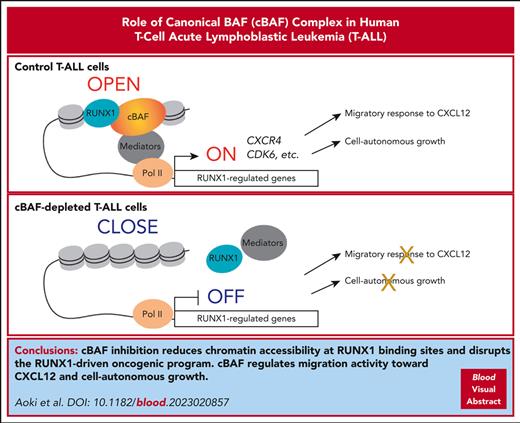
cBAF inhibition reduces chromatin accessibility mainly at RUNX1 binding sites and disrupts the RUNX1-driven oncogenic program in T-ALL.
cBAF regulates migration activity toward CXCL12 and cell-autonomous growth in T-ALL cells, thus representing a promising therapeutic target.
Visual Abstract
Acute leukemia cells require bone marrow microenvironments, known as niches, which provide leukemic cells with niche factors that are essential for leukemic cell survival and/or proliferation. However, it remains unclear how the dynamics of the leukemic cell–niche interaction are regulated. Using a genome-wide CRISPR screen, we discovered that canonical BRG1/BRM-associated factor (cBAF), a variant of the switch/sucrose nonfermenting chromatin remodeling complex, regulates the migratory response of human T-cell acute lymphoblastic leukemia (T-ALL) cells to a niche factor CXCL12. Mechanistically, cBAF maintains chromatin accessibility and allows RUNX1 to bind to CXCR4 enhancer regions. cBAF inhibition evicts RUNX1 from the genome, resulting in CXCR4 downregulation and impaired migration activity. In addition, cBAF maintains chromatin accessibility preferentially at RUNX1 binding sites, ensuring RUNX1 binding at these sites, and is required for expression of RUNX1-regulated genes, such as CDK6; therefore, cBAF inhibition negatively impacts cell proliferation and profoundly induces apoptosis. This anticancer effect was also confirmed using T-ALL xenograft models, suggesting cBAF as a promising therapeutic target. Thus, we provide novel evidence that cBAF regulates the RUNX1-driven leukemic program and governs migration activity toward CXCL12 and cell-autonomous growth in human T-ALL.
Introduction
Acute leukemias, including acute myeloid leukemia (AML), B-cell acute lymphoblastic leukemia (B-ALL), and T-cell acute lymphoblastic leukemia (T-ALL), are aggressive neoplasms arising from hematopoietic cells.1 Developments in chemotherapy and stem cell transplantation have improved event-free survival of patients with these diseases; however, the prognosis of relapsed or treatment-refractory acute leukemias remains poor.2,3 Therefore, development of novel therapeutic strategies for these leukemias are needed. Similar to normal hematopoietic stem cells (HSCs), acute leukemia cells reside in the bone marrow and are in contact with and maintained by special microenvironments, known as niches, during leukemogenesis and even after treatment in most of the cases.4-9 Bone marrow niches provide leukemic cells with niche factors critical for leukemic cell survival and/or proliferation.4,6,10 In murine AML and T-ALL models, Cxcl12 deletion in bone marrow niche cells led to impairment of leukemogenesis.4,10 In xenograft models bearing human acute leukemia, knockdown or knockout of CXCR4 in leukemic cells or blocking CXCR4 impairs leukemic cell growth and prolongs the survival of mice.4,11-13 These results indicate that CXCL12 is one of the most important niche factors for acute leukemias. However, it is not fully understood how the dynamics of the leukemic cell–niche interaction are regulated.
Switch/sucrose nonfermenting (SWI/SNF) complex is 1 of the 4 major families of nucleosome remodeling complexes in mammals, namely SWI/SNF, imitation switch, chromodomain helicase DNA binding, and INO80, all of which use energy from adenosine triphosphate (ATP) hydrolysis to reposition or eject nucleosomes, allowing factors involving transcription, replication, and DNA repair to access genomic DNA.14-17 The SWI/SNF complex is thought to exist as 3 main variants: the ARID1A/ARID1B-containing canonical BRG1/BRM-associated factor (cBAF), the ARID2-containing polybromo-associated BAF, and the BRD9-containing noncanonical BAF (ncBAF).18 All variants contain 1 of the 2 ATPase subunits, SMARCA4 and SMARCA2.18 Pharmacological inhibition of BRD9 or knockdown of SMARCA4 downregulates MYC expression by decreasing the occupancy of hematopoietic transcription factors (TFs) at MYC distal enhancer sites, which impairs leukemic cell growth in AML.19,20 In prostate cancer, degradation of SMARCA4/2 rapidly compacts cis-regulatory elements bound by TFs required for cancer cell proliferation and inhibits tumor growth.21 Thus, SWI/SNF inhibition impairs tumor cell growth in a cell-autonomous manner and represents an emerging attractive therapeutic strategy against cancers that are dependent on SWI/SNF.
Here, using a genome-wide CRISPR screen, we discovered that cBAF is essential for the migration activity of human T-ALL cells toward CXCL12. Additionally, cBAF is required for cell-autonomous growth of T-ALL cells. Mechanistically, cBAF inhibition impaired the accessibility at RUNX1 binding sites, abolished RUNX1 binding to its target regions, and downregulated the expression of RUNX1-regulated genes. Thus, we provide evidence that cBAF involves the T-ALL oncogenic program and that this vulnerability can be exploited for a novel therapeutic approach.
Methods
Detailed methodology is provided in the supplemental Methods (available on the Blood website).
Ethics statement
This study was approved by the Ethics Committee of Kyoto University Graduate School and Faculty of Medicine (G1030), the Animal Research Committee of Institute for Life and Medical Sciences, Kyoto University (Y-20-1-2), and the Animal Research Committee of Graduate School of Medicine, Kyoto University (Medkyo22269). All experiments performed in this study conformed to the Declaration of Helsinki and ethical guidelines of Kyoto University. All patients participating in this study signed informed consent.
Cell culture
Jurkat, KOPT-K1, CCRF-CEM, and HPB-ALL cells were cultured in RPMI-1640 supplemented with 10% fetal bovine serum (FBS). TALL-1 cells were cultured in RPMI-1640 supplemented with 15% FBS. Cells from patient-derived xenografts (PDXs) were cultured in minimum essential medium α, supplemented with 5% FBS, 5% human AB serum, human interleukin-7 (hIL-7; 10 ng/mL), human Fms-related tyrosine kinase 3 ligand (20 ng/mL), hIL-2 (100 ng/mL), and human stem cell factor (50 ng/mL).
Lentivirus production
Lentiviruses were produced using 293FT cells and Lipofectamine LTX (Thermo Fisher Scientific, 15338-100).
CRISPR screening
CRISPR screening was performed using the human v3 library.22-24 Cas9-expressing Jurkat cells were transduced with the genome-wide guide RNA lentiviral supernatant, selected with puromycin, and used in migration assays.
Results
A CRISPR screen identifies the cBAF complex as an essential factor for human T-ALL migration toward CXCL12
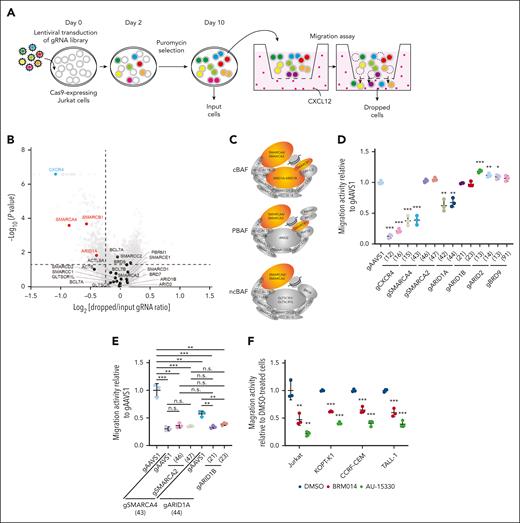
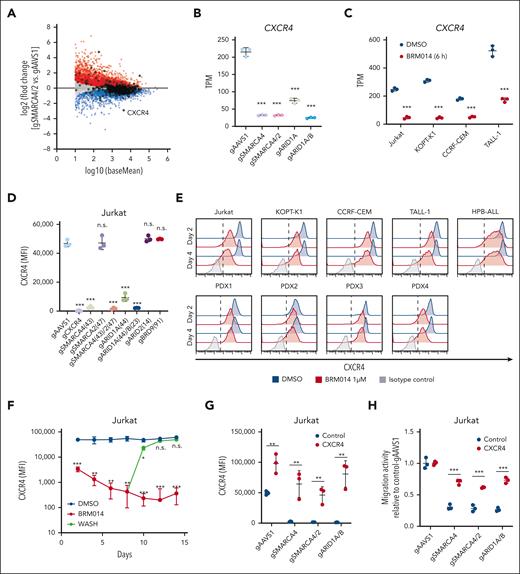
To identify genes involved in the regulation of CXCL12 responsiveness of T-ALL cells, we conducted a CRISPR-Cas9 screen22-24 using a transwell migration assay with Jurkat cells (Figure 1A-B; supplemental Tables 1 and 2). We identified that CXCR4 (Rank 1), which encodes the main physiological receptor for CXCL12, and genes related to the Rho pathway, including RIC8A (Rank 2) and RHOA (Rank 60), were highly depleted from the migrated cells, confirming the robustness of our screen (supplemental Figure 1). Among other top hits, we focused on SMARCA4 (Rank 83), which encodes a core ATPase of SWI/SNF chromatin remodeling complex. ARID1A (Rank 446), encoding a component of cBAF, and SMARCB1 (Rank 76), encoding a component of cBAF and polybromo-associated BAF, were also significantly depleted, suggesting that among the 3 SWI/SNF variants, cBAF is most likely to be required for migration (Figure 1B-C).
A genome-wide CRISPR-Cas9 knockout screen identified the cBAF complex as an essential factor for leukemic cell migration toward CXCL12. (A) Screening strategy to identify genes essential for leukemic cell migration toward CXCL12. (B) Volcano plot showing the gRNA ratio (dropped/input) vs P value obtained from MAGeCK analysis. Depleted and nondepleted genes coding SWI/SNF subunits are highlighted in red and black, respectively. CXCR4, highlighted in blue, is a positive control. (C) Subunits of cBAF, PBAF, and ncBAF. Subunits identified as screening hits are highlighted in orange. (D-E) Migration activity of Jurkat cells in which the indicated genes are knocked out individually (D) or in combination (E). Cells expressing gAAVS1 were used as a control. Data are shown as mean ± SD (n = 3). (F) Migration activity of BRM014- or AU-15330–treated cells. Cells were treated at 1 μM for 3 days and then used for migration assays. Data are normalized to those of DMSO-treated cells and are shown as mean ± SD (n = 3). Two-tailed Student t test was used to assess statistical significance in panels D, E, and F (∗∗∗P < .001; ∗∗P < .01; ∗P < .05; n.s., not significant). DMSO, dimethyl sulfoxide; gRNA, guide RNA; MAGeCK, model-based analysis of genome-wide CRISPR-Cas9 knockout; PBAF, polybromo-associated BAF; SD, standard deviation.
A genome-wide CRISPR-Cas9 knockout screen identified the cBAF complex as an essential factor for leukemic cell migration toward CXCL12. (A) Screening strategy to identify genes essential for leukemic cell migration toward CXCL12. (B) Volcano plot showing the gRNA ratio (dropped/input) vs P value obtained from MAGeCK analysis. Depleted and nondepleted genes coding SWI/SNF subunits are highlighted in red and black, respectively. CXCR4, highlighted in blue, is a positive control. (C) Subunits of cBAF, PBAF, and ncBAF. Subunits identified as screening hits are highlighted in orange. (D-E) Migration activity of Jurkat cells in which the indicated genes are knocked out individually (D) or in combination (E). Cells expressing gAAVS1 were used as a control. Data are shown as mean ± SD (n = 3). (F) Migration activity of BRM014- or AU-15330–treated cells. Cells were treated at 1 μM for 3 days and then used for migration assays. Data are normalized to those of DMSO-treated cells and are shown as mean ± SD (n = 3). Two-tailed Student t test was used to assess statistical significance in panels D, E, and F (∗∗∗P < .001; ∗∗P < .01; ∗P < .05; n.s., not significant). DMSO, dimethyl sulfoxide; gRNA, guide RNA; MAGeCK, model-based analysis of genome-wide CRISPR-Cas9 knockout; PBAF, polybromo-associated BAF; SD, standard deviation.
To validate the variant specificity, we individually knocked out essential components of each variant along with CXCR4, SMARCA4, and SMARCA2 (supplemental Figure 2A), and tested their migration activity (Figure 1D). SMARCA4 and CXCR4 knockouts showed marked impairment of migration activity, whereas ARID1A knockout showed a significant but milder defect. In contrast, SMARCA2, ARID1B, ARID2, and BRD9 knockouts did not show impairment. Because SMARCA4 and ARID1A are known to be functionally redundant with SMARCA2 and ARID1B, respectively, we generated SMARCA4/2 and ARID1A/B double knockouts. SMARCA4/2 knockout had a phenotype similar to that of SMARCA4 knockout, indicating that SMARCA2 is dispensable in this context (Figure 1E). In contrast, ARID1A/B knockout exhibited a severer defect than ARID1A knockout to a level equivalent to that of SMARCA4 knockout, indicating that ARID1A and ARID1B are functionally redundant but ARID1B contributes less (Figure 1E). These findings indicate that cBAF, rather than the other SWI/SNF variants, is required for the migratory response of Jurkat cells.
Additionally, we tested whether BRM014, a SMARCA4/2 ATPase inhibitor,25 and AU-15330,21 a proteolysis-targeting chimera (PROTAC) degrader of SMARCA4/2, inhibit migration activity in various human T-ALL cell lines (supplemental Figure 2B). All 4 cell lines treated with BRM014 or AU-15330 showed significant impairment in migration activity (Figure 1F). Taken together, our CRISPR screen identified that cBAF is essential for human T-ALL cell migration toward CXCL12.
cBAF is required for the maintenance of the chromatin accessibility at RUNX1 binding sites, RUNX1 binding to the chromatin, and the expression of RUNX1-regulated genes
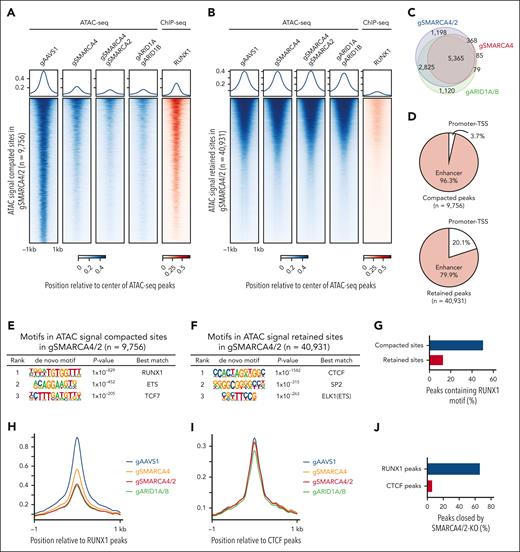
We next profiled the effects of SMARCA4, SMARCA4/2, and ARID1A/B knockouts and BRM014 treatment on physical chromatin accessibility using assay for transposase-accessible chromatin followed by sequencing (ATAC-seq). We detected ∼50 000 nonredundant ATAC-seq peaks (supplemental Table 3). Of these, a significant loss in chromatin accessibility was detected at ∼10 000 sites in SMARCA4/2 knockout, whereas the remaining ∼40 000 genomic sites showed little to no change (Figure 2A-B). In SMARCA4 single knockout, ∼6 000 genomic sites showed a significant loss of chromatin accessibility and were mostly (97.2%) included in the compacted sites of SMARCA4/2 knockout, indicating that SMARCA4 plays a predominant role at these sites (Figure 2A-C). Changes in chromatin accessibility induced by 6-hour BRM014 treatment were mostly similar to those induced by 8-day treatment or SMARCA4/2 knockout, indicating that pharmacological inhibition of SMARCA4/2 phenocopies the genetic deletion and results in a rapid and durable loss of accessibility (supplemental Figure 3A-C). Similar to SMARCA4/2 knockout, ARID1A/B knockout showed significant loss at ∼10 000 sites, which were substantially overlapped with the compacted sites in SMARCA4/2 knockout (Figure 2A-C), indicating that the changes in chromatin accessibility induced by SMARCA4/2 perturbation are mainly due to a defect in cBAF. Consistent with the previous finding that the cBAF complex is preferentially bound to cell-specific enhancer sites,26-30 >95% of the sites compacted by SMARCA4/2 knockout were located in distal regions, whereas ∼80% of the retained sites were located in distal regions (Figure 2D).
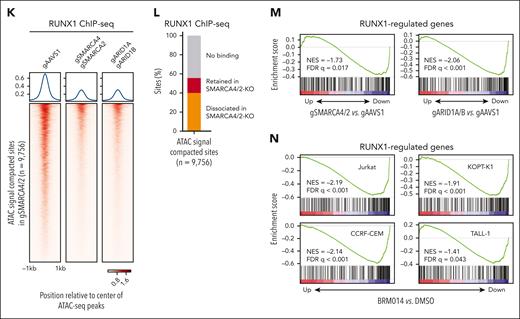
Inhibition of cBAF activity impairs accessibility at RUNX1 binding sites, RUNX1 binding to the chromatin, and inhibits the RUNX1 transcriptional program. (A-B) ATAC-seq read-density heat maps from the indicated knockout Jurkat cells and ChIP-seq read-density heat maps of RUNX1 from Jurkat cells at genomic sites that are compacted (A, 9756 sites) or retained (B, 40 931 sites) in SMARCA4/2 knockout cells compared with control cells (gAAVS1). (C) Venn diagram showing the overlap between genomic sites compacted in the indicated knockouts. (D) Pie charts showing the fractions of promoter- and enhancer-associated compacted (top) and retained (bottom) sites identified in SMARCA4/2 knockout cells. (E-F) Top 3 motifs enriched within genomic sites compacted (E) or retained (F) in SMARCA4/2 knockout cells (HOMER, hypergeometric test, ranked by P value). (G) The proportions of the compacted and retained sites containing the RUNX1 motif. (H-I) Changes in chromatin accessibility at RUNX1 (H) and CTCF (I) binding sites in the indicated knockouts. (J) The proportion of RUNX1 and CTCF peaks compacted by SMARCA4/2 knockout. (K) ChIP-seq read-density heat maps of RUNX1 in the indicated knockout and control Jurkat cells at the compacted sites. (L) The proportion in the compacted sites relative to RUNX1 binding. (M-N) GSEA on transcriptomes of SMARCA4/2 and ARID1A/B knockout Jurkat cells (M) and BRM014-treated human T-ALL cell lines (N). Cells were treated with BRM014 at 1 μM for 6 hours (N). An in-house gene set consisting of RUNX1-regulated genes (supplemental Table 4) was used in panels M and N. GSEA, gene set enrichment analysis.
Inhibition of cBAF activity impairs accessibility at RUNX1 binding sites, RUNX1 binding to the chromatin, and inhibits the RUNX1 transcriptional program. (A-B) ATAC-seq read-density heat maps from the indicated knockout Jurkat cells and ChIP-seq read-density heat maps of RUNX1 from Jurkat cells at genomic sites that are compacted (A, 9756 sites) or retained (B, 40 931 sites) in SMARCA4/2 knockout cells compared with control cells (gAAVS1). (C) Venn diagram showing the overlap between genomic sites compacted in the indicated knockouts. (D) Pie charts showing the fractions of promoter- and enhancer-associated compacted (top) and retained (bottom) sites identified in SMARCA4/2 knockout cells. (E-F) Top 3 motifs enriched within genomic sites compacted (E) or retained (F) in SMARCA4/2 knockout cells (HOMER, hypergeometric test, ranked by P value). (G) The proportions of the compacted and retained sites containing the RUNX1 motif. (H-I) Changes in chromatin accessibility at RUNX1 (H) and CTCF (I) binding sites in the indicated knockouts. (J) The proportion of RUNX1 and CTCF peaks compacted by SMARCA4/2 knockout. (K) ChIP-seq read-density heat maps of RUNX1 in the indicated knockout and control Jurkat cells at the compacted sites. (L) The proportion in the compacted sites relative to RUNX1 binding. (M-N) GSEA on transcriptomes of SMARCA4/2 and ARID1A/B knockout Jurkat cells (M) and BRM014-treated human T-ALL cell lines (N). Cells were treated with BRM014 at 1 μM for 6 hours (N). An in-house gene set consisting of RUNX1-regulated genes (supplemental Table 4) was used in panels M and N. GSEA, gene set enrichment analysis.
We next examined TF motifs that were enriched at retained and compacted sites in SMARCA4/2 knockout and identified DNA binding motifs for CTCF and RUNX1 as top hits, respectively (Figure 2E-F). CTCF is known to function as a transcriptional activator, suppressor, and insulator. Consistent with our finding, it has been shown that the accessibility at CTCF binding sites is regulated by imitation switch rather than SWI/SNF.31 RUNX1 is a member of the RUNX family of TFs, predominantly expressed in hematopoietic lineage cells and plays central roles in embryonic and adult hematopoiesis32,33 and leukemogenesis in T-ALL.34,35 Approximately half of the compacted sites contain RUNX1 motifs (Figure 2G). In agreement with the motif analysis, chromatin immunoprecipitation followed by sequencing (ChIP-seq) analysis revealed the significant binding of RUNX1 at the compacted sites (Figure 2A-B). RUNX1 binding sites are shown to be co-occupied by ETS1 at a juxtaposed position, thereby forming a composite binding site.36 Consistent with this observation, we also found that ETS was ranked second after RUNX1 (Figure 2E).
To confirm the enrichment of these motifs, we performed a reciprocal analysis and found that accessibility at the RUNX1, but not CTCF, binding sites were reduced by the knockouts (Figure 2H-J). Inhibition of SWI/SNF activity by BRM014 showed comparable patterns of chromatin accessibility at both RUNX1 and CTCF binding sites (supplemental Figure 3D-E).
The above results strongly suggest that inhibition of cBAF activity has a global impact on RUNX1 binding to the chromatin and RUNX1-regulated gene expression. As expected, ChIP-seq analysis revealed a significant decrease in RUNX1 binding at the compacted sites in SMARCA4/2 and ARID1A/B knockouts (Figure 2K-L). We next performed RNA sequencing analysis of the knockouts and cells treated with BRM014. A gene set consisting of RUNX1-regulated genes (supplemental Table 4) was negatively enriched in these knockouts (Figure 2M). This enrichment was consistently observed in all 4 T-ALL cell lines treated with BRM014 for 6 hours (Figure 2N). The transcriptomic changes in SMARCA4/2 and ARID1A/B knockouts were highly concordant (R = 0.59), and these knockouts showed significantly overlapped differentially expressed genes (supplemental Figure 4A-B). The changes by 8-day BRM014 treatment were highly correlated with those induced by SMARCA4/2 knockout (R = 0.84; supplemental Figure 4C-D), confirming that BRM014 treatment mimics SMARCA4/2 deletion at the transcriptome level as well. Among the genes downregulated by 8-day BRM014 treatment, around one-third of the genes were acutely downregulated by 6-hour treatment (supplemental Figure 4E-F). None of the RUNX family genes were downregulated at the RNA level by SWI/SNF perturbation (supplemental Figure 5A-B). RUNX1 protein was also expressed at the same level as control cells in Jurkat cells when treated with BRM014 for up to 6 hours (supplemental Figure 5C). However, RUNX1 protein levels were slightly decreased in cells treated with BRM014 for ≥12 hours and in the knockout cells (supplemental Figure 5C-D). Taken together, these results indicate that loss of accessibility at the RUNX1 binding sites by cBAF inhibition led to the eviction of RUNX1 from the genome, which caused the downregulation of RUNX1-regulated genes.
CXCR4 downregulation by cBAF inhibition results in impaired migration of T-ALL cells
We next investigated the mechanism by which the cBAF complex regulates migration activity. Among the genes identified as essential for migration activity, CXCR4 showed the largest reduction in expression by SMARCA4/2 knockout (Figure 3A). SMARCA4 and ARID1A/B knockouts also downregulated CXCR4 expression, whereas ARID1A knockout showed a partial reduction (Figure 3B). BRM014 treatment markedly decreased CXCR4 expression in all 4 T-ALL cell lines tested (Figure 3C). Consistent with the messenger RNA level, cell surface expression of CXCR4 was also decreased in SMARCA4, SMARCA4/2, and ARID1A/B knockout cells (Figure 3D). In contrast, ARID2 and BRD9 knockouts did not affect the CXCR4 expression level, further supporting the notion that cBAF plays a primary role in this context. BRM014 treatment also downregulated cell surface CXCR4 expression in all human T-ALL cell lines and PDX cells tested (Figure 3E; supplemental Figure 6). Interestingly, BRM014 withdrawal rapidly restored cell surface CXCR4 expression to its original level (Figure 3F), indicating that continuous inhibition of cBAF is required for durable downregulation of CXCR4 expression.
Decreased expression of CXCR4 by cBAF inhibition impairs leukemic cell migration toward CXCL12. (A) MA plot showing differentially expressed genes in SMARCA4/2 knockout Jurkat cells. Significantly (adjusted P < .05) upregulated and downregulated genes are highlighted in red and blue, respectively. Genes identified in the migration screen (Figure 1B) are circled in black. (B-C) CXCR4 expression levels in the indicated knockout Jurkat cells (B) and BRM014-treated T-ALL cell lines (C) by RNA-seq analysis. Data are shown as mean ± SD (n = 3). (D) Cell surface CXCR4 expression on the indicated knockout Jurkat cells. Expression was determined by flow cytometry. Data are shown as mean ± SD (n = 3). (E) Cell surface CXCR4 expression on T-ALL cell lines and T-ALL PDX cells treated with BRM014 for 2 or 4 days. Expression was determined by flow cytometry. (F) Cell surface CXCR4 expression on Jurkat cells after BRM014 withdrawal. Expression was determined by flow cytometry. (G) Cell surface CXCR4 expression on the indicated knockout Jurkat cells with exogenous CXCR4 or control cDNA expression. Data are shown as mean ± SD (n = 3). (H) Migration activity toward CXCL12 of the indicated knockout Jurkat cells with exogenous CXCR4 expression. AAVS1 knockout cells were used as a control. Data are shown as mean ± SD (n = 3). Two-tailed Student t tests were used to assess statistical significance in panels B, C, D, F, G, and H (∗∗∗P < .001; ∗∗P < .01; ∗P < .05; n.s., not significant). MA, log ratio and mean average; RNA-seq, RNA sequencing; SD, standard deviation.
Decreased expression of CXCR4 by cBAF inhibition impairs leukemic cell migration toward CXCL12. (A) MA plot showing differentially expressed genes in SMARCA4/2 knockout Jurkat cells. Significantly (adjusted P < .05) upregulated and downregulated genes are highlighted in red and blue, respectively. Genes identified in the migration screen (Figure 1B) are circled in black. (B-C) CXCR4 expression levels in the indicated knockout Jurkat cells (B) and BRM014-treated T-ALL cell lines (C) by RNA-seq analysis. Data are shown as mean ± SD (n = 3). (D) Cell surface CXCR4 expression on the indicated knockout Jurkat cells. Expression was determined by flow cytometry. Data are shown as mean ± SD (n = 3). (E) Cell surface CXCR4 expression on T-ALL cell lines and T-ALL PDX cells treated with BRM014 for 2 or 4 days. Expression was determined by flow cytometry. (F) Cell surface CXCR4 expression on Jurkat cells after BRM014 withdrawal. Expression was determined by flow cytometry. (G) Cell surface CXCR4 expression on the indicated knockout Jurkat cells with exogenous CXCR4 or control cDNA expression. Data are shown as mean ± SD (n = 3). (H) Migration activity toward CXCL12 of the indicated knockout Jurkat cells with exogenous CXCR4 expression. AAVS1 knockout cells were used as a control. Data are shown as mean ± SD (n = 3). Two-tailed Student t tests were used to assess statistical significance in panels B, C, D, F, G, and H (∗∗∗P < .001; ∗∗P < .01; ∗P < .05; n.s., not significant). MA, log ratio and mean average; RNA-seq, RNA sequencing; SD, standard deviation.
To confirm that CXCR4 is responsible, we performed a rescue experiment by first introducing human CXCR4 complementary DNA (cDNA) into Jurkat-Cas9 cells and then knocking out SMARCA4, SMARCA4/2, or ARID1A/B. The expression of CXCR4 was maintained above the wild-type level in these cDNA-rescued knockout cells (Figure 3G). In the migration assay, exogenous CXCR4 expression nearly fully rescued the impaired migration of the knockouts (Figure 3H). However, the rescue was incomplete, suggesting a minor contribution of other mechanisms that are affected by cBAF inhibition. Together, these results demonstrate that cBAF inhibition results in the loss of CXCR4 expression, leading to functional impairment of migration in human T-ALL cells.
RUNX1-cBAF–mediated enhancer activity is required for CXCR4 expression
Given that cBAF inhibition affects RUNX1 binding and the expression of RUNX1-regulated genes, RUNX1 may act as a TF for CXCR4 expression in human T-ALL cells, and cBAF plays a pivotal role in RUNX1 binding to CXCR4 enhancers. As hypothesized, RUNX1, but not ETS1, knockout decreased CXCR4 expression (Figure 4A; supplemental Figure 7A). Because Jurkat cells predominantly express RUNX1 but also express RUNX3 at a lower level (supplementary Figure 5A-B and Choi et al35), we examined RUNX1/3 double knockout and found a more profound decrease in CXCR4 expression than that of RUNX1 single knockout to a level comparable with that of SMARCA4/2 and ARID1A/B knockouts (Figure 4A). Consistent with this, RUNX1/3 knockout hampered migration (Figure 4B). RUNX3 knockout neither decreased CXCR4 expression nor affected migration activity. Thus, RUNX1 plays a major role in this context. It is known that GATA3 and TAL1 co-occupy the genomic sites with RUNX1, and these 3 TFs cooperatively regulate the target genes.34 In agreement with this, GATA3 and TAL1 binding was also enriched at the sites compacted upon cBAF inhibition (supplemental Figure 7B-C). Although TAL1 and GATA3 are required for proliferation,34 these knockouts did not downregulate CXCR4 expression (supplemental Figure 7A). These results indicate that CXCR4 expression is predominantly regulated by the cBAF-RUNX1 axis.
Inhibition of cBAF activity hampers RUNX1 binding to the CXCR4 enhancers and downregulates CXCR4 expression. (A-B) Cell surface CXCR4 expression (A) and migration activity (B) of the indicated knockout Jurkat cells. Expression was determined by flow cytometry. Cells expressing gAAVS1 were used as a control. Data are shown as mean ± SD (n = 3). (C) ATAC-seq and ChIP-seq data in a 35-kb region including CXCR4 enhancer regions (SE [intron 1], E1 [−13.5 kb], E2 [−16.2 kb], E3 [−18.1 kb], and E4 [−18.6 kb]) and an NR3C1 motif (see “Discussion”). (D-E) Cell surface expression of CXCR4 (D) and migration activity (E) of Jurkat cells in which the indicated RUNX1 binding sites were disrupted by the CRISPR-Cas9 technology. Data are shown as mean ± SD (n = 3). E1 disruption also significantly decreased CXCR4 expression but the change was negligible (D). (F-H) ChIP-qPCR analysis of RUNX1 (F, H) and H3K27ac (G) at the CXCR4 SE and E4 enhancers in the indicated knockout Jurkat cells (F, G) and Jurkat cells treated with DMSO and BRM014 at 1 μM for 6 hours (H). The CTCF binding element 9-kb upstream of SMARCC1 was used as a control region. Data are shown as mean ± SD (n = 3-4). Two-tailed Student t test was used to assess statistical significance in panels A, B, D, E, F, G, and H (∗∗∗P < .001; ∗∗P < .01; ∗P < .05). DMSO, dimethyl sulfoxide; qPCR, quantitative polymerase chain reaction; SD, standard deviation.
Inhibition of cBAF activity hampers RUNX1 binding to the CXCR4 enhancers and downregulates CXCR4 expression. (A-B) Cell surface CXCR4 expression (A) and migration activity (B) of the indicated knockout Jurkat cells. Expression was determined by flow cytometry. Cells expressing gAAVS1 were used as a control. Data are shown as mean ± SD (n = 3). (C) ATAC-seq and ChIP-seq data in a 35-kb region including CXCR4 enhancer regions (SE [intron 1], E1 [−13.5 kb], E2 [−16.2 kb], E3 [−18.1 kb], and E4 [−18.6 kb]) and an NR3C1 motif (see “Discussion”). (D-E) Cell surface expression of CXCR4 (D) and migration activity (E) of Jurkat cells in which the indicated RUNX1 binding sites were disrupted by the CRISPR-Cas9 technology. Data are shown as mean ± SD (n = 3). E1 disruption also significantly decreased CXCR4 expression but the change was negligible (D). (F-H) ChIP-qPCR analysis of RUNX1 (F, H) and H3K27ac (G) at the CXCR4 SE and E4 enhancers in the indicated knockout Jurkat cells (F, G) and Jurkat cells treated with DMSO and BRM014 at 1 μM for 6 hours (H). The CTCF binding element 9-kb upstream of SMARCC1 was used as a control region. Data are shown as mean ± SD (n = 3-4). Two-tailed Student t test was used to assess statistical significance in panels A, B, D, E, F, G, and H (∗∗∗P < .001; ∗∗P < .01; ∗P < .05). DMSO, dimethyl sulfoxide; qPCR, quantitative polymerase chain reaction; SD, standard deviation.
To explore the molecular mechanisms how the cBAF-RUNX1 axis regulates CXCR4 expression, we analyzed our ATAC-seq, as well as RUNX1 and H3K27ac ChIP-seq data, focusing on a region surrounding the CXCR4 gene. We found 4 RUNX1-bound putative enhancers upstream of the CXCR4 transcription start site and 1 in intron 1, which is located within the region reported as the CXCR4 super enhancer (SE) in B-cell lymphoma37 (Figure 4C). SMARCA4/2 and ARID1A/B knockouts led to reduced accessibility and RUNX1 binding at these sites (Figure 4C). Among them, disruption of RUNX1 motifs in SE and E4 resulted in a significant reduction in CXCR4 expression (Figure 4D). Sequential double disruption of SE and E4 resulted in a more profound reduction in CXCR4 expression and severer migration impairment than single disruption (Figure 4D-E). These results indicate that SE and E4 are the major functional enhancers in which RUNX1 drives CXCR4 expression. ChIP followed by quantitative polymerase chain reaction confirmed significant reductions in RUNX1 binding to the SE and E4 elements upon SMARCA4/2 or ARID1A/B disruption (Figure 4F), which were accompanied by depletion of the H3K27ac marks (Figure 4G). RUNX1 dissociation was also confirmed at the acute phase of cBAF inhibition (Figure 4H). Taken together, these results indicate that cBAF activity is required for RUNX1 binding to CXCR4 enhancers and the maintenance of CXCR4 expression in human T-ALL cells.
cBAF inhibition impairs human T-ALL growth in a cell-autonomous manner
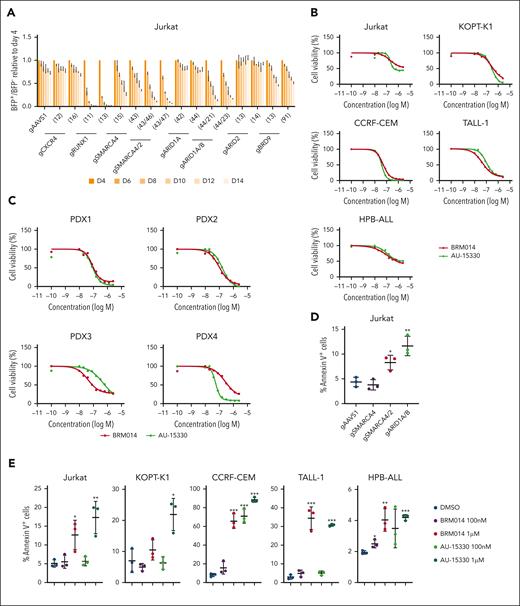
It has been shown that RUNX1 knockdown inhibits cell-autonomous growth in T-ALL cells.34,35 Given that cBAF inhibition broadly affects RUNX1-regulated gene expression, the cBAF complex may also play an essential role in cell proliferation. We performed a competition-based proliferation assay and found that SMARCA4/2 and ARID1A/B knockouts, as well as RUNX1 knockout, were rapidly outcompeted by nontransduced cells (Figure 5A). SMARCA4 single knockout was also outcompeted, albeit at a slower rate. Although ARID2 knockout showed normal proliferation, BRD9 knockout showed a weak but significant growth defect. These results suggest that cBAF is the major contributor to the proliferation of Jurkat cells, whereas ncBAF plays a minor role. CXCR4 knockout did not result in a growth disadvantage (Figure 5A). Moreover, exogenous expression of CXCR4 did not rescue the impaired growth of SMARCA4/2 and ARID1A/B knockouts (supplemental Figure 8), suggesting that CXCR4 is not involved in in vitro cell proliferation. Consistent with the results of the genetic perturbation, treatment of human T-ALL cell lines with BRM014 or AU-15330 inhibited growth with average 50% inhibitory concentration values of 102 nM and 124 nM, respectively (Figure 5B). We also observed that both compounds suppressed the growth of T-ALL PDX cells with average 50% inhibitory concentration values of 100 nM and 136 nM, respectively (Figure 5C). SMARCA4/2 and ARID1A/B, but not SMARCA4, knockouts increased the percentage of annexin V–positive cells (Figure 5D). Treatment with BRM014 or AU-15330 also increased the percentage of annexin V–positive cells in all human T-ALL cell lines tested in a dose-dependent manner (Figure 5E). Collectively, these data indicate that cBAF activity is required for cell proliferation and survival of human T-ALL cells in a cell-autonomous manner.
cBAF inhibition impairs leukemic growth in a cell-autonomous manner. (A) Fourteen-day competitive coculture assay of Jurkat cells transduced with gRNAs targeting the indicated genes. Data are shown as mean ± SD (n = 3). (B-C) Drug sensitivity assay of BRM014 and AU-15330 in the indicated T-ALL cell lines (B) and T-ALL PDX cells (C). Data are shown as mean (n = 3). (D-E) Percentage of annexin V–positive in the indicated knockout Jurkat cells (D) and human T-ALL cell lines treated with BRM014 or AU-15330 (E). Data are shown as mean ± SD (n = 3). Two-tailed Student t test was used to assess statistical significance in panels D and E (∗∗∗P < .001; ∗∗P < .01; ∗P < .05). gRNA, guide RNA; SD, standard deviation.
cBAF inhibition impairs leukemic growth in a cell-autonomous manner. (A) Fourteen-day competitive coculture assay of Jurkat cells transduced with gRNAs targeting the indicated genes. Data are shown as mean ± SD (n = 3). (B-C) Drug sensitivity assay of BRM014 and AU-15330 in the indicated T-ALL cell lines (B) and T-ALL PDX cells (C). Data are shown as mean (n = 3). (D-E) Percentage of annexin V–positive in the indicated knockout Jurkat cells (D) and human T-ALL cell lines treated with BRM014 or AU-15330 (E). Data are shown as mean ± SD (n = 3). Two-tailed Student t test was used to assess statistical significance in panels D and E (∗∗∗P < .001; ∗∗P < .01; ∗P < .05). gRNA, guide RNA; SD, standard deviation.
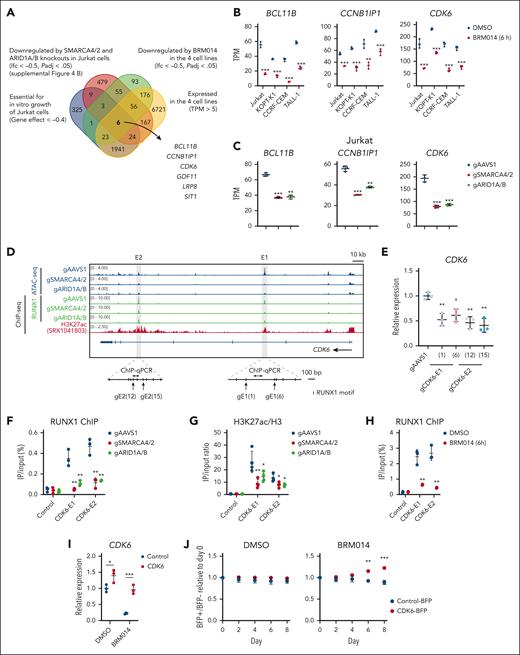
CDK6 downregulation by the inhibition of the cBAF-RUNX1 axis results in impaired T-ALL growth
We next investigated the mechanism by which cBAF inhibition impairs the growth of human T-ALL cells in relation to RUNX1. RUNX1 knockdown causes MYC downregulation in T-ALL.35 SWI/SNF inhibition also causes MYC downregulation in AML and prostate cancer.20,21 We found that 3 of 4 T-ALL cell lines treated with BRM014 showed significant downregulation of MYC and a negative enrichment of MYC targets (supplemental Figure 9A-B). SMARCA4/2 knockout Jurkat also showed the same pattern (supplemental Figure 9C-D). However, BRM014-treated KOPT-K1 and ARID1A/B knockout Jurkat showed neither MYC downregulation nor MYC target enrichment (supplemental Figure 9A-D). Instead, in Jurkat cells, BRD9 knockout, but not ARID2 knockout, significantly downregulated MYC expression, suggesting that ncBAF regulates MYC expression (supplemental Figure 9E). These results suggest that the MYC dysregulation may not solely explain cBAF dependency in T-ALL.
To identify common factors regulated by RUNX1 and cBAF in T-ALL, we set the selection criteria summarized in Figure 6A and identified 6 genes (Figure 6A-C; supplemental Figure 9F-G). Among them, CCNB1IP1, CDK6, and BCL11B were confirmed to be expressed downstream of RUNX1 (supplemental Figure 10A). We then examined whether RUNX1 is directly involved in the expression of these genes. At the CCNB1IP1 locus, it is known that a noncoding region located 280-kb upstream of the transcription start site functions as an enhancer.38 However, we observed neither RUNX1 enrichment nor a change in chromatin accessibility in this or surrounding regions (supplemental Figure 10B), suggesting that CCNB1IP1 is unlikely to be directly regulated by RUNX1 or cBAF. Next, we analyzed the CDK6 locus and found 2 strong RUNX1-binding signals, which were weakened by SMARCA4/2 and ARID1A/B knockouts and accompanied by H3K27ac enrichment and loss of ATAC-seq enrichment in intron 3 (E1) and intron 5 (E2) (Figure 6D). Disruption of RUNX1 motifs in these candidate enhancers led to significant reduction in CDK6 expression (Figure 6E), indicating that these CDK6 enhancers are functional. For the BCL11B locus, distal enhancers located 1-Mb downstream have been characterized in T-ALL cells.39 We found several RUNX1 peaks (E1, E2, E3, and E4) that coincided with H3K27ac marks, loss of chromatin accessibility, and RUNX1 binding in SMARCA4/2 and ARID1A/B knockouts (supplemental Figure 11A). Disruption of RUNX1 motifs in these candidate enhancers led to significant reduction in BCL11B expression (supplemental Figure 11B), indicating that these BCL11B enhancers are functional. We performed ChIP followed by quantitative polymerase chain reaction analyses and found significant reductions in RUNX1 binding at the enhancer regions of CDK6 and BCL11B upon SMARCA4/2 and ARID1A/B knockouts, which were accompanied by significant depletion of H3K27ac marks (Figure 6F-G and supplemental Figure 11C-D). Acute inhibition by 6-hour BRM014 treatment also significantly reduced RUNX1 binding to CDK6-E1 and E2 elements (Figure 6H). Taken together, these results indicate that cBAF activity is required for RUNX1-mediated expression of CDK6 and BCL11B. Finally, we introduced human CDK6 and BCL11B cDNA into Jurkat cells and then treated these cells with BRM014. Exogenous CDK6, but not BCL11B, significantly rescued the impaired growth by BRM014 treatment (Figure 6I-J and supplemental Figure 11E-F). Collectively, these findings indicate that CDK6 downregulation by cBAF inhibition is one of the causes of the impaired growth. Other essential genes that are regulated by cBAF may also contribute to the growth inhibitory effect of cBAF inhibition in combination with CDK6.
cBAF inhibition prevents RUNX1 from binding CDK6 enhancers, downregulates its expression, and affects cell-autonomous growth. (A) Venn diagram showing the overlap among Jurkat dependency genes, genes downregulated in SMARCA4/2 and ARID1A/B knockouts, genes downregulated in all 4 T-ALL cell lines treated with BRM014, and genes commonly expressed in the 4 T-ALL cell lines. The 6 genes, namely BCL11B, CCNB1IP1, CDK6, GDF11, LRP8, and SIT1, were identified. (B-C) BCL11B, CCNB1IP1, and CDK6 expression levels in BRM014-treated human T-ALL cell lines (B) and the indicated knockout Jurkat cells (C) by RNA-seq analysis. Data are shown as mean ± SD (n = 3). (D) ATAC-seq and ChIP-seq data in a 250-kb region including CDK6 enhancers. (E) CDK6 expression levels in Jurkat cells in which RUNX1 binding sites were disrupted by the CRISPR-Cas9 technology, determined by RT-qPCR. Data are shown as mean ± SD (n = 3). (F-H) ChIP-qPCR analysis of RUNX1 (F-H) and H3K27ac (G) at the CDK6 E1 and E2 in the indicated knockout Jurkat cells (F-G) and Jurkat cells treated with DMSO and BRM014 at 1 μM for 6 hours (H). Values in the control are identical with those in Figure 4F-H because the assays were performed in parallel. Data are shown as mean ± SD (n = 3-4). (I) CDK6 expression levels of DMSO- or BRM014-treated Jurkat cells with exogenous CDK6 or control cDNA expression. Data are shown as mean ± SD (n = 3). (J) Eight-day competitive coculture assay of Jurkat cells expressing exogenous CDK6 or control cDNA with control Jurkat cells under DMSO (left) or BRM014 (1μM) treatment (right). Data are shown as mean ± SD (n = 3). Two-tailed Student t test was used to assess statistical significance in panels B, C, E, F, G, H, I, and J (∗∗∗P < .001; ∗∗P < .01; ∗P < .05). cDNA, complementary DNA; DMSO, dimethyl sulfoxide; qPCR, quantitative polymerase chain reaction; RNA-seq, RNA sequencing; SD, standard deviation.
cBAF inhibition prevents RUNX1 from binding CDK6 enhancers, downregulates its expression, and affects cell-autonomous growth. (A) Venn diagram showing the overlap among Jurkat dependency genes, genes downregulated in SMARCA4/2 and ARID1A/B knockouts, genes downregulated in all 4 T-ALL cell lines treated with BRM014, and genes commonly expressed in the 4 T-ALL cell lines. The 6 genes, namely BCL11B, CCNB1IP1, CDK6, GDF11, LRP8, and SIT1, were identified. (B-C) BCL11B, CCNB1IP1, and CDK6 expression levels in BRM014-treated human T-ALL cell lines (B) and the indicated knockout Jurkat cells (C) by RNA-seq analysis. Data are shown as mean ± SD (n = 3). (D) ATAC-seq and ChIP-seq data in a 250-kb region including CDK6 enhancers. (E) CDK6 expression levels in Jurkat cells in which RUNX1 binding sites were disrupted by the CRISPR-Cas9 technology, determined by RT-qPCR. Data are shown as mean ± SD (n = 3). (F-H) ChIP-qPCR analysis of RUNX1 (F-H) and H3K27ac (G) at the CDK6 E1 and E2 in the indicated knockout Jurkat cells (F-G) and Jurkat cells treated with DMSO and BRM014 at 1 μM for 6 hours (H). Values in the control are identical with those in Figure 4F-H because the assays were performed in parallel. Data are shown as mean ± SD (n = 3-4). (I) CDK6 expression levels of DMSO- or BRM014-treated Jurkat cells with exogenous CDK6 or control cDNA expression. Data are shown as mean ± SD (n = 3). (J) Eight-day competitive coculture assay of Jurkat cells expressing exogenous CDK6 or control cDNA with control Jurkat cells under DMSO (left) or BRM014 (1μM) treatment (right). Data are shown as mean ± SD (n = 3). Two-tailed Student t test was used to assess statistical significance in panels B, C, E, F, G, H, I, and J (∗∗∗P < .001; ∗∗P < .01; ∗P < .05). cDNA, complementary DNA; DMSO, dimethyl sulfoxide; qPCR, quantitative polymerase chain reaction; RNA-seq, RNA sequencing; SD, standard deviation.
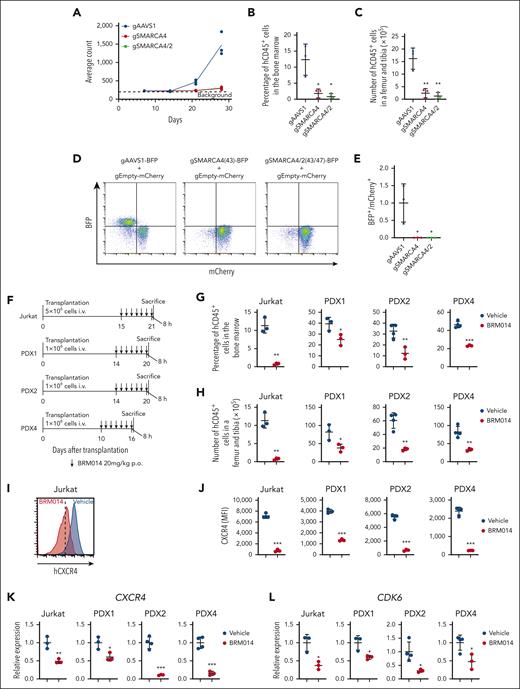
cBAF is crucial for leukemic cell growth in in vivo human T-ALL models
The above findings suggest that cBAF inhibition could impair human T-ALL growth in vivo and may represent a new therapeutic opportunity. Bioluminescence imaging of luciferase-expressing Jurkat-derived xenografts showed a markedly impaired leukemic cell expansion by SMARCA4 and SMARCA4/2 knockouts (Figure 7A and supplemental Figure 12). The percentage and number of human CD45+ cells were highly decreased in the bone marrow (Figure 7B-C). We next performed an in vivo competition assay and found that SMARCA4 or SMARCA4/2 knockout cells were remarkably outcompeted by control cells (Figure 7D-E). Importantly, we obtained comparable results using KOPT-K1 cells (supplemental Figure 13). Together, these data indicate that the cBAF complex is required for human T-ALL growth in vivo.
The cBAF complex is required for the growth of human T-ALL in vivo. (A) Quantification of luminescence signals of mice transplanted with luciferase-labeled Jurkat cells in which the indicated genes were knocked out. Data are shown as mean (n = 3). (B-C) Percentage (B) and number (C) of human CD45+ cells in the bone marrow of mice transplanted with the indicated knockout Jurkat cells at day 28 after transplantation. Data are shown as mean ± SD (n = 3). (D-E) Ratio of BFP+ cells to mCherry+ cells in the bone marrow of mice transplanted with a 1:1 mixture of BFP+ indicated knockout Jurkat cells and mCherry+ control Jurkat cells at day 21 after transplantation. Data are normalized to those of the AAVS1 knockout cells and are shown as mean ± SD (n = 3) in panel E. (F) Treatment schedule for the Jurkat xenograft model and PDX models. Mice were euthanized 8 hours after the last treatment and used for the subsequent analyses (data shown in panels G-L). (G-H) Percentage (G) and number (H) of human CD45+ cells in the bone marrow of vehicle- and BRM014-treated xenograft models. Data are shown as mean ± SD (n = 3-4). (I-J) Cell surface CXCR4 expression on human CD45+ cells in the bone marrow of vehicle or BRM014-treated xenograft models. Data are shown as mean ± SD (n = 3-4) in J. (K-L) Relative gene expression of CXCR4 (K) and CDK6 (L) in human CD45+ cells in the bone marrow of vehicle- and BRM014-treated xenograft models, as determined by RT-qPCR. Data are shown as mean ± SD (n = 3-4). Two-tailed Student t test was used to assess statistical significance in panels B, C, E, G, H, J, K, and L (∗∗∗P < .001; ∗∗P < .01; ∗P < .05). BFP, blue fluorescent protein; RT-qPCR, reverse transcription quantitative polymerase chain reaction; SD, standard deviation.
The cBAF complex is required for the growth of human T-ALL in vivo. (A) Quantification of luminescence signals of mice transplanted with luciferase-labeled Jurkat cells in which the indicated genes were knocked out. Data are shown as mean (n = 3). (B-C) Percentage (B) and number (C) of human CD45+ cells in the bone marrow of mice transplanted with the indicated knockout Jurkat cells at day 28 after transplantation. Data are shown as mean ± SD (n = 3). (D-E) Ratio of BFP+ cells to mCherry+ cells in the bone marrow of mice transplanted with a 1:1 mixture of BFP+ indicated knockout Jurkat cells and mCherry+ control Jurkat cells at day 21 after transplantation. Data are normalized to those of the AAVS1 knockout cells and are shown as mean ± SD (n = 3) in panel E. (F) Treatment schedule for the Jurkat xenograft model and PDX models. Mice were euthanized 8 hours after the last treatment and used for the subsequent analyses (data shown in panels G-L). (G-H) Percentage (G) and number (H) of human CD45+ cells in the bone marrow of vehicle- and BRM014-treated xenograft models. Data are shown as mean ± SD (n = 3-4). (I-J) Cell surface CXCR4 expression on human CD45+ cells in the bone marrow of vehicle or BRM014-treated xenograft models. Data are shown as mean ± SD (n = 3-4) in J. (K-L) Relative gene expression of CXCR4 (K) and CDK6 (L) in human CD45+ cells in the bone marrow of vehicle- and BRM014-treated xenograft models, as determined by RT-qPCR. Data are shown as mean ± SD (n = 3-4). Two-tailed Student t test was used to assess statistical significance in panels B, C, E, G, H, J, K, and L (∗∗∗P < .001; ∗∗P < .01; ∗P < .05). BFP, blue fluorescent protein; RT-qPCR, reverse transcription quantitative polymerase chain reaction; SD, standard deviation.
We next asked whether BRM014 impairs human T-ALL growth in vivo using Jurkat-derived xenografts and PDXs (Figure 7F). The percentage and number of human CD45+ cells were significantly decreased in the bone marrow of BRM014-treated mice (Figure 7G-H). In addition, cell surface expression of CXCR4 on human CD45+ cells was significantly decreased by the treatment (Figure 7I-J). CXCR4 and CDK6 expression was also significantly decreased in human CD45+ cells (Figure 7K-L). Collectively, these findings indicate that BRM014 monotherapy inhibits T-ALL cell growth in vivo and suggest that cBAF inhibition represents a promising therapeutic approach for human T-ALL.
Discussion
The functional reciprocity between TFs and chromatin remodeling complexes has been reported in several contexts.20,21,40,41 Depletion of TFs results in a decrease of SWI/SNF complex binding at TF-binding sites and loss of chromatin accessibility, leading to inactivation of enhancer activity. Conversely, when SWI/SNF function is disturbed, local chromatin accessibility decreases, and TFs that have bound these sites are no longer able to bind and exert transcriptional activation. The latter has recently been shown extensively in prostate cancer, underscoring the therapeutic potential of targeting SWI/SNF.21 In leukemic cells, RUNX1 has been shown to physically interact with subunits of the SWI/SNF complex and recruits the complex to target gene promoters in Jurkat cells.42 In a murine T-ALL model, Runx1 deficiency results in reduced chromatin accessibility at the Notch1-bound Myc enhancer.35 These and other reports collectively suggest that SWI/SNF functions to maintain open accessibility at RUNX1 binding sites and protect the integrity of the RUNX1-mediated leukemic program. However, the exact role of the SWI/SNF complex, variant specificity, and therapeutic potential of SWI/SNF inhibition against T-ALL has remained unclear. In this study, we have demonstrated that cBAF is specifically required to maintain accessibility at RUNX1-bound enhancers and protect the integrity of the RUNX1-driven leukemic program. cBAF inhibition resulted in the downregulation of RUNX1-regulated genes, including CXCR4 and CDK6, thereby eliciting antitumor activity.
At the CXCR4 locus, cBAF inhibition impaired accessibility of multiple open chromatin regions, of which 2 regions were shown to function as RUNX1-bound enhancers. Interestingly, a NR3C1 binding site adjacent to the RUNX1 binding site in the SE region was recently reported to be recurrently mutated in B-cell lymphoma.37 This mutation affects the recruitment of NR3C1, which functions as a transcriptional repressor.37 Therefore, this SE modulates CXCR4 expression with 2 counteracting transcriptional activities. The accessibility at the NR3C1 binding site in this region did not seem to be affected by cBAF inhibition (Figure 4C), suggesting that cBAF inhibition does not affect NR3C1 binding. In such circumstances, cBAF inhibition might tip the balance of the enhancer activity toward active gene suppression rather than just weakening expression, which would cause strong CXCR4 downregulation. In fact, CXCR4 was markedly downregulated among the genes whose expressions were affected by SMARCA4/2 knockout. One caveat of cBAF inhibition would be that enhancer function will be restored soon after cBAF inhibition is terminated (Figure 3F and Iurlaro et al40). For a durable anticancer effect, continuous suppression of the remodeling activity would be critical. Nevertheless, given that BRM014 treatment substantially represses CXCR4 expression in xenograft models (Figure 7I-J), it may disrupt the interaction between leukemic cells and bone marrow niches, resulting in impaired leukemic cell growth.
Human T-ALL can be divided into several subgroups based on genetic and transcriptional characteristics.43 The largest subgroup expresses RUNX1-TAL1-GATA3 and would most benefit from cBAF inhibition as shown in this study. A subgroup that overexpresses TLX1 or TLX3 is known to have RUNX1-regulated genes downregulated44; however, a representative cell line of this subgroup, HPB-ALL, is sensitive to a pan-RUNX inhibitor, AI-10-104, indicating that RUNX1 still plays an essential role in proliferation.35 Consistent with this, we have shown that HPB-ALL is sensitive to BRM014 and AU-15330. In contrast, RUNX1 plays a tumor suppressor role in early T-cell precursor–ALL, which often carry RUNX1 loss-of-function mutations. AI-10-104 does not inhibit the growth of cells derived from this subtype35; therefore, early T-cell precursor–ALL is less likely to respond to cBAF blockade.
We have shown that BCL11B is regulated by cBAF and RUNX1. Previous reports showed using human T-ALL cells that BCL11B is an accessory subunit of the cBAF complex45,46; therefore, BCL11B downregulation by cBAF inhibition may further disturb the function of the complex. Importantly, BCL11B loss-of-function mutations are detected in some T-ALL cases.43,45BCL11B mutation status may affect the effect of cBAF inhibition in human T-ALL cells.
The effect of SWI/SNF inhibition on normal hematopoiesis has been demonstrated by pharmacological and genetic approaches.47,48 In the pharmacological inhibition using BRM014, lymphocyte counts and hemoglobin levels are slightly decreased but neutrophil, monocyte, and platelet counts are unchanged.48 In addition, it is noteworthy that the number of phenotypic HSCs in the bone marrow remains stable even after BRM014 treatment.48 These findings suggest that there exists a therapeutic window for cBAF inhibition in human T-ALL. However, it has been reported that Smarca4 knockout disturbs hematopoietic differentiation, most severely in the T-cell lineage,47 cBAF is essential for the differentiation of activated CD8+ T cells into T effector cells,49 and ncBAF is known to be required for Foxp3 expression in murine regulatory T cells.50 How SWI/SNF inhibition influences the function of immune cells and whether it affects anti–T-ALL immunity in vivo should be investigated in future studies.
In summary, cBAF involves the RUNX1-driven leukemic program by maintaining chromatin accessibility of RUNX1-bound active enhancer regions. cBAF inhibition impairs leukemic cell migration toward CXCL12 and cell-autonomous growth. Further progress in cBAF inhibitor/PROTAC development could potentially open the prospect of new therapeutic approaches for T-ALL. The efficacy and safety of cBAF inhibitors/PROTACs for T-ALL therapy will need to be further assessed in future preclinical studies.
Acknowledgments
The authors thank members of the Single-cell Genome Information Analysis Core at the Institute for the Advanced Study of Human Biology, Kyoto University, for their support; and Y. Odan for her secretarial assistance.
This work was supported by KAKENHI from the Japan Society for the Promotion of Science (21K08393 [K. Aoki], 22K16320 [Y.O.], 20H00528 [J.T.], 21K19405 [J.T.], 19H05656 [S.O.], and 21H04959 [K.Y.]), Japan Agency for Medical Research and Development Project for Promotion of Cancer Research and Therapeutic Evolution (22ama221505h0001 [J.T.]), and Takeda Science Foundation Grant (K. Aoki and K.Y.).
Authorship
Contribution: K. Aoki and K.Y. conceived the project and designed the experiments; K. Aoki, M.H., Y.T., G.N., A.U., Y.O., S.S., T.M., H.K., and I.K. performed the experiments, analyzed the results, and produced the figures; Y.O. and S.O. performed assay for transposase-accessible chromatin followed by sequencing and RNA sequencing; K. Akahane, T.I., and A.T.-K. provided the study materials; K. Aoki, M.H., Y.T., G.N., Y.O., and K.Y. wrote the manuscript; and all authors approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kazunari Aoki, Stem Cell Genetics, Institute for Life and Medical Sciences, Kyoto University, 53 Shogoin-kawahara-cho, Sakyo-ku, Kyoto 606-8507, Japan; email: aoki.kazunari.4c@kyoto-u.ac.jp; and Kosuke Yusa, Stem Cell Genetics, Institute for Life and Medical Sciences, Kyoto University, 53 Shogoin-kawahara-cho, Sakyo-ku, Kyoto 606-8507, Japan; email: k.yusa@infront.kyoto-u.ac.jp.
References
Author notes
All sequencing-based data are available at GEO under accession GSE227275.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![Inhibition of cBAF activity hampers RUNX1 binding to the CXCR4 enhancers and downregulates CXCR4 expression. (A-B) Cell surface CXCR4 expression (A) and migration activity (B) of the indicated knockout Jurkat cells. Expression was determined by flow cytometry. Cells expressing gAAVS1 were used as a control. Data are shown as mean ± SD (n = 3). (C) ATAC-seq and ChIP-seq data in a 35-kb region including CXCR4 enhancer regions (SE [intron 1], E1 [−13.5 kb], E2 [−16.2 kb], E3 [−18.1 kb], and E4 [−18.6 kb]) and an NR3C1 motif (see “Discussion”). (D-E) Cell surface expression of CXCR4 (D) and migration activity (E) of Jurkat cells in which the indicated RUNX1 binding sites were disrupted by the CRISPR-Cas9 technology. Data are shown as mean ± SD (n = 3). E1 disruption also significantly decreased CXCR4 expression but the change was negligible (D). (F-H) ChIP-qPCR analysis of RUNX1 (F, H) and H3K27ac (G) at the CXCR4 SE and E4 enhancers in the indicated knockout Jurkat cells (F, G) and Jurkat cells treated with DMSO and BRM014 at 1 μM for 6 hours (H). The CTCF binding element 9-kb upstream of SMARCC1 was used as a control region. Data are shown as mean ± SD (n = 3-4). Two-tailed Student t test was used to assess statistical significance in panels A, B, D, E, F, G, and H (∗∗∗P < .001; ∗∗P < .01; ∗P < .05). DMSO, dimethyl sulfoxide; qPCR, quantitative polymerase chain reaction; SD, standard deviation.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/143/7/10.1182_blood.2023020857/4/m_blood_bld-2023-020857-gr4.jpeg?Expires=1770171435&Signature=MJdrAhjU~UdhNCctbBviOa2lfuC4870GBCGIGl5LJWYhGSv9NUhpcopOLoPCM7INlMKCKz6uGi~1D1leWQ1afhMmsmTAGCNTGhxuHQBuddwqJpnJeipFMrzLg2lmPGZOJB~chZiJdYM01kqQHfhpMU47p2krDedTIRcxtCx1RPCW8jEqZrXE2gwAy7Xjosneb6kpRl1qwy94SyOwXxvs92hameJrAsGTzy~9WedWi7Z8psOCR2~dmdNqp1mNgNhNaA7B2dsXhfPL6NkIBAyHRXAVdouBrHlM~7j90xJqHLTP9HTdM-9VZkJNUHTP6-FvjmLwPdcHK3SWub7F2J0gHQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal