FKBP12F36V-HA-UBTF-TD genomic occupancy experiment. (B) Tornado plots depicting the merged genomic occupancy (n = 3 replicates) at significantly depleted HA: FKBP12F36V-HA-UBTF-TD target regions (n = 266 regions, FDR < 0.5) after treatment with dTAG-13 for 3 days. Occupancy for KMT2A and menin are also shown. (C) Heatmap of the top depleted regions and their closest genes (n = 29). Heatmap colors depict row normalized enrichment. (D) Genomic tracks of merged coverage (n = 3) of HA: FKBP12F36V-HA-UBTF-TD cells treated with DMSO (black) or dTAG-13 (red) for HA, KMT2A, menin, H3K27ac (27ac), or H3K4me3 (me3). (E) Volcano plot of differentially expressed genes in FKBP12F36V-HA-UBTF-TD cells treated with DMSO or dTAG-13. Top targets (29 genes from panel C) are annotated in red. (F) mRNA levels of top target genes from panel C as compared with nontargets. (G) Upstream regulator analysis from ingenuity pathway analysis. Top targets from panel B were used for the prediction. FDR, false discovery rate; DMSO, dimethyl sulfoxide.
UBTF-TD genomic localization is required for gene activation. (A) Experimental design of FKBP12F36V-HA-UBTF-TD genomic occupancy experiment. (B) Tornado plots depicting the merged genomic occupancy (n = 3 replicates) at significantly depleted HA: FKBP12F36V-HA-UBTF-TD target regions (n = 266 regions, FDR < 0.5) after treatment with dTAG-13 for 3 days. Occupancy for KMT2A and menin are also shown. (C) Heatmap of the top depleted regions and their closest genes (n = 29). Heatmap colors depict row normalized enrichment. (D) Genomic tracks of merged coverage (n = 3) of HA: FKBP12F36V-HA-UBTF-TD cells treated with DMSO (black) or dTAG-13 (red) for HA, KMT2A, menin, H3K27ac (27ac), or H3K4me3 (me3). (E) Volcano plot of differentially expressed genes in FKBP12F36V-HA-UBTF-TD cells treated with DMSO or dTAG-13. Top targets (29 genes from panel C) are annotated in red. (F) mRNA levels of top target genes from panel C as compared with nontargets. (G) Upstream regulator analysis from ingenuity pathway analysis. Top targets from panel B were used for the prediction. FDR, false discovery rate; DMSO, dimethyl sulfoxide.
UBTF-TD leukemias are sensitive to menin inhibition
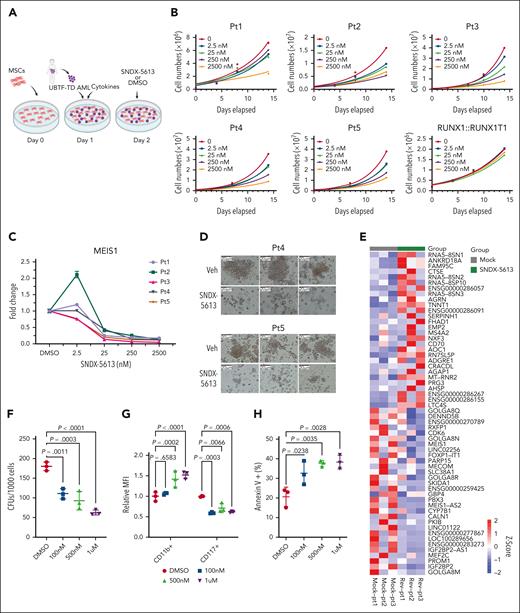
The above molecular findings suggested that UBTF-TD leukemias may be sensitive to menin inhibitors like other HOX-dysregulated leukemias.33 This was also indicated by a compound screen in our cbCD34+ model, consisting of 160 epigenetic compounds and 32 compounds commonly used to treat AML, in which we found that UBTF-TD–expressing cells were sensitive to the first-generation menin inhibitor MI-2-2 (supplemental Table 1). Furthermore, other compounds that directly interfere with KMT2A or components of the COMPASS complex also scored as possible hits (eg, MI-503). We also observed that treatment of the UBTF-TD CD34+ cell model with SNDX-5613 (revumenib), a clinical-grade menin inhibitor, led to a variable reduction of UBTF-TD and KMT2A binding (supplemental Figure 5A-C). In contrast to dTAG-13, SNDX-5613 also markedly reduced menin binding, as previously reported for menin inhibitors in KMT2A-rearranged AMLs.13 SNDX-5613 has shown promising results in preclinical models of NPM1-mutated KMT2A-r13,14 and is currently in phase 1/2 clinical trials16,34 for refractory KMT2A-r or NPM1-mut leukemias, and preclinical studies have recently also shown efficacy in NUP98-rearranged (NUP98-r) leukemias, which also share a similar molecular signature to UBTF-TD AMLs.4,35,36 Using an ex vivo culture system with 5 patient samples harboring UBTF-TDs (Figure 5A; supplemental Table 2), we found a dose-dependent inhibition of growth with SNDX-5613 treatment in all 5 UBTF-TD samples (Figure 5B), with an average inhibitory concentration 50 of 19.8 nM (supplemental Figure 6A). In contrast, a primary AML with a RUNX1::RUNX1T1 fusion, a subtype without HOX gene dysregulation, showed no response to SNDX-5613 (Figure 5B). Decreased cell growth in UBTF-TD samples was accompanied by a dose-dependent decrease in MEIS1 mRNA levels (Figure 5C), which could be maintained over time (supplemental Figure 6B-C). SNDX-5613 also resulted in decreased UBTF-TD/KMT2A/menin interactions (supplemental Figure 6D). Furthermore, primary AML samples treated with SNDX-5613 had reduced colony-forming capacity compared with the vehicle-treated samples (Figure 5D). We assessed the transcriptional consequence on 3 of these primary AMLs and found that treatment with SNDX-5613 for 7 days induced global gene expression changes (Figure 5E), including a reduction in the expression of genes like MEIS1, PBX3, IGF2BP2, PROM1, and MEF2C.37-40 We also observed that SNDX-5613 reduced the colony-forming potential of a UBTF-TD PDX model (Figure 5F). These cells also displayed decreased stem-cell marker CD117 (KIT) expression, increased CD11b expression, and increased cell death as measured by annexin V+ cells (Figure 5G-H).
UBTF-TD leukemias are sensitive to menin inhibitors. (A) Schema for in vitro culture of primary UBTF-TD AMLs. (B) Primary tumors from 5 individual patients (Pt1-5) harboring a UBTF-TD alteration treated with menin inhibitor SNDX-5613 in vitro. Primary sample from a single patient with a RUNX1::RUNX1T1 alteration was used as a control. (C) MEIS1 mRNA steady-state levels in patient cells treated with SNDX-5613. (D) CFU capacity of UBTF-TD leukemias after exposure to SNDX-5613. Cells from Pt4 and Pt5 were treated with SNDX-5613 (250nM, SFEMII media) for 12 days and then plated in methylcellulose (#H4435, STEMCELL Technologies). (E) Heatmap depicting mRNA levels of the top 30 differentially expressed genes after 7 days of treatment with SNDX-5613. The divergent color map depicts row normalized expression. (F) CFU of UBTF-TD PDX cells plated in methylcellulose (H4435, STEMCELL) and treated with DMSO or SNDX-5613 (100nM, 500nM, 1μM). (G) Immunophenotyping of cells from panel F for stem-cell marker CD117 and CD11b. (H) Annexin V+ staining of cells from panel F. For panels F and G, statistical significances were calculated using a 1-way analysis of variance test with Dunnett’s multiple comparison adjustment using DMSO as control. CFU, colony forming unit; DMSO, dimethyl sulfoxide.
UBTF-TD leukemias are sensitive to menin inhibitors. (A) Schema for in vitro culture of primary UBTF-TD AMLs. (B) Primary tumors from 5 individual patients (Pt1-5) harboring a UBTF-TD alteration treated with menin inhibitor SNDX-5613 in vitro. Primary sample from a single patient with a RUNX1::RUNX1T1 alteration was used as a control. (C) MEIS1 mRNA steady-state levels in patient cells treated with SNDX-5613. (D) CFU capacity of UBTF-TD leukemias after exposure to SNDX-5613. Cells from Pt4 and Pt5 were treated with SNDX-5613 (250nM, SFEMII media) for 12 days and then plated in methylcellulose (#H4435, STEMCELL Technologies). (E) Heatmap depicting mRNA levels of the top 30 differentially expressed genes after 7 days of treatment with SNDX-5613. The divergent color map depicts row normalized expression. (F) CFU of UBTF-TD PDX cells plated in methylcellulose (H4435, STEMCELL) and treated with DMSO or SNDX-5613 (100nM, 500nM, 1μM). (G) Immunophenotyping of cells from panel F for stem-cell marker CD117 and CD11b. (H) Annexin V+ staining of cells from panel F. For panels F and G, statistical significances were calculated using a 1-way analysis of variance test with Dunnett’s multiple comparison adjustment using DMSO as control. CFU, colony forming unit; DMSO, dimethyl sulfoxide.
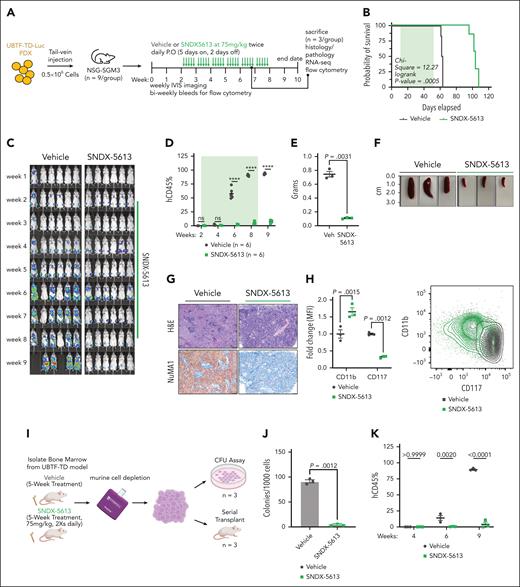
We next assessed the in vivo efficacy of menin inhibition using the UBTF-TD PDX model previously tested in vitro (Figure 6A). Treatment with SNDX-5613 significantly increased the survival of these mice compared to vehicle controls (vehicle: median survival = 62.5 days, SNDX-5613: median survival = 104 days, log-rank P value = .00005) (Figure 6B). This was accompanied by a decrease in tumor burden as measured by total body luminescence and human chimerism in the peripheral blood (Figure 6C-D; supplemental Figure 7A). To accurately compare the molecular and phenotypic changes associated with SNDX-5613 treatment, we sacrificed a subset of the mice from the vehicle and SNDX-5613 groups after 5 weeks of treatment (n = 3 per group). SNDX-treated mice had smaller spleens and less spleen infiltration (Figure 6E-G). Evaluation of cells in the bone marrow from these mice showed that the transplanted cells had increased CD11b and reduced CD117 expression, suggesting that SNDX-5613 promotes myeloid differentiation in UBTF-TD leukemias (Figure 6H). Human cells isolated from these mice also displayed increased mRNA levels of myeloid differentiation genes (MPO, ITGAM) and decreased mRNA levels of hematopoietic stem cell–associated genes (KIT, PBX3) (supplemental Figure 7B). Cells harvested from the bone marrow of recipient mice treated with SNDX-5613 had markedly reduced in vitro colony-forming capacity and secondary reconstitution potential in vivo compared with cells harvested from vehicle-treated primary mice (Figure 6I-K). Collectively, these data show that menin inhibition is a viable therapeutic option for AMLs with UBTF-TD alterations that induce differentiation and reduce their leukemic potential.
Preclinical model of UBTF-TD leukemia shows in vivo sensitivity to menin inhibitor SNDX-5613. (A) Schematic of in vivo SNDX-5613 treatment of UBTF-TD PDX. (B) Kaplan-Meier curves of UBTF-TD PDX model treated with vehicle or SNDX-5613 (n = 6 per group). Treatment period is shaded in green. (C) IVIS images of mice treated with vehicle or SNDX-5613 over time. Treatment period is shaded in green. Luminescence is row normalized to each time point. (D) Human CD45 chimerism (% of live) in the peripheral blood of mice from panel B. Treatment period is shaded in green (∗∗∗∗ indicates P value < .0001, 2-stage step-up Benjamini, Krieger, and Yekutieli test). (E) Spleen weight of mice harvested after 5 weeks of treatment with SNDX-5613 or vehicle (n = 3 per group) (2-tailed unpaired t test). (F) Spleen size of mice from panel E. (G) H&E and human NuMA1 IHC staining of spleens from panel F. (H) Flow cytometry analysis of bone marrow isolated from mice from panel D (P values are calculated with 2-tailed unpaired t test). MFI of CD11b-APC/Cy7 and CD117-PE/Cy7 (left panel) and representative flow plot (right panel) are shown (P values are calculated with 2-tailed unpaired t test). (I) Schematic of serial transplant experiment. Cells from each group in panel E were plated onto methylcellulose (#H4435, STEMCELL Technologies) (n = 3) or serially transplant into NSG-SGM3 mice (n = 3). (J) CFU capacity of cells from panel I (P value was calculated using an unpaired, 2-tailed t test). (K) Peripheral blood chimerism of mice from panel I. P values were calculated using 2-stage step-up Benjamini, Krieger, and Yekutieli test. CFU, colony-forming unit; IVIS, in vivo imaging system; H&E, hematoxylin and eosin; IHC, immunohistochemistry; MFI, mean fluorescent intensity; P.O, per os.
Preclinical model of UBTF-TD leukemia shows in vivo sensitivity to menin inhibitor SNDX-5613. (A) Schematic of in vivo SNDX-5613 treatment of UBTF-TD PDX. (B) Kaplan-Meier curves of UBTF-TD PDX model treated with vehicle or SNDX-5613 (n = 6 per group). Treatment period is shaded in green. (C) IVIS images of mice treated with vehicle or SNDX-5613 over time. Treatment period is shaded in green. Luminescence is row normalized to each time point. (D) Human CD45 chimerism (% of live) in the peripheral blood of mice from panel B. Treatment period is shaded in green (∗∗∗∗ indicates P value < .0001, 2-stage step-up Benjamini, Krieger, and Yekutieli test). (E) Spleen weight of mice harvested after 5 weeks of treatment with SNDX-5613 or vehicle (n = 3 per group) (2-tailed unpaired t test). (F) Spleen size of mice from panel E. (G) H&E and human NuMA1 IHC staining of spleens from panel F. (H) Flow cytometry analysis of bone marrow isolated from mice from panel D (P values are calculated with 2-tailed unpaired t test). MFI of CD11b-APC/Cy7 and CD117-PE/Cy7 (left panel) and representative flow plot (right panel) are shown (P values are calculated with 2-tailed unpaired t test). (I) Schematic of serial transplant experiment. Cells from each group in panel E were plated onto methylcellulose (#H4435, STEMCELL Technologies) (n = 3) or serially transplant into NSG-SGM3 mice (n = 3). (J) CFU capacity of cells from panel I (P value was calculated using an unpaired, 2-tailed t test). (K) Peripheral blood chimerism of mice from panel I. P values were calculated using 2-stage step-up Benjamini, Krieger, and Yekutieli test. CFU, colony-forming unit; IVIS, in vivo imaging system; H&E, hematoxylin and eosin; IHC, immunohistochemistry; MFI, mean fluorescent intensity; P.O, per os.
Discussion
UBTF has a well-established role in rRNA biogenesis and nucleoli formation1,2 but only recently have alterations in UBTF been recognized in leukemias. In particular, UBTF tandem duplications have been characterized as subtype-defining genomic alterations in childhood AML associated with HOX gene dysregulation.4,5,11,12 Recent studies have now shown that UBTF-TDs are not restricted to pediatric AML but also occur in ∼ 3% of adult AML patients aged 18 to 60 years (median age = 37 years).9,10UBTF alterations have also recently been described in B-cell acute lymphoblastic leukemia, although these are structural variations leading to focal deletions of exons 18 to 21, resulting in UBTF::ATXN7L3 fusions.6-8 The recent identification of novel UBTF alterations in hematological malignancies highlights a critical need to understand how UBTF alterations drive leukemogenesis.
At the transcriptional level, UBTF-TD AMLs are similar to other well-defined AML subtypes, including NUP98-rearranged (NUP98-r, ie, NUP98::NSD1), NPM1 mutations, DEK::NUP214, KMT2A-rearranged (KMT2A-r), and KMT2A partial tandem duplication (KMT2A-PTD) AMLs.4,12 The similarities in the transcriptional profiles of these AML subtypes suggest a shared mechanism. Similar to AMLs with NPM1 mutations or KMT2A fusion oncoproteins,16 we demonstrate that UBTF-TD, KMT2A, and menin co-occupy genomic regions that are transcriptionally dysregulated in UBTF-TD leukemias (Figures 1 and 2). Our dTAG strategy demonstrates that UBTF-TD is necessary to maintain leukemic phenotypes. Considering that UBTF-WT is primarily localized to rDNA in normal cbCD34+ cells and not to genomic regions dysregulated in UBTF-TD AMLs, like the HOXA or HOXB clusters, these collective data suggest that aberrant localization of UBTF is a key event in the pathogenesis of these leukemias and reflects a gain-of-function activity for the mutant UBTF.
Disrupting the KMT2A interaction with its cofactor, menin, has antileukemic activity in NUP98-r, mutant NPM1, and KMT2A-r AMLs.35,41,42 Based on the similarity of HOX gene expression profiles and the presented mechanistic studies, we determined that UBTF-TD AML cells are also dependent on the KMT2A/menin complex. Here, we show that degradation of UBTF-TD minimally affects the genomic binding of KMT2A and menin, yet disrupting the menin-KMT2A interaction with SNDX-5613 reduces binding of menin and has variable affects on genome binding of UBTF-TD and KMT2A. These data suggest that the integrity of the KMT2A-menin complex is critical to maintaining the molecular properties of cells transformed by UBTF-TD, and the presence of UBTF-TD is necessary for leukemia maintenance and blocking myeloid differentiation. Although the mechanism by which UBTF-TD interacts with the genome is currently not clear, our data and the previous observations that HOXA and HOXB genes are expressed in normal CD34+ cells,43 along with KMT2A binding to these regions in CD34+ cells,44 suggest that recruitment of UBTF-TD could be mediated by KMT2A or its interacting partners.
From a clinical perspective, these data demonstrate that UBTF-TD AMLs can be recognized as a new menin inhibitor–responsive leukemia subtype in future clinical studies. This could be critical to overcoming the dismal outcomes for children with UBTF-TD AMLs. Recent studies that identified UBTF-TD at a relatively high incidence in adults with AML also broadened the impact of these findings. These data also support the idea that other AML subtypes with similar HOX gene expression profiles, such as AMLs with DEK::NUP214, may also be sensitive to menin inhibitors.
Here, we propose a mechanism where the tandem duplications within exon 13 of UBTF observed in AML result in a gain-of-function activity defined by aberrant genomic localization. UBTF-TD/KMT2A/menin co-occupy target genes that characterize the UBTF-TD molecular signature and present a potential therapeutic opportunity through disruption of the KMT2A-menin interaction by clinically available menin inhibitors, whose efficacy was confirmed in preclinical models in this study. Therefore, we propose menin inhibition as a therapeutic strategy for UBTF-TD leukemias, which can be a key advance in the clinical management of patients with these AMLs, considering their refractoriness to conventional chemotherapy.
Acknowledgments
The authors thank the Hematological Malignancies Program within the St. Jude Comprehensive Cancer Center and the Center for In Vivo Imaging and Therapeutics (Melissa Johnson and Jamila Moore) for help and support with the preclinical in vivo studies.
The work was funded by the American Lebanese and Syrian Associated Charities of St. Jude Children's Research Hospital and funds from the National Institutes of Health (NIH), including NIH, National Heart, Lung, and Blood Institute grant 5F32HL154636-02 (J.M.B.), and KiKa program grant 329 (O.H.). The studies were also funded by the Jane Coffin Childs Fund (J.M.B.). J.M.K. holds a career award for medical scientists from the Burroughs Welcome Fund. Support was also provided by shared resources through the St. Jude Comprehensive Cancer Center (P30-CA21765), Flow cytometry and Cell Sorting, Comparative Pathology Core, and Genome Sequencing (Hartwell Center).
The content of this article does not necessarily represent the official views of the NIH and is solely the responsibility of the authors.
Authorship
Contribution: J.M.K. and O.H. conceptualized the study; J.M.B., M.R., M.U., R.H., S.A., B.A., T.W., I.I., B.X., T.-C.C., W.R., O.H., and J.M.K. performed the methodology; B.X., T.-C.C, and W.R. were responsible for software; J.M.B., M.R., R.H., B.A., and R.K. validated the data; M.B., B.X., T.-C.C., W.R., M.P.W., J.M., M.R, A.K.-H., R.M., and M.A. conducted formal analysis; J.M.B., M.R., R.H., B.A., M.E.T., E.X., C.R., J.E.R. and R.K. conducted investigation; O.H. and J.M.K. provided resources; G.S., M.P.W., and T.W. curated the data; J.M.B. wrote the original draft; all authors were involved in review and editing of the manuscript; J.M.B. and M.R. visualized the data; O.H. and J.M.K. supervised the study; J.M.B., M.R., S.A., I.I., O.H., and J.M.K. were responsible for project administration; and J.M.B., O.H., and J.M.K. acquired funding.
Conflict-of-interest disclosure: J.E.R. has been a consultant for Biomea Fusion. The remaining authors declare no competing financial interests.
Correspondence: Jeffery M. Klco, Department of Pathology, St. Jude Children’s Research Hospital, Mail Stop 342, Room D4047B, 262 Danny Thomas Place, Memphis, TN 38105-3678; email: jeffery.klco@stjude.org; and Olaf Heidenreich, Princess Maxima Center for Pediatric Oncology, Heidelberglaan 25, 3584 CS Utrecht, The Netherlands; email: o.t.heidenreich@prinsesmaximacentrum.nl.
References
Author notes
∗J.M.B. and M.R. contributed equally to this study as first authors.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal