Key Points
AML cells have increased mitochondrial mass, low respiratory chain complex activities, and low spare reserve capacity compared with normal cells.
AML cells have heightened sensitivity to inhibitors of the respiratory chain complexes and oxidative stressors.
Abstract
Mitochondrial respiration is a crucial component of cellular metabolism that can become dysregulated in cancer. Compared with normal hematopoietic cells, acute myeloid leukemia (AML) cells and patient samples have higher mitochondrial mass, without a concomitant increase in respiratory chain complex activity. Hence these cells have a lower spare reserve capacity in the respiratory chain and are more susceptible to oxidative stress. We therefore tested the effects of increasing the electron flux through the respiratory chain as a strategy to induce oxidative stress and cell death preferentially in AML cells. Treatment with the fatty acid palmitate induced oxidative stress and cell death in AML cells, and it suppressed tumor burden in leukemic cell lines and primary patient sample xenografts in the absence of overt toxicity to normal cells and organs. These data highlight a unique metabolic vulnerability in AML, and identify a new therapeutic strategy that targets abnormal oxidative metabolism in this malignancy.
Introduction
Oxidative metabolism is a critical mitochondrial process that generates intracellular energy and metabolic intermediates necessary to maintain and increase cellular biomass. To meet their energy and biosynthetic requirements, cells metabolize substrates such as glucose, glutamine, and fatty acids to generate electrons that flow into respiratory chain complexes.1-4 Electrons are passed along this respiratory chain, with oxygen as the final acceptor. During this process, protons are pumped across the inner mitochondrial membrane, establishing an electrochemical gradient across the membrane. The energy stored in this gradient is used to drive adenosine triphosphate (ATP) production. In cancer cells, the requirement of energy and biosynthetic precursors is higher; therefore, oxidative metabolism is also frequently amplified to meet these demands.5-7
Mitochondrial biogenesis is a reflection of energy, metabolic, and signaling requirements of a cell.8 In response to physiological, metabolic, and genetic signals, cells can modulate mitochondrial biogenesis and mass to alter the energy produced through oxidative phosphorylation (OXPHOS).9,10 We demonstrated that inhibiting mitochondrial translation reduced the levels of mitochondrially translated respiratory chain proteins, decreased oxygen consumption, and preferentially induced cell death in acute myeloid leukemia (AML) cells compared with normal hematopoietic cells. These effects were observed by both inhibiting the mitochondrial ribosome with the small molecule tigecycline or through knocking down the mitochondrial elongation factor EF-Tu/TUFM.11 The heightened sensitivity of AML cells to the inhibition of mitochondrial translation was associated with greater mitochondrial mass, higher oxygen consumption, and a greater reliance on OXPHOS for survival compared with normal hematopoietic cells.11 As such, this study highlighted a unique metabolic vulnerability that could be exploited therapeutically in this disease.
Here, we further explored the unique mitochondrial characteristics of AML cells. In comparison with normal hematopoietic cells, we demonstrated that a subset of AML cells had increased mitochondrial mass. This occurred without a corresponding increase in activity of respiratory chain complexes, including mitochondrial DNA encoded subunits. As a result, the spare reserve capacity of these complexes was lower. Low spare reserve capacity may impede the ability of cells to cope with oxidative stress. Accordingly, we demonstrated that increasing the electron flux through the respiratory chain in AML cells preferentially increased oxidative stress and induced cell death, in comparison with normal hematopoietic cells.
Materials and methods
See supplemental material on the Blood Web site for additional methods.
Primary AML and normal hematopoietic cells
Primary human AML samples were isolated from peripheral blood or marrow samples from consenting patients with AML, who had at least 80% malignant cells among low-density cells. AML cells were isolated by Ficoll density centrifugation. Except where otherwise noted, primary normal hematopoietic cells refer to normal mononuclear cells obtained from healthy consenting volunteers donating peripheral blood stem cells (PBSCs) for allogeneic stem cell transplantation after granulocyte-colony stimulating factor (G-CSF) mobilization. Normal human bone marrow was obtained from Stem Cell Technologies (Vancouver, BC). Normal CD34+ cells were isolated from primary normal hematopoietic cells using the Human CD34 selection kit (StemCell Technologies). Primary cells were cultured at 37°C in Iscove modified Dulbecco medium and were supplemented with 20% fetal bovine serum and appropriate antibiotics. The University Health Network and Mount Sinai Hospital institutional review boards approved the collection and use of human tissue for this study. All animal studies were performed according to the regulations of the Canadian Council on Animal Care and with the approval of the Ontario Cancer Institute animal ethics review board. AML patient information and per-sample experimental results are provided in supplemental Tables.
Oxygen consumption rate and spare reserve capacity
Measurement of oxygen consumption rate (OCR) was performed using a Seahorse XF96 analyzer (Seahorse Bioscience, North Billerica, MA). After treatment, cells were resuspended with unbuffered medium and were seeded at 1 × 105 cells/well (cell lines) or 1 × 106 cells/well (primary cells) in XF96 plates. Cells were equilibrated in the unbuffered medium for 45 minutes at 37°C in a CO2-free incubator before being transferred to the XF96 analyzer. Basal OCR and the change in oxygen consumption were measured after drug treatment.
Spare reserve capacity of the mitochondrial respiratory chain was measured by treating cells with oligomycin and FCCP (carbonyl cyanide p-trifluoromethoxyphenylhydrazone) in succession. Oxygen consumption was measured as previously described. The spare reserve capacity of individual respiratory complexes was determined by treating cells with complex inhibitors. The concentrations of rotenone, antimycin, and oligomycin required to reduce OCR by 50% (EC50) and the concentration of sodium azide required to reduce oxygen consumption by 25% (EC25) were determined.
Determination of ROS generation
Intracellular reactive oxygen species (ROS) were detected by staining cells with MitoSOX (5 μM) and followed by flow cytometric analysis as described.11 Briefly, cells were stained with MitoSOX in Hanks balanced salt solution buffer at 37°C for 30 minutes, and then resuspended in binding buffer with Annexin V to identify viable cells and to assess their reactive oxygen intermediate levels. For detection of the progenitor population in PBSCs and primary AML samples, CD34-PE-Cy7 (clone 8G12) and CD38-PE-Cy5 (clone H1T2) were used. Data were analyzed with FlowJo version 7.7.1 (TreeStar).
Results
AML cells have increased mitochondrial mass and biogenesis factors
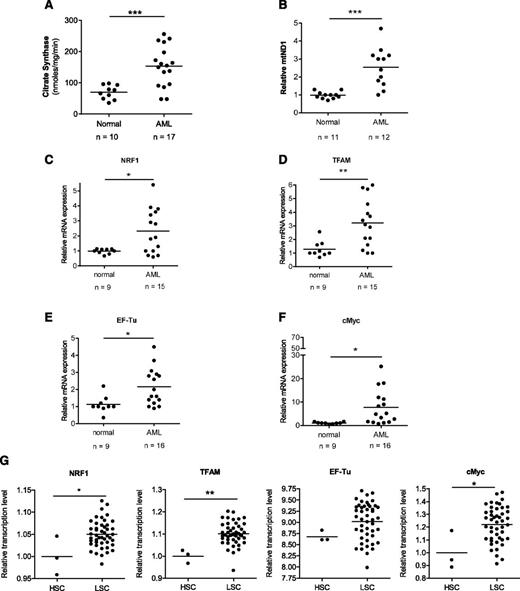
To extend our previous studies, we sought to better understand the mitochondrial characteristics of AML cells. Compared with normal hematopoietic cells, primary AML samples showed increased mitochondrial mass, as measured by both the activity of the mitochondrial matrix enzyme citrate synthase and mitochondrial DNA copy number (Figure 1A-B). We also measured mRNA expression of factors known to regulate mitochondrial biogenesis positively, such as nuclear respiratory factor 1 (NRF1), transcription factor A mitochondrial (TFAM), and EF-Tu, along with c-Myc, a positive regulator of these genes and mitochondrial biogenesis.12 Compared with normal hematopoietic cells, a subset of primary AML samples had increased mRNA expression of these genes (Figure 1C-F). In addition, we also demonstrated a higher mRNA expression of NRF1, TFAM, and c-Myc in functionally defined AML stem cells compared with normal hematopoietic stem cells (Figure 1G). Increased mitochondrial mass and mRNA expression of the aforementioned genes in the primary AML samples occurred across French-American-British subtypes, cytogenetic risk groups, and known molecular mutations (supplemental Tables 1 and 2). In addition, we analyzed the expression of the earlier mitochondrial biogenesis factors using a public dataset of 283 primary AML samples.13 Similar to the previously mentioned findings, a subset of AML patients had increased expression of mitochondrial biogenesis factors (supplemental Figure 1). Thus, our findings suggest that increased mitochondrial biogenesis in AML is a downstream consequence of multiple dysregulated pathways.
Primary AML samples have increased mitochondrial mass and mRNA level of mitochondrial biogenesis regulators. (A) Citrate synthase activity as a marker of mitochondrial mass was determined in primary normal hematopoietic cells (G-CSF mobilized peripheral blood mononuclear cells) and AML samples. (B) Mitochondrial DNA copy number was determined in primary normal hematopoietic and AML samples. DNA was extracted from cells and mRNA levels of the mitochondrial ND1 gene (mtND1) relative to human globulin (HGB) were measured by qRT-PCR. (C-F) Expression of NRF1, TFAM, EF-Tu, and c-Myc mRNA was measured in primary normal hematopoietic and AML samples. Expression was determined by qRT-PCR using 18s RNA as an internal standard. (G) Expression of NRF1, TFAM, EF-Tu, and c-Myc mRNA in functionally defined AML stem cells (LSC) vs normal hematopoietic stem cells (HSC) (G-CSF mobilized peripheral blood mononuclear cells). Data were derived from the publically accessible data set GSE 30377, achieved on the Gene Expression Omnibus. In all panels, *P < .05; **P < .01; ***P < .001 as determined by the unpaired Student t test.
Primary AML samples have increased mitochondrial mass and mRNA level of mitochondrial biogenesis regulators. (A) Citrate synthase activity as a marker of mitochondrial mass was determined in primary normal hematopoietic cells (G-CSF mobilized peripheral blood mononuclear cells) and AML samples. (B) Mitochondrial DNA copy number was determined in primary normal hematopoietic and AML samples. DNA was extracted from cells and mRNA levels of the mitochondrial ND1 gene (mtND1) relative to human globulin (HGB) were measured by qRT-PCR. (C-F) Expression of NRF1, TFAM, EF-Tu, and c-Myc mRNA was measured in primary normal hematopoietic and AML samples. Expression was determined by qRT-PCR using 18s RNA as an internal standard. (G) Expression of NRF1, TFAM, EF-Tu, and c-Myc mRNA in functionally defined AML stem cells (LSC) vs normal hematopoietic stem cells (HSC) (G-CSF mobilized peripheral blood mononuclear cells). Data were derived from the publically accessible data set GSE 30377, achieved on the Gene Expression Omnibus. In all panels, *P < .05; **P < .01; ***P < .001 as determined by the unpaired Student t test.
An increase in mitochondrial mass can be indicative of larger and/or more numerous mitochondria. Therefore, we evaluated the size and number of mitochondria in normal hematopoietic cells and primary AML samples by transmission electron microscopy. Mitochondria in AML cells were generally larger than those in normal hematopoietic cells, although they were fewer in number (supplemental Figures 2 and 3). As such, total mitochondrial area was higher in most primary AML samples compared with normal hematopoietic cells (supplemental Figure 3).
Respiratory chain complex activity does not increase concomitantly with mitochondrial mass in AML
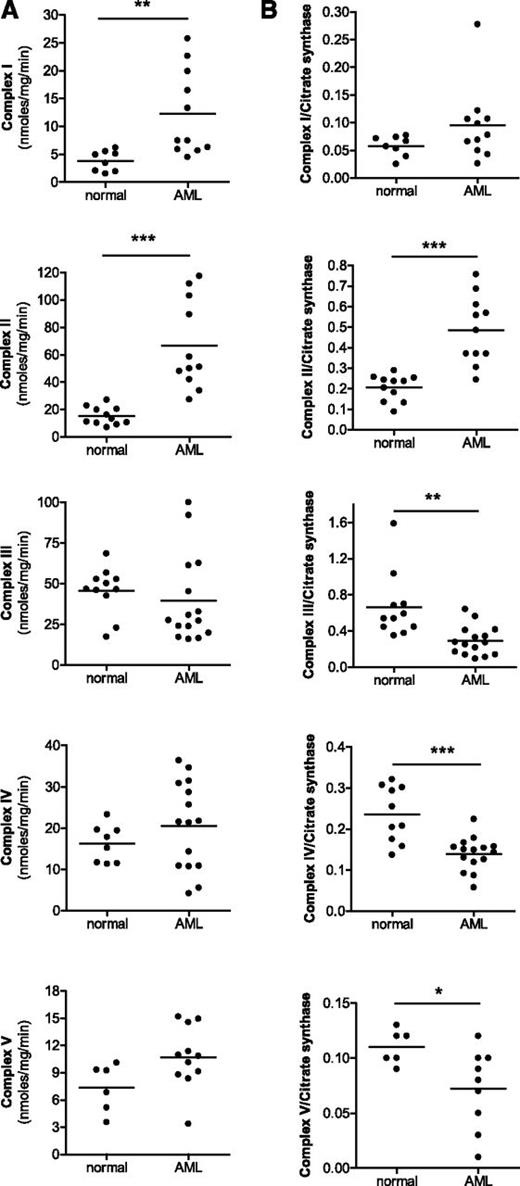
Next, we compared the activity of mitochondrial respiratory chain complexes in AML and normal hematopoietic cells (Figure 2) as well as in solid tumor cell lines (supplemental Figure 4). When normalized for total protein concentration, the enzymatic activity of complexes I and II were higher in AML cell lines and primary AML patient samples compared with normal hematopoietic cells or solid tumor cell lines. However, the activity of complexes III, IV, and V were similar between cell types (Figure 2A; supplemental Figure 4G-H upper panel). When viewed relative to mitochondrial mass, AML cell lines and primary AML samples had substantially lower activities of respiratory complexes III, IV, and V compared with normal hematopoietic cells (Figure 2B; supplemental Figure 4G-H lower panel). The activity of complex I was similar between AML and normal hematopoietic cells. In contrast, the activity of respiratory complex II was higher in primary AML cells compared with normal hematopoietic. Of note, complex II is the only complex that is comprised exclusively of subunits encoded by the nuclear genome. Thus, taken together, the increased mitochondrial mass observed in AML cell lines (supplemental Figure 4) and primary AML samples (Figure 1) was not accompanied by a corresponding increase in the activities of respiratory complexes III, IV, and V.
Activities of respiratory chain complexes do not increase in primary AML samples in parallel with mitochondrial mass. The activities of respiratory complexes I-V were measured in isolated mitochondria from primary normal hematopoietic (G-CSF mobilized peripheral blood mononuclear cells) and primary AML samples. (A) Complex activity was normalized to total protein concentration. (B) Complex activity was normalized to mitochondrial mass using citrate synthase activity. *P < .05; **P < .001; ***P < .0001 as determined by unpaired Student t test.
Activities of respiratory chain complexes do not increase in primary AML samples in parallel with mitochondrial mass. The activities of respiratory complexes I-V were measured in isolated mitochondria from primary normal hematopoietic (G-CSF mobilized peripheral blood mononuclear cells) and primary AML samples. (A) Complex activity was normalized to total protein concentration. (B) Complex activity was normalized to mitochondrial mass using citrate synthase activity. *P < .05; **P < .001; ***P < .0001 as determined by unpaired Student t test.
Genetic modulation of mitochondrial mass does not concomitantly change respiratory chain complex activity
To further explore the relationship between mitochondrial mass and respiratory complex activity, we manipulated mitochondrial mass genetically by knocking down Myc or TFAM. As a genetic model to manipulate mitochondrial mass and metabolism, we employed P493-6 cells with inducible c-Myc knockdown (− Myc).12 Previously, we and others have used P493-6 cell system to evaluate the effects of genetically altering mitochondrial biogenesis.11,12,14 P493-6 cells expressing c-Myc (+ Myc) had increased mitochondrial mass (supplemental Figure 5A-B), as well as increased expression of TFAM, NRF1, and EF-Tu compared with − Myc cells (supplemental Figure 5C). By transmission electron microscopy analysis, + Myc cells also had larger mitochondria compared with − Myc cells, although the number of mitochondria per cell was the same (supplemental Figure 6C). When normalized for total protein concentration, enzymatic activities of complexes II and V were significantly higher in + Myc cells compared with − Myc cells, and activities of the other complexes did not differ (supplemental Figure 5D). However, when viewed relative to mitochondrial mass, + Myc cells had lower activities of respiratory complexes I, III, IV, and V compared with − Myc cells (supplemental Figure 5E). Similar to AML cells, the lack of increase in activity compared with elevated mitochondrial mass was higher for complexes III and IV.
We also measured expression levels of COX-1 (mitochondrially encoded cytochrome c oxidase I) and COX-2 (mitochondrially encoded cytochrome c oxidase II) subunits of respiratory complex IV encoded by the mitochondrial genome, as well as those of COX-4 (cytochrome c oxidase subunit IV), a subunit of respiratory complex IV encoded by the nuclear genome. Consistent with our previous findings, despite alterations in mitochondrial mass, the expression of COX-1, COX-2, and COX-4 proteins did not differ between + Myc and − Myc cells when normalized for total cellular protein (supplemental Figure 6). Likewise, COX-1 and COX-2 mRNA levels showed little difference between + Myc and − Myc cells, although COX-4 mRNA levels were slightly decreased (supplemental Figure 6). Thus these results further support our findings that the regulation of respiratory chain activity can be dissociated from the regulation of mitochondrial mass.
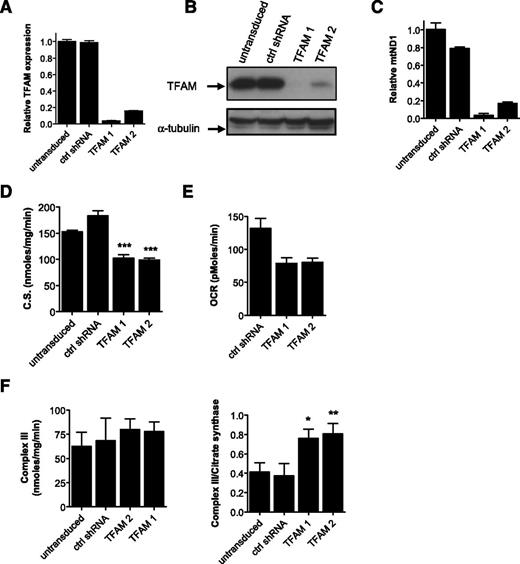
As an additional genetic approach, we knocked down TFAM in OCI-AML-2 and K562 cells using shRNA in lentiviral vectors. Target knockdown was confirmed by qRT-PCR and immunoblotting (Figure 3A-B). Knockdown of TFAM decreased mitochondrial mass (Figure 3C-D) and oxygen consumption (Figure 3E; supplemental Figure 7). However, despite the decrease in mitochondrial mass after TFAM knockdown, the activity of respiratory chain complex III did not change when normalized for total protein content. In fact, complex III activity increased on TFAM knockdown when viewed relative to mitochondrial mass (Figure 3F; supplemental Figure 7).
Genetic inhibition of mitochondrial biogenesis factor TFAM rescues effects of oxidative stress. (A-B) OCI-AML-2 cells were infected with TFAM targeting shRNAs or control sequences in lentiviral vectors. Four days posttransduction, TFAM mRNA expression relative to 18S was made by qRT-PCR (A) and TFAM protein expression was determined by immunoblotting (B). (C) DNA was extracted from cells, and quantitative PCR was used to measure levels of ND1 relative to HGB. ND1/HGB ratio is shown relative to control cells. (D) Citrate synthase activity as a marker of mitochondrial mass was determined in TFAM knockdown clones. (E) Basal OCR was shown after 1 hour of incubation in cell chambers. (F) Activity of complex III was measured in control and TFAM knockdown cells. Left panel shows complex activity was normalized to total protein concentration. Right panel shows complex activity was normalized to mitochondrial mass using citrate synthase activity. Data represent the mean complex activity ± standard deviation (SD) from representative experiments performed in triplicate. TFAM knockdown experiments in AML cells were repeated twice. In all panels, *P < .05; **P < .001; ***P < .0001 as determined by the unpaired Student t test.
Genetic inhibition of mitochondrial biogenesis factor TFAM rescues effects of oxidative stress. (A-B) OCI-AML-2 cells were infected with TFAM targeting shRNAs or control sequences in lentiviral vectors. Four days posttransduction, TFAM mRNA expression relative to 18S was made by qRT-PCR (A) and TFAM protein expression was determined by immunoblotting (B). (C) DNA was extracted from cells, and quantitative PCR was used to measure levels of ND1 relative to HGB. ND1/HGB ratio is shown relative to control cells. (D) Citrate synthase activity as a marker of mitochondrial mass was determined in TFAM knockdown clones. (E) Basal OCR was shown after 1 hour of incubation in cell chambers. (F) Activity of complex III was measured in control and TFAM knockdown cells. Left panel shows complex activity was normalized to total protein concentration. Right panel shows complex activity was normalized to mitochondrial mass using citrate synthase activity. Data represent the mean complex activity ± standard deviation (SD) from representative experiments performed in triplicate. TFAM knockdown experiments in AML cells were repeated twice. In all panels, *P < .05; **P < .001; ***P < .0001 as determined by the unpaired Student t test.
AML cells have low spare reserve capacity in their respiratory chain complexes
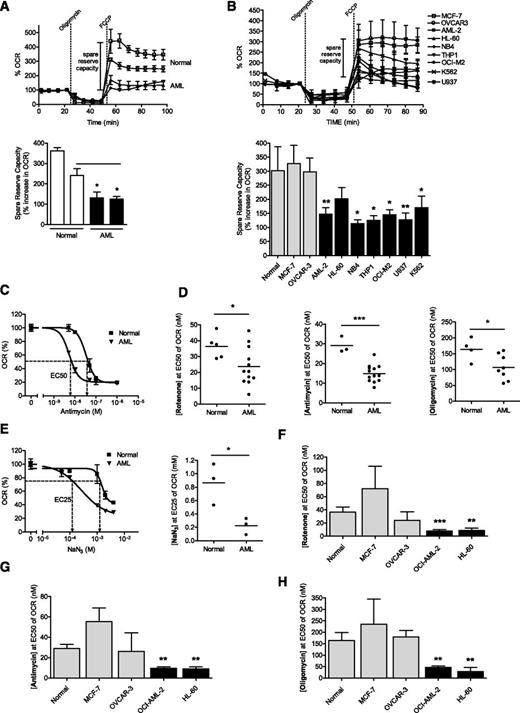
To understand the functional implications of these findings, we evaluated spare reserve capacity in AML cell lines, primary AML samples, primary normal hematopoietic cells, and solid tumor cell lines. Spare reserve capacity reflects the difference between basal and maximal respiratory rate, and this capacity was determined by measuring OCR after treatment with oligomycin to block ATP synthesis and FCCP to uncouple ATP synthesis from the electron transport chain.15-17 The spare reserve capacity in primary AML samples and cell lines was lower than that in normal hematopoietic cells or solid tumor cell lines (Figure 4A-B; summary of results in supplemental Table 3).
Primary AML cells and leukemic cell lines have lower spare reserve capacity in their respiratory chain enzymes than normal hematopoietic cells. Spare reserve capacity measured by OCR of primary AML samples and normal hematopoietic cells (A) and leukemic cell lines and solid tumor cell lines (B) after the sequential addition of oligomycin and FCCP. (C) Primary AML and normal hematopoietic cells were treated with increasing concentrations of antimycin and changes in oxygen consumption were measured. A representative graph is shown. Primary AML and normal hematopoietic cells (D-E) and leukemia, MCF-7 breast, and OVCAR-3 ovarian cancer cells (F-H) were treated with increasing concentrations of inhibitors of complex I (rotenone) (D,F), complex III (antimycin) (D,G), or complex V (oligomycin) (D,H). The concentration of the complex inhibitor required to reduce OCR by 50% (EC50) was determined. Data for cell lines represent the mean complex activity ± SD from representative experiments performed in triplicate. Experiments with cell lines were performed at least 3 times. (E) Primary AML cells and normal hematopoietic cells were treated with increasing concentrations of complex IV inhibitor, sodium azide (NaN3), and changes in oxygen consumption were measured. A representative graph is shown. The concentration of NaN3 required to reduce OCR by 25% (EC25) was determined. In all panels, *P < .05; **P < .001; ***P < .0001 as determined by the unpaired Student t test.
Primary AML cells and leukemic cell lines have lower spare reserve capacity in their respiratory chain enzymes than normal hematopoietic cells. Spare reserve capacity measured by OCR of primary AML samples and normal hematopoietic cells (A) and leukemic cell lines and solid tumor cell lines (B) after the sequential addition of oligomycin and FCCP. (C) Primary AML and normal hematopoietic cells were treated with increasing concentrations of antimycin and changes in oxygen consumption were measured. A representative graph is shown. Primary AML and normal hematopoietic cells (D-E) and leukemia, MCF-7 breast, and OVCAR-3 ovarian cancer cells (F-H) were treated with increasing concentrations of inhibitors of complex I (rotenone) (D,F), complex III (antimycin) (D,G), or complex V (oligomycin) (D,H). The concentration of the complex inhibitor required to reduce OCR by 50% (EC50) was determined. Data for cell lines represent the mean complex activity ± SD from representative experiments performed in triplicate. Experiments with cell lines were performed at least 3 times. (E) Primary AML cells and normal hematopoietic cells were treated with increasing concentrations of complex IV inhibitor, sodium azide (NaN3), and changes in oxygen consumption were measured. A representative graph is shown. The concentration of NaN3 required to reduce OCR by 25% (EC25) was determined. In all panels, *P < .05; **P < .001; ***P < .0001 as determined by the unpaired Student t test.
The previously cited studies measured the reserve capacity in the respiratory chain as a whole. We next sought to measure the spare reserve capacity in individual respiratory chain complexes. For these studies, we focused on respiratory chain complexes I, III, IV, and V. Respiratory chain complex II was not tested in these assays, as its activity was higher in AML cells than in normal cells when viewed relative to mitochondrial mass. AML cell lines, primary AML samples, normal hematopoietic cells, and solid tumor cell lines, were treated with increasing concentrations of rotenone, antimycin, sodium azide (NaN3), or oligomycin to inhibit complexes I, III, IV, and V, respectively. After treatment, oxygen consumption was measured, and the concentration of complex inhibitor required to reduce oxygen consumption by 50% (EC50) was determined (Figure 4C-H). Of note, when evaluating complex IV, we determined the concentration of NaN3 required the reduction of oxygen consumption by 25% (EC25), as we could not inhibit 50% of oxygen consumption in normal hematopoietic cells, consistent with previously described results.18 In these assays, greater sensitivity to the complex inhibitor reflects lower spare reserve capacity in the respiratory complex. Compared with normal hematopoietic cells and solid tumor cell lines, AML cell lines and primary AML samples had less spare reserve capacity in complexes I, III, IV, and V (Figure 4C-H). It is important to note that complexes III and IV demonstrated the most striking differences in spare reserve capacity between AML and normal hematopoietic cells (Figure 4D-E), and the activity of these complexes was lowest in AML cells when normalized for mitochondrial mass (Figure 2B). As complex III showed the most striking difference in spare reserve capacity, and because we could block at least 50% of oxygen consumption in AML cells using the complex III inhibitor antimycin, we focused further studies on this complex.
As a genetic approach to investigate the relationship between mitochondrial mass and spare reserve capacity, we knocked down Myc in P493-6 and measured changes in reserve capacity. Despite reductions in mitochondrial mass (supplemental Figure 5A-B), activity of complex III was unchanged (supplemental Figure 5D) and spare reserve capacity in complex III increased (supplemental Figure 5F).
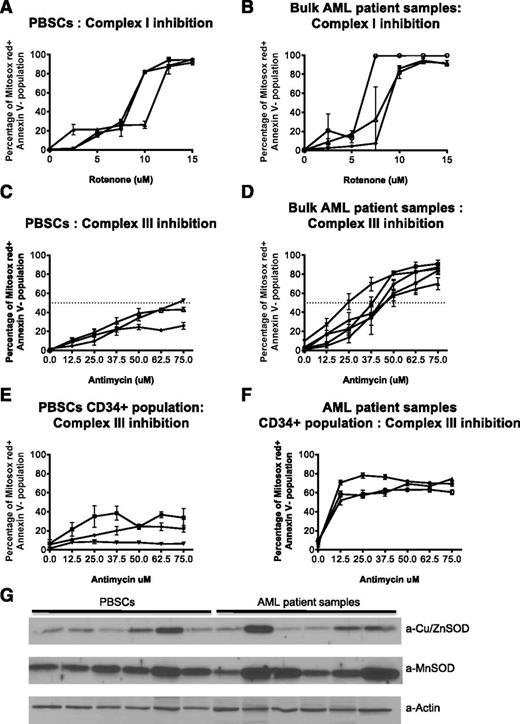
As an additional strategy to assess the reserve capacity in the respiratory complexes in primary AML and normal cells, we treated primary AML and normal hematopoietic PBSCs with increasing concentrations of the complex I inhibitor rotenone and the complex III inhibitor antimycin. We then measured mitochondrial ROS production by flow cytometry. Primary AML cells were equally sensitive to rotenone-induced ROS production compared with normal hematopoietic cells (Figure 5A-B). In contrast, compared with normal hematopoietic cells, a subset of primary AML cells (Figure 5C-F) were more sensitive to antimycin-induced mitochondrial ROS production. We also investigated sensitivity toward antimycin in bone marrow cells from AML patients and normal volunteers. Similar to the earlier results obtained with the peripheral blood samples, AML cells from patients’ bone marrow showed increased sensitivity toward antimycin (supplemental Figure 8). Of note, there was no difference in levels of the major mitochondrial antioxidants MnSOD and Cu/ZnSOD between primary AML and normal cells (Figure 5G). Thus, these results provide further evidence that reserve capacity is reduced in a subset of primary AML cells at the level of complex III.
Primary AML cells have increased sensitivity to complex III inhibition. (A-D) PBSCs (A,C) and AML patient samples (B,D) were treated with the indicated concentrations of rotenone or antimycin to block complex I and III, respectively. After 2 hours (rotenone) or 4 hours (antimycin) of treatment, cells were stained with 5 μM MitoSOX Red. After 30 minutes, the stain was replaced with Annexin V to detect apoptotic cells and cells were analyzed using a BD FACS Canto II flow cytometer with a High Throughput Sampler. Data represent the mean value of triplicates ± SD. Each curve represents a patient/normal sample. (E-F) For detection of the progenitor population, CD34-PE-Cy7 (Clone 8G12) and CD38-PE-Cy5 (Clone HIT2) antibodies were also added with mitosox. (G) Immunoblots of cell lysates from PBSCs and AML patients, probed with the indicated antibodies against SOD1 (Cu/ZnSOD), present in the intermembrane space as well as cytoplasm, and SOD2 (MnSOD), present in the matrix. Lower panel shows actin as a loading control. 30 μg of total protein loaded in each lane.
Primary AML cells have increased sensitivity to complex III inhibition. (A-D) PBSCs (A,C) and AML patient samples (B,D) were treated with the indicated concentrations of rotenone or antimycin to block complex I and III, respectively. After 2 hours (rotenone) or 4 hours (antimycin) of treatment, cells were stained with 5 μM MitoSOX Red. After 30 minutes, the stain was replaced with Annexin V to detect apoptotic cells and cells were analyzed using a BD FACS Canto II flow cytometer with a High Throughput Sampler. Data represent the mean value of triplicates ± SD. Each curve represents a patient/normal sample. (E-F) For detection of the progenitor population, CD34-PE-Cy7 (Clone 8G12) and CD38-PE-Cy5 (Clone HIT2) antibodies were also added with mitosox. (G) Immunoblots of cell lysates from PBSCs and AML patients, probed with the indicated antibodies against SOD1 (Cu/ZnSOD), present in the intermembrane space as well as cytoplasm, and SOD2 (MnSOD), present in the matrix. Lower panel shows actin as a loading control. 30 μg of total protein loaded in each lane.
AML cells are more sensitive to mitochondrial oxidative stress
In addition to being sensitive to inhibitors of the respiratory chain, we hypothesized that AML cells would be more vulnerable to mitochondrial oxidative stress. Toward this end, we increased electron flux through the respiratory chain by treating AML cells with increasing concentrations of the fatty acid palmitate to increase the production of oxidative metabolites. Treatment with palmitate increased levels of the trichloracetic acid (TCA) cycle component succinate, decreased spare reserve capacity, increased mitochondrial ROS production, and induced cell death in AML cells (Figure 6A-B; supplemental Figure 9). Palmitate-induced cell death appeared ROS-dependent, as pretreatment with the ROS scavenger N-acetylcysteine blocked cell death (Figure 6C). Further supporting the proposed mechanism, reductions in spare reserve capacity after treating cells with the respiratory chain complex III inhibitors antimycin and myxothiazol enhanced ROS production after palmitate treatment (supplemental Figure 10). Of note, the combination of palmitate with cytarabine or daunorubicin, the standard chemotherapeutic agents used in the treatment of AML, produced primarily additive cytotoxicity toward AML cells (supplemental Figure 11).
Low spare reserve capacity renders AML cells sensitive to oxidative metabolic stress by palmitate, and this sensitivity can be rescued by genetically inhibiting fatty acid oxidation pathway. (A) Leukemic cells and MCF-7 cells were treated with increasing concentrations of palmitate for 72 hours. Cell viability and growth were measured by Cell Titer Fluor viability assay. (B) OCI-AML-2 and HL-60 cells were treated with increasing concentrations of palmitate for 24 hours. ROS production was measured by staining with MitoSOX and flow cytometry. (C) OCI-AML-2 cells were treated with increasing concentrations of palmitate for 72 hours in the presence and absence of N-acetylcysteine. Cell growth and viability was measured by Cell Titer Fluor viability assay. (D-F) OCI-AML-2 cells were infected with lentiviral vectors containing shRNAs targeting CPT1a or noncellular targets (control). A total of 6 days postinfection, CPT1a mRNA expression relative to 18s RNA was analyzed by qRT-PCR (D) and CPT1a protein expression was determined by immunoblotting (E). Cell growth and viability were measured by Cell Titer Flo after treating cells with palmitate for 72 hours (F). (G) Infected OCI-AML-2 cells were treated with increasing concentrations of palmitate for 24 hours. ROS production was measured by staining with MitoSOX and flow cytometry. (H-K) OCI-AML-2 cells were infected with lentiviral vectors containing shRNAs targeting PPARα or noncellular targets (control). Four days postinfection, PPARα mRNA expression relative to 18s RNA was analyzed by qRT-PCR (H). Cell growth and viability were measured by Cell Titer Flo after treating cells with palmitate for 72 hours (I). Infected OCI-AML-2 cells were treated with increasing concentrations of palmitate. 72 hours after treatment, cell viability was measured by Annexin V/PI staining (J). Infected OCI-AML-2 cells were treated with increasing concentrations of palmitate for 24 hours. ROS production was measured by staining with MitoSOX and flow cytometry (K). In all panels, error bars represent mean ± SD of independent/representative experiments. *P < .05; **P < .001 as determined by Tukey’s test after 1-way analysis of variance, comparing to controls. CPT1a and PPARα knockdown experiments were repeated twice.
Low spare reserve capacity renders AML cells sensitive to oxidative metabolic stress by palmitate, and this sensitivity can be rescued by genetically inhibiting fatty acid oxidation pathway. (A) Leukemic cells and MCF-7 cells were treated with increasing concentrations of palmitate for 72 hours. Cell viability and growth were measured by Cell Titer Fluor viability assay. (B) OCI-AML-2 and HL-60 cells were treated with increasing concentrations of palmitate for 24 hours. ROS production was measured by staining with MitoSOX and flow cytometry. (C) OCI-AML-2 cells were treated with increasing concentrations of palmitate for 72 hours in the presence and absence of N-acetylcysteine. Cell growth and viability was measured by Cell Titer Fluor viability assay. (D-F) OCI-AML-2 cells were infected with lentiviral vectors containing shRNAs targeting CPT1a or noncellular targets (control). A total of 6 days postinfection, CPT1a mRNA expression relative to 18s RNA was analyzed by qRT-PCR (D) and CPT1a protein expression was determined by immunoblotting (E). Cell growth and viability were measured by Cell Titer Flo after treating cells with palmitate for 72 hours (F). (G) Infected OCI-AML-2 cells were treated with increasing concentrations of palmitate for 24 hours. ROS production was measured by staining with MitoSOX and flow cytometry. (H-K) OCI-AML-2 cells were infected with lentiviral vectors containing shRNAs targeting PPARα or noncellular targets (control). Four days postinfection, PPARα mRNA expression relative to 18s RNA was analyzed by qRT-PCR (H). Cell growth and viability were measured by Cell Titer Flo after treating cells with palmitate for 72 hours (I). Infected OCI-AML-2 cells were treated with increasing concentrations of palmitate. 72 hours after treatment, cell viability was measured by Annexin V/PI staining (J). Infected OCI-AML-2 cells were treated with increasing concentrations of palmitate for 24 hours. ROS production was measured by staining with MitoSOX and flow cytometry (K). In all panels, error bars represent mean ± SD of independent/representative experiments. *P < .05; **P < .001 as determined by Tukey’s test after 1-way analysis of variance, comparing to controls. CPT1a and PPARα knockdown experiments were repeated twice.
To determine whether the effects of palmitate were mediated through mitochondrial fatty acid oxidation, we knocked down CPT1a (carnitine palmitoyltransferase 1a [liver]) and PPARα through lentiviral vector-mediated shRNA in OCI-AML-2 cells. CPT1a is a transmembrane protein of the mitochondrial outer membrane, which converts long-chain acyl-CoA such as palmitoyl to acyl carnitine, which enters the mitochondrial matrix and undergoes fatty acid oxidation.19 PPARα is a transcription factor that positively regulates β-fatty acid oxidation.20 Thus CPT1a and PPARα knockdown would prevent palmitate oxidation and entry of electrons through the respiratory chain. Consistent with the proposed mechanism, knockdown of CPT1a and PPARα abrogated the effects of palmitate on cell viability and mitochondrial ROS production (Figure 6D-K). Similar results were obtained for CPT1a knockdown in K562 cells (supplemental Figure 12).
As an alternate approach to induce oxidative stress by promoting electron flux through the respiratory chain, we treated cells with the cell-permeable TCA cycle component dimethyl succinate. Similar to the effects of palmitate, dimethyl succinate increased mitochondrial ROS production, and induced cell death in AML cells (supplemental Figure 13). Further supporting our proposed mechanism, sensitivity of AML cells to dimethyl succinate was increased by shifting metabolism toward OXPHOS by culturing AML cells in increased galactose-containing medium (supplemental Figure 13C-D). In addition, reducing dependence on mitochondrial metabolism by knockdown of Myc rendered cells resistant to dimethyl succinate (supplemental Figure 13F).
Palmitate induces oxidative stress in primary AML cells in vitro and in vivo
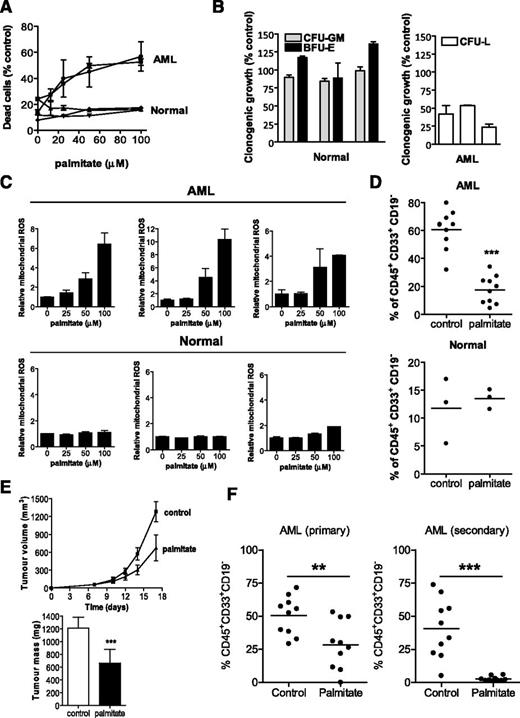
Next, we evaluated the effects of increasing oxidative stress on primary AML samples and normal hematopoietic cells. Similar to the abovementioned results using cell lines, primary AML cells were more sensitive to palmitate treatment than normal hematopoietic cells (Figure 7A). In addition, pretreatment of primary AML samples with palmitate reduced their clonogenic growth in colony-formation assays and reduced their ability to engraft immune-deficient mice, suggesting a selective effect on the AML progenitor population. In contrast, treatment of normal hematopoietic cells with palmitate did not inhibit their clonogenic growth or their ability to engraft mice (Figure 7B). Palmitate treatment increased ROS production in primary AML samples and had no effect on ROS production in normal hematopoietic cells (Figure 7C). Taken together, these results further support that promoting electron flux through the respiratory chain can target AML progenitor cells by increasing oxidative stress.
Palmitate demonstrates therapeutic efficacy on AML growing in vitro and in vivo. (A) CD34+ AML cells, normal bulk hematopoietic cells, and CD34+ normal hematopoietic cells were treated with increasing concentrations of palmitate (stock concentration of 2 mM palmitate conjugated with 0.17 mM bovine serum albumin). A total of 24 hours after treatment, cell viability was measured by Annexin V/PI staining. (B) Primary AML (n = 3) and normal hematopoietic cells (n = 3) were treated with 50 μM palmitate for 24 hours and were plated in clonogenic growth assays. The number of resultant colonies was counted, including CFU-GM, BFU-E, and CFU-L colony-forming units. The mean percentage of colonies obtained ± SD compared with buffer control–treated cells is shown. (C) Normal hematopoietic cells and primary AML samples were treated with increasing concentrations of palmitate. After 4 hours of treatment, levels of ROS were measured by staining with MitoSOX and flow cytometry. (D) Primary AML and Lin− CD34+-enriched human cord blood cells were treated with 50 μM palmitate or buffer control for 24 hours. After treatment, equal cell numbers were injected into the right femurs of irradiated NOD/SCID mice preconditioned with anti-CD122. Eight weeks later, the percentage of human CD45+CD33+CD19− cells in the noninjected femurs was measured by FACS. ***P < .0001 as determined by the unpaired Student t test. (E) NOD/SCID mice were injected subcutaneously with OCI- AML-2 leukemia cells. After tumors were palpable (day 7), mice were treated with palmitate or vehicle control as described in Materials and Methods. Tumor volume was measured with time and tumor mass was measured at the end of the experiment. Data represent mean ± SD. **P < .001, by Student t test. (F) Sublethally irradiated NOD/SCID mice preconditioned with anti-mouse CD122 were injected intrafemorally with primary AML cells. Six days after injection, mice were treated with palmitate or vehicle control as described in Materials and methods. Engraftment of human AML cells into the mouse marrow was assessed by determining the percentage of human CD45+CD33+CD19− cells by flow cytometry. *P < .01 by Student t test.
Palmitate demonstrates therapeutic efficacy on AML growing in vitro and in vivo. (A) CD34+ AML cells, normal bulk hematopoietic cells, and CD34+ normal hematopoietic cells were treated with increasing concentrations of palmitate (stock concentration of 2 mM palmitate conjugated with 0.17 mM bovine serum albumin). A total of 24 hours after treatment, cell viability was measured by Annexin V/PI staining. (B) Primary AML (n = 3) and normal hematopoietic cells (n = 3) were treated with 50 μM palmitate for 24 hours and were plated in clonogenic growth assays. The number of resultant colonies was counted, including CFU-GM, BFU-E, and CFU-L colony-forming units. The mean percentage of colonies obtained ± SD compared with buffer control–treated cells is shown. (C) Normal hematopoietic cells and primary AML samples were treated with increasing concentrations of palmitate. After 4 hours of treatment, levels of ROS were measured by staining with MitoSOX and flow cytometry. (D) Primary AML and Lin− CD34+-enriched human cord blood cells were treated with 50 μM palmitate or buffer control for 24 hours. After treatment, equal cell numbers were injected into the right femurs of irradiated NOD/SCID mice preconditioned with anti-CD122. Eight weeks later, the percentage of human CD45+CD33+CD19− cells in the noninjected femurs was measured by FACS. ***P < .0001 as determined by the unpaired Student t test. (E) NOD/SCID mice were injected subcutaneously with OCI- AML-2 leukemia cells. After tumors were palpable (day 7), mice were treated with palmitate or vehicle control as described in Materials and Methods. Tumor volume was measured with time and tumor mass was measured at the end of the experiment. Data represent mean ± SD. **P < .001, by Student t test. (F) Sublethally irradiated NOD/SCID mice preconditioned with anti-mouse CD122 were injected intrafemorally with primary AML cells. Six days after injection, mice were treated with palmitate or vehicle control as described in Materials and methods. Engraftment of human AML cells into the mouse marrow was assessed by determining the percentage of human CD45+CD33+CD19− cells by flow cytometry. *P < .01 by Student t test.
To assess the in vivo antileukemic efficacy of oxidative stress, we first used a leukemia xenograft model with the OCI-AML-2 cells. Mice were treated with palmitate or vehicle control for 11 days after tumors became palpable. Compared with vehicle control, palmitate decreased tumor mass and volume without any gross or histologic changes to the organs at necropsy (Figure 7D-E; supplemental Figure 14).
As an additional approach to assess the in vivo antileukemic efficacy of palmitate, we evaluated palmitate in mice engrafted with cells from primary AML samples. Primary AML cells were injected intrafemorally into irradiated NOD/SCID mice preconditioned with anti-CD122. Compared with vehicle control, 2 of 3 AML patient samples treated with palmitate decreased human leukemic burden in the mouse bone marrow without altering renal or liver functions (supplemental Figure 14G). Furthermore, engrafted AML cells harvested from the bone marrow of palmitate-treated primary mice had a dampened ability to engraft NOD/SCID mice in secondary transplant experiments (Figure 7F; supplemental Figure 15). Thus, our results suggest that overwhelming the respiratory chain displays antileukemic activity, including the ability to target AML stem and progenitor cells.
Discussion
In this study, we showed that AML cells have increased mitochondrial mass without a corresponding increase in the activity of the respiratory chain enzymes that contain mitochondrially encoded subunits (Complexes I, III, IV, and V). As a result, AML cells display lower spare reserve capacity in their respiratory chain compared with normal hematopoietic cells, a potential metabolic vulnerability.
Low spare reserve capacity in AML cells suggests that a subset of patients might benefit from strategies that target the OXPHOS chain. The antidiabetic agent and a known inhibitor of complex I, metformin, has been evaluated in preclinical and clinical studies of solid tumors and has shown promising results.21 This agent could potentially also be evaluated in AML. In addition, a potent inhibitor of respiratory complex I, IACS-1131, selectively induced death in a subset of AML cells and patient samples preferentially over normal hematopoietic cells.22 Adding to the potential value of targeting the OXPHOS chain, a report by Lagadinou et al23 demonstrated that AML cells and stem cells cannot upregulate glycolysis after the inhibition of OXPHOS.
Interestingly, some of our results in AML appear consistent with the metabolic consequences of aging. In studies of rat neuron cells, respiratory rates increase, spare reserve capacity declines, and mitochondrial mass increases with aging.24-27 The cause of the decline in the spare reserve capacity in these aging rat cells is unclear, but may relate to the accumulation of nitric oxide that damages respiratory complexes.10,27 The aging mitochondria and dysfunctional respiratory complexes lead to increased ROS production, which results in further damage to the mitochondria. In addition, aging mitochondria accumulate mitochondrial DNA damage that can also impair respiratory complex activity and increase ROS production.25,28,29 Thus, we speculate that the increased demands on mitochondrial activity in AML lead to premature aging with a resultant decline in spare reserve capacity of the respiratory chain.
Previous studies have demonstrated that AML cells have increased rates of fatty acid oxidation.2 As such, attention has focused on inhibiting fatty acid oxidation as a therapeutic strategy for AML. In contrast, we demonstrated that AML cells were vulnerable to strategies that promote oxidative metabolism and increase electron flux through the respiratory chain. Using palmitate and dimethyl succinate, we increased cellular levels of TCA cycle substrates and electron flux through the respiratory chain. This induced oxidative stress in AML cells, triggering cell death. This work is not meant to suggest that patients with AML should be placed on high-fat diets. Rather, it highlights that strategies that promote electron flux through the respiratory chain may offer a new therapeutic strategy for some AML patients. Through its ability to increase mitochondrial mass and β-oxidation,30,31 the PPARα agonist bezafibrate may include antileukemic activity. Alternatively, promoting OXPHOS flux with compounds such as 2-deoxyglucose, 3-bromopyruvate,32,33 or dichloroacetic acid34 may also selectively induce death in a subset of AML cells, and it could be evaluated alone or in combination with standard chemotherapeutic agents.
In conclusion, AML cells have dysregulated mitochondrial biogenesis and metabolism. These abnormalities highlight new vulnerabilities and potential novel therapeutic strategies for the treatment of this disease. Thus, although the genetic heterogeneity of AML may be difficult to target therapeutically, the resultant metabolic dysfunction may be more amenable to therapeutic intervention.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Jill Flewelling for administrative assistance and Aisha Shamas-Din for help with preparing the final manuscript.
This work was supported by the Canadian Stem Cell Network, the Leukemia and Lymphoma Society, the National Institutes of Health (National Cancer Institute 1R01CA157456), The Ontario Ministry of Research and Innovation, the Princess Margaret Hospital Foundation, and the Ministry of Long Term Health and Planning in the Province of Ontario. This work was also supported by the Barbara Baker chair in Leukemia and Related Diseases (A.D.S.). D.V.J is a Fonds de recherche du Québec—Santé postdoctoral scholar.
Authorship
Contribution: S.S. and D.V.J. designed the study, collected and analyzed data, and wrote the paper; S.P., T.E.C., W.X., M.S., B.J., R.H., M.G., X.W., Y.J., M.A.S., F.-H.L., N.M., R.L., and S.X. performed experiments and collected and analyzed data; C.A.G. analyzed data and helped write the manuscript; P.J.M., L.Z.P., and J.E.D. contributed technical expertise and resources; I.M.R. and M.D.M. contributed patient material; and A.D.S. designed the study and wrote the paper. All authors reviewed and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.S. is Clark Smith Brain Tumour Centre, Southern Alberta Cancer Research Institute, Calgary, AB, Canada.
Correspondence: Aaron D. Schimmer, Princess Margaret Cancer Centre, Room 7-116, 610 University Ave, Toronto, ON, Canada M5G 2M9; e-mail: aaron.schimmer@utoronto.ca.
References
Author notes
S.S. and D.V.J. contributed equally to this study.