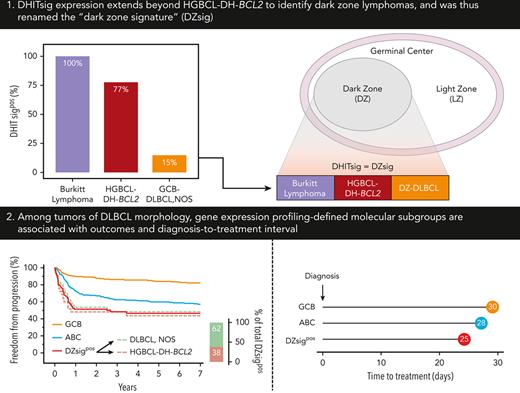
Key Points
DHITsig expression extends beyond HGBCL-DH-BCL2 to identify dark zone lymphomas and was thus renamed the “DZsig.”
DZsig refines cell-of-origin classification by identifying patients within GCB-DLBCL with inferior outcomes and shorter time to treatment.
Abstract
Molecular heterogeneity of diffuse large B-cell lymphoma (DLBCL) underlies the variable outcomes achieved with immunochemotherapy. However, outcomes of gene expression profiling (GEP)–defined molecular subgroups in a real-world DLBCL population remain unknown. Here we examined the prevalence and outcomes of molecular subgroups in an unselected population of 1149 patients with de novo DLBCL in British Columbia, Canada. Evaluable biopsies were profiled by fluorescence in situ hybridization (FISH), immunohistochemistry, and digital GEP to assign cell-of-origin and the so-called “double-hit signature” (DHITsig)—a signature originally described as being characteristic for high-grade B-cell lymphoma with MYC and BCL2 rearrangements (HGBCL-DH-BCL2). DHITsig was expressed in 21% of 431 germinal center B-cell-like (GCB)–DLBCL and all 55 Burkitt lymphomas examined. Reflecting this latter finding, DHITsig has been renamed the “dark zone signature” (DZsig). DZsigpos-DLBCL, non-DZsigpos GCB-DLBCL and activated B-cell-like (ABC)–DLBCL were associated with a 2 year overall survival of 57%, 89%, and 71%, respectively. 62% of DZsigpos tumors were negative for HGBCL-DH-BCL2 by FISH, but were associated with outcomes similar to HGBCL-DH-BCL2. A small group of HGBCL-DH-BCL2 that lacked DZsig expression had different molecular features compared with DZsig-expressing HGBCL-DH-BCL2 and were associated with favorable outcomes comparable to DLBCL, not otherwise specified. DZsigpos and ABC-DLBCL had a shorter diagnosis-to-treatment interval (DTI) than GCB-DLBCL, with this metric being associated with outcome. In conclusion, DZsig expression extends beyond HGBCL-DH-BCL2 and captures a poor-prognosis DLBCL subgroup with short DTI, including patients unidentifiable by routine FISH testing, that should be considered for treatment intensification or novel therapies in prospective trials.
Introduction
Although 60% to 70% of patients with diffuse large B-cell lymphoma (DLBCL) are cured with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) immunochemotherapy, patients who develop relapsed or refractory disease after frontline treatment have poor outcomes.1,2 Heterogeneity in treatment response provoked intense efforts over the past 2 decades to identify prognostic and predictive biomarkers to refine therapeutic strategies. Cell-of-origin (COO) classification created the foundations for this endeavor, using gene expression profiling (GEP) to dichotomize the majority of DLBCL tumors into the germinal center B-cell–like (GCB-DLBCL) and activated B-cell–like (ABC-DLBCL) subgroups,3 with ∼15% to 25% higher 3 year progression-free survival in GCB-DLBCL vs ABC-DLBCL following R-CHOP.4-6 Although GCB-DLBCL constitutes the majority of DLBCL in the Western world,4,6 COO distribution varies globally,7,8 and some studies suggest that the proportion of ABC-DLBCL increases with age.9 Recent large-scale studies identified unique patterns of recurrent genetic alterations, both within and beyond COO, which further refine molecular subtypes of DLBCL, offering the potential for personalized therapy based on targetable biology.10-13 Despite this progress, these genetics based classifications do not fully account for the variable responses following R-CHOP, and with >30% of tumors unclassified (UNC), genetic classification is yet to be optimized for routine clinical application.12
Approximately 8% of DLBCL are reclassified to high-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements (HGBCL-DH), based on the detection of gene rearrangements typically using fluorescence in situ hybridization (FISH).14 The homogenous mutational landscape of HGBCL-DH with BCL2 rearrangement, irrespective of BCL6 rearrangement (HGBCL-DH-BCL2), has led to its recognition as a distinct entity in the latest classification systems.15,16 In contrast, the heterogeneity of HGBCL with MYC and BCL6 rearrangements (HGBCL-DH-BCL6) has resulted in relegation of this group to a provisional entity in the International Consensus Classification15 and reversion to DLBCL, not otherwise specified (NOS) or HGBCL, NOS, according to cytomorphological features in the 5th edition of the World Health Organization Classification of Haematolymphoid Tumors.16 Retrospective analyses reported poor outcomes of HGBCL-DH with R-CHOP17-19 prompting many centers to use dose-intensive regimens, although randomized data supporting this approach are lacking.
We recently described a unifying gene expression signature in HGBCL-DH-BCL2, which was translated into a digital GEP-based assay (DLBCL90) that can be applied to routine formalin-fixed, paraffin-embedded (FFPE) biopsies.20 Double-hit signature (DHITsig) expression extends beyond HGBCL-DH-BCL2 to identify a subset of DLBCL tumors that lack MYC and/or BCL2 rearrangements by FISH,20,21 ∼20% of which harbor cryptic rearrangements.21 Although rearrangements are the only structural variants (SVs) incorporated in the definition of HGBCL-DH, an atypical double-hit (atypical-DH) category has been proposed that comprises any other combination of SVs (rearrangement or copy number variation) in MYC and BCL2 and/or BCL6, with some reports suggesting outcomes comparable with HGBCL-DH.22-24 However, unlike HGBCL-DH-BCL2, atypical-DH tumors are rarely DHITsig positive (DHITsigpos).25 The majority of DHITsigpos tumors are classified into the LymphGen EZB genetic subtype, and although DHITsig expression within the EZB subtype is associated with inferior outcomes, limited genetic differences separate DHITsigpos and DHITsig-negative (DHITsigneg) EZB tumors.12 Potential mechanisms contributing to DHITsig expression beyond genetic alterations remain unknown. In this study, the spectrum of germinal center (GC) aggressive B-cell lymphomas that express this signature was explored by assessing Burkitt lymphoma (BL).
Most studies examining clinical outcomes of molecular subgroups in DLBCL were conducted using cohorts selected out of population registries and/or compiled from clinical trials. Real-world outcomes on a population level have not been systematically examined to date. Furthermore, previous analyses were often restricted to biopsies with a matched source of frozen biopsy material6 or incisional/excisional biopsies large enough for tissue microarray (TMA) construction,19 which excludes core biopsies despite their increasing trend in use.26,27 Here, we report the molecular determinants of real-world outcomes in a population-based DLBCL cohort from British Columbia (BC), Canada, treated under uniform provincial guidelines by profiling all evaluable diagnostic FFPE biopsies. We further examined the association between GEP–defined molecular subgroups and diagnosis-to-treatment interval (DTI), a metric that affects patient outcomes and clinical trial inclusion in DLBCL.28,29
Materials and methods
Patient cohorts
The study cohort comprised all patients diagnosed with de novo DLBCL, NOS or HGBCL-DH with DLBCL morphology in BC between 2005 and 2010, identified using the BC Cancer Lymphoid Cancer Database (supplemental Table 1, available on the Blood website). Biopsies from previously defined DLBCL6 and BL30 cohorts were used in extended molecular analyses (supplemental Tables 2 and 3). All cohort characteristics are described in the supplemental Methods. This study was approved by The University of British Columbia-BC Cancer Research Ethics Board in accordance with the Declaration of Helsinki.
Biopsy evaluation, FISH, and immunohistochemistry (IHC)
All available diagnostic biopsies underwent central pathology review to confirm the diagnosis and to select a representative area for TMA construction as previously described.6 Bone/bone marrow (BM) biopsies and biopsies with tumor surface area of <2 mm2 were excluded from research analysis. MYC, BCL2, and BCL6 FISH were performed on TMA or whole tissue section for small biopsies unsuitable for TMA. IHC was performed on TMA sections using commercially available antibodies. IHC positivity was defined as ≥30% (CD10, BCL6, and MUM1),31 ≥40% (MYC), and ≥50% (BCL2) staining of tumor cells.17 Clinical IHC and FISH results were used to complete missing data when available. Further details are in the supplemental Methods.
GEP and molecular subtyping
Digital GEP was performed on RNA extracted from FFPE material using the DLBCL90 assay on the nCounter platform (NanoString Technologies, Seattle, WA) to assign COO and DHITsig status and to determine MYC and BCL2 messenger RNA expression levels, as previously described.20,21 Gene expression subgroups were assigned hierarchically, with COO taking precedence over DHITsig status for ABC tumors. GCB and UNC tumors that were DHITsigpos were assigned to the DHITsigpos group, whereas DHITsigind and DHITsigneg tumors were assigned to their respective COO subgroups (supplemental Figure 1). MYC rearrangement partner identification by sequencing is described in the supplemental Methods.
DTI analysis
DTI was defined as the interval between the date of the definitive DLBCL diagnostic biopsy and the date on which the first immunochemotherapy cycle was administered, identified using the BC Cancer Pharmacy database.
Statistical analysis
Baseline patient clinical and tumor pathological characteristics were compared using χ2 or Fisher exact tests. Survival endpoints are defined in the supplemental Methods. Kaplan-Meier analysis was used to estimate survival rates and groups were compared using the log-rank test. Cox proportional hazard models were used to assess the association between GEP–defined molecular subgroups and clinical outcomes after adjusting for known prognostic factors. DTI of molecular subgroups were compared using negative binomial regression. Statistical analyses were performed using GraphPad Prism (version 9.3.1, GraphPad Software, San Diego, CA) or R statistical computing software (version 4.2.0; R Core Team 2022).
Results
Characteristics of the study population
Of the 1149 patients in the study population (supplemental Table 1), 902 had an evaluable clinical diagnostic biopsy (supplemental Figure 3A), 649 of which were included on TMA. Biopsies were unevaluable if they were not available (n = 34), had insufficient remaining material for research analysis (n = 159), or were obtained from bone/BM (n = 54; where sample processing precludes the molecular profiling performed in this study). Clinical FISH and IHC data were used for unevaluable biopsies when available. Assay result availability of the study cohort is shown in supplemental Figure 4.
GEP data were available for 804 patients in the study population, including 629 of the 866 patients treated with R-CHOP (supplemental Figure 3A-B). Comparison of the baseline clinical characteristics between patients with available GEP data and the entire study population showed that patients with GEP data were broadly representative of the study population, except for a significantly lower proportion of patients with stage III/IV disease and >1 extranodal site (Table 1, left panel). In patients treated with R-CHOP, the proportion of patients with >1 extranodal site was the only significant difference (Table 1, right panel). There was no significant difference in outcomes between the total study population and patients with available GEP data, although patients with missing GEP data had significantly inferior outcomes than patients with available GEP data (supplemental Figure 5A-B). Among the reasons for missing GEP data, patients with insufficient material or with a bone/BM biopsy were associated with inferior outcomes compared with patients with available GEP data (supplemental Figure 5C-D).
Comparison of baseline clinical characteristics between the entire study population and patients with evaluable GEP data
| Characteristic . | Entire cohort . | R-CHOP-treated patients . | ||||
|---|---|---|---|---|---|---|
| ALL . | With GEP data (DLBCL90) . | P . | ALL . | With GEP data (DLBCL90) . | P . | |
| Total (n) | 1149 | 804 | 866 | 629 | ||
| Age, median (range, y) | 69 (19-96) | 69 (20-96) | .36 | 66 (19-93) | 66 (20-93) | .72 |
| Age > 60 y (n, %) | 824 (72) | 565 (70) | .51 | 573 (66) | 409 (65) | .66 |
| Female sex (n, %) | 500 (44) | 351 (44) | .96 | 381 (44) | 274 (44) | .87 |
| Stage (n, %) | .02 | .08 | ||||
| I/II | 436 (41) | 345 (46) | 369 (44) | 296 (49) | ||
| III/IV | 633 (59) | 402 (54) | 471 (56) | 313 (51) | ||
| NA | 80 | 57 | 26 | 20 | ||
| PS (n, %) | .07 | .15 | ||||
| 0-1 | 593 (57) | 447 (61) | 523 (64) | 403 (68) | ||
| 2-4 | 445 (43) | 280 (39) | 289 (36) | 188 (32) | ||
| NA | 111 | 77 | 54 | 38 | ||
| LDH (n, %) | .28 | .23 | ||||
| Normal | 467 (50) | 348 (53) | 386 (52) | 299 (55) | ||
| Elevated | 459 (50) | 305 (47) | 357 (48) | 240 (45) | ||
| NA | 223 | 151 | 123 | 90 | ||
| Extranodal sites (n, %) | .04 | .04 | ||||
| 0-1 | 800 (75) | 591 (79) | 633 (75) | 487 (80) | ||
| ≥2 | 269 (25) | 156 (21) | 207 (25) | 122 (20) | ||
| NA | 80 | 57 | 26 | 20 | ||
| B symptoms (n, %) | .16 | .21 | ||||
| No | 677 (64) | 502 (68) | 555 (66) | 422 (70) | ||
| Yes | 375 (36) | 240 (32) | 280 (34) | 184 (30) | ||
| NA | 97 | 62 | 31 | 23 | ||
| Bulky disease ≥10 cm (n, %) | .28 | .34 | ||||
| No | 727 (71) | 531 (73) | 567 (71) | 425 (73) | ||
| Yes | 296 (29) | 192 (27) | 237 (29) | 158 (27) | ||
| NA | 126 | 81 | 62 | 46 | ||
| IPI risk group (n, %) | .07 | .13 | ||||
| Low (0-1) | 272 (29) | 225 (34) | 247 (33) | 207 (38) | ||
| Low-intermediate (2) | 215 (23) | 157 (24) | 180 (24) | 134 (25) | ||
| High-intermediate (3) | 216 (23) | 141 (21) | 170 (23) | 113 (21) | ||
| High (4, 5) | 235 (25) | 136 (21) | 158 (21) | 92 (17) | ||
| NA | 211 | 145 | 111 | 83 | ||
| HGBCL-DH status | .93 | .95 | ||||
| Non-HGBCL-DH | 781 (92) | 700 (92) | 607 (92) | 553 (93) | ||
| HGBCL-DH-BCL2 | 53 (6) | 44 (6) | 42 (6) | 36 (6) | ||
| HGBCL-DH-BCL6 | 15 (2) | 14 (2) | 8 (1) | 8 (1) | ||
| NA | 300 | 46 | 209 | 32 | ||
| Treatment regimen (n, %) | .74 | |||||
| R-CHOP | 866 (79) | 629 (82) | ||||
| R-CHOP-HDMTX | 6 (0.5) | 4 (0.5) | ||||
| R-CEOP | 54 (5) | 38 (5) | ||||
| CEOP/CHOP∗ | 2 (0.2) | 0 | ||||
| Palliative | 158 (14) | 89 (12) | ||||
| Intensive | 7 (0.6) | 4 (0.5) | ||||
| Patient refused | 10 (0.9) | 7 (0.9) | ||||
| Unknown | 46 | 33 | ||||
| Characteristic . | Entire cohort . | R-CHOP-treated patients . | ||||
|---|---|---|---|---|---|---|
| ALL . | With GEP data (DLBCL90) . | P . | ALL . | With GEP data (DLBCL90) . | P . | |
| Total (n) | 1149 | 804 | 866 | 629 | ||
| Age, median (range, y) | 69 (19-96) | 69 (20-96) | .36 | 66 (19-93) | 66 (20-93) | .72 |
| Age > 60 y (n, %) | 824 (72) | 565 (70) | .51 | 573 (66) | 409 (65) | .66 |
| Female sex (n, %) | 500 (44) | 351 (44) | .96 | 381 (44) | 274 (44) | .87 |
| Stage (n, %) | .02 | .08 | ||||
| I/II | 436 (41) | 345 (46) | 369 (44) | 296 (49) | ||
| III/IV | 633 (59) | 402 (54) | 471 (56) | 313 (51) | ||
| NA | 80 | 57 | 26 | 20 | ||
| PS (n, %) | .07 | .15 | ||||
| 0-1 | 593 (57) | 447 (61) | 523 (64) | 403 (68) | ||
| 2-4 | 445 (43) | 280 (39) | 289 (36) | 188 (32) | ||
| NA | 111 | 77 | 54 | 38 | ||
| LDH (n, %) | .28 | .23 | ||||
| Normal | 467 (50) | 348 (53) | 386 (52) | 299 (55) | ||
| Elevated | 459 (50) | 305 (47) | 357 (48) | 240 (45) | ||
| NA | 223 | 151 | 123 | 90 | ||
| Extranodal sites (n, %) | .04 | .04 | ||||
| 0-1 | 800 (75) | 591 (79) | 633 (75) | 487 (80) | ||
| ≥2 | 269 (25) | 156 (21) | 207 (25) | 122 (20) | ||
| NA | 80 | 57 | 26 | 20 | ||
| B symptoms (n, %) | .16 | .21 | ||||
| No | 677 (64) | 502 (68) | 555 (66) | 422 (70) | ||
| Yes | 375 (36) | 240 (32) | 280 (34) | 184 (30) | ||
| NA | 97 | 62 | 31 | 23 | ||
| Bulky disease ≥10 cm (n, %) | .28 | .34 | ||||
| No | 727 (71) | 531 (73) | 567 (71) | 425 (73) | ||
| Yes | 296 (29) | 192 (27) | 237 (29) | 158 (27) | ||
| NA | 126 | 81 | 62 | 46 | ||
| IPI risk group (n, %) | .07 | .13 | ||||
| Low (0-1) | 272 (29) | 225 (34) | 247 (33) | 207 (38) | ||
| Low-intermediate (2) | 215 (23) | 157 (24) | 180 (24) | 134 (25) | ||
| High-intermediate (3) | 216 (23) | 141 (21) | 170 (23) | 113 (21) | ||
| High (4, 5) | 235 (25) | 136 (21) | 158 (21) | 92 (17) | ||
| NA | 211 | 145 | 111 | 83 | ||
| HGBCL-DH status | .93 | .95 | ||||
| Non-HGBCL-DH | 781 (92) | 700 (92) | 607 (92) | 553 (93) | ||
| HGBCL-DH-BCL2 | 53 (6) | 44 (6) | 42 (6) | 36 (6) | ||
| HGBCL-DH-BCL6 | 15 (2) | 14 (2) | 8 (1) | 8 (1) | ||
| NA | 300 | 46 | 209 | 32 | ||
| Treatment regimen (n, %) | .74 | |||||
| R-CHOP | 866 (79) | 629 (82) | ||||
| R-CHOP-HDMTX | 6 (0.5) | 4 (0.5) | ||||
| R-CEOP | 54 (5) | 38 (5) | ||||
| CEOP/CHOP∗ | 2 (0.2) | 0 | ||||
| Palliative | 158 (14) | 89 (12) | ||||
| Intensive | 7 (0.6) | 4 (0.5) | ||||
| Patient refused | 10 (0.9) | 7 (0.9) | ||||
| Unknown | 46 | 33 | ||||
P values derived from Fisher exact test except for median age where Mann-Whitney test was used. P < .05 have been highlighted in bold.
HDMTX, high-dose methotrexate; IPI, international prognostic index; LDH, lactate dehydrogenase; NA, not available; non-HGBCL-DH, no double-hit lymphoma; PS, performance status; R-CEOP, rituximab, cyclophosphamide, etoposide, vincristine, and prednisone.
Rituximab intentionally omitted.
DHITsig expression extends beyond HGBCL-DH-BCL2 to identify dark zone (DZ) lymphomas
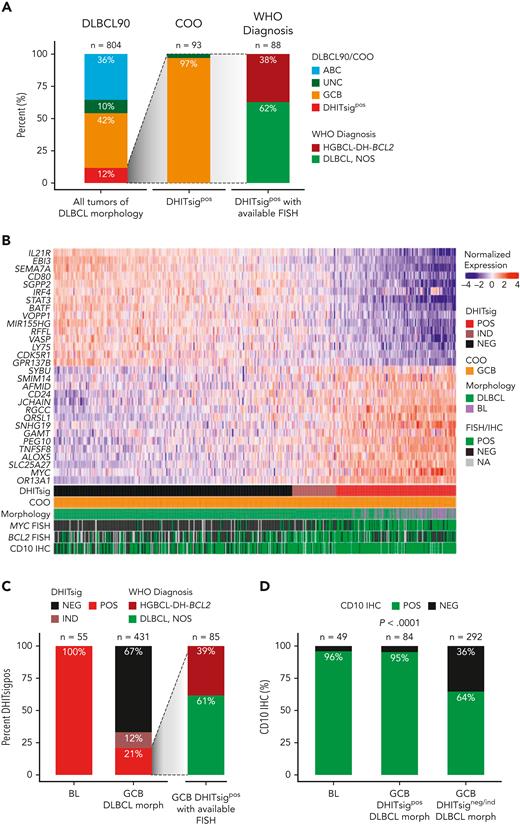
Expression of DHITsig was observed in 93 of 804 (12%) of all tumors of DLBCL morphology (Figure 1A), with 90 (97%) and 3 (3%) of DHITsigpos tumors having GCB and UNC COO, respectively. HGBCL-DH-BCL2, determined using FISH, made up only 33 of 88 (38%) of DHITsigpos tumors with available FISH data. The observation that the majority are not HGBCL-DH-BCL2, along with the alignment of DHITsig with GC DZ gene expression20 prompted assessment of DHITsig expression in the archetypical DZ lymphoma, BL. All 55 BLs profiled were DHITsigpos (Figure 1B-C) and both BL and DHITsigpos GCB tumors showed near-universal expression of the GC marker CD10 (Figure 1D). Altogether, DHITsigpos identifies a group of aggressive B-cell lymphomas of GC origin with a common gene expression phenotype which reflects the GC DZ, extending beyond HGBCL-DH-BCL2. These observations motivated us to rename the DHITsig to the DZ signature (DZsig) and will henceforth be referred to as such.
DHITsig expression extends beyond HGBCL-DH-BCL2 to identify DZ lymphomas. (A) Characterization of DHITsigpos tumors with DLBCL morphology. Left bar: GEP-based classification of all tumors with DLBCL morphology in the study population using the DLBCL90 assay (Nanostring), based on the algorithm in supplemental Figure 1. Central bar: COO classification of DHITsigpos tumors. Right bar: HGBCL-DH-BCL2 status of DHITsigpos tumors as determined by FISH. Tumors negative for HGBCL-DH-BCL2 are classified as DLBCL, NOS according to WHO HAEM5. (B) Heat map showing normalized expression of DHITsig genes in 55 BL tumors from an independent cohort and 431 germinal-center B-cell-like tumors with DLBCL morphology (GCB-DLBCL) from the study population. (C) Proportion of DHITsigpos tumors in BL (left bar) vs GCB-DLBCL (central bar). All profiled BL tumors were DHITsigpos. Right bar: proportion of DHITsigpos GCB-DLBCL tumors that are HGBCL-DH-BCL2 vs DLBCL, NOS. (D) Proportion of tumors positive for the germinal center marker CD10 by IHC in BL and GCB-DLBCL in the presence (GCB-DLBCL DHITsigpos) or absence of DHITsig (GCB-DLBCL DHITsigneg/ind), compared with χ2 test. IND, indeterminate; morph, morphology; NA, not available; NEG, negative; POS, positive; WHO HAEM5, 5th edition of the World Health Organization Classification of Haematolymphoid Tumors.
DHITsig expression extends beyond HGBCL-DH-BCL2 to identify DZ lymphomas. (A) Characterization of DHITsigpos tumors with DLBCL morphology. Left bar: GEP-based classification of all tumors with DLBCL morphology in the study population using the DLBCL90 assay (Nanostring), based on the algorithm in supplemental Figure 1. Central bar: COO classification of DHITsigpos tumors. Right bar: HGBCL-DH-BCL2 status of DHITsigpos tumors as determined by FISH. Tumors negative for HGBCL-DH-BCL2 are classified as DLBCL, NOS according to WHO HAEM5. (B) Heat map showing normalized expression of DHITsig genes in 55 BL tumors from an independent cohort and 431 germinal-center B-cell-like tumors with DLBCL morphology (GCB-DLBCL) from the study population. (C) Proportion of DHITsigpos tumors in BL (left bar) vs GCB-DLBCL (central bar). All profiled BL tumors were DHITsigpos. Right bar: proportion of DHITsigpos GCB-DLBCL tumors that are HGBCL-DH-BCL2 vs DLBCL, NOS. (D) Proportion of tumors positive for the germinal center marker CD10 by IHC in BL and GCB-DLBCL in the presence (GCB-DLBCL DHITsigpos) or absence of DHITsig (GCB-DLBCL DHITsigneg/ind), compared with χ2 test. IND, indeterminate; morph, morphology; NA, not available; NEG, negative; POS, positive; WHO HAEM5, 5th edition of the World Health Organization Classification of Haematolymphoid Tumors.
The prevalence and real-world outcomes of GEP-defined subgroups in DLBCL
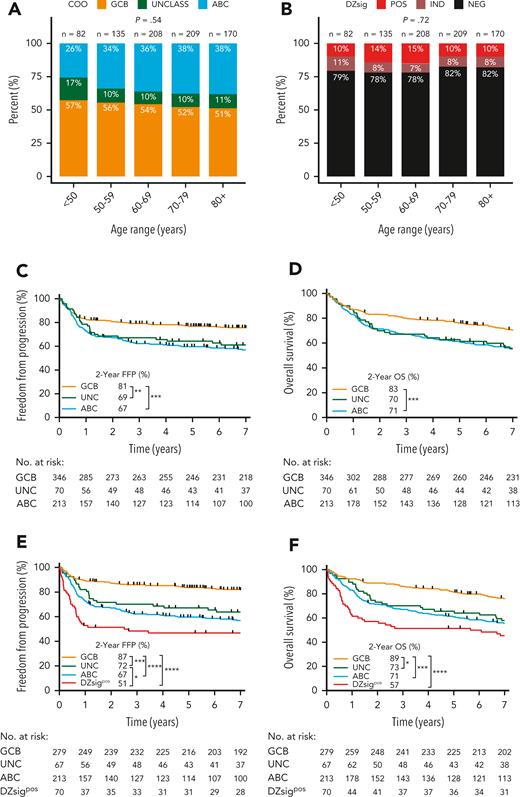
Among patients with DLBCL with GEP data, the proportion of COO subtypes was 54% GCB, 11% UNC, and 36% ABC. There was no significant difference in the proportion of COO subtypes across age groups (<50 years, 50-60, 60-70, 70-80, >80 years; Figure 2A) at diagnosis. Similarly, there was no significant difference in the proportion of DZsigpos across age groups (Figure 2B).
Proportion by age group and clinical outcomes of DLBCL patients in the study population according to GEP–defined molecular subgroups using the DLBCL90 assay. (A) COO subgroup distribution according to age group at diagnosis. (B) DZsig distribution according to age group at diagnosis. P values of categorical comparisons across age groups were derived from χ2 test. (C-D) FFP and OS according to COO classification in patients treated with curative-intent R-CHOP. (E-F) FFP and OS in patients treated with R-CHOP after removing patients with DZsig positive (DZsigpos) DLBCL from the GCB and unclassified COO (UNCLASS) subgroups, to form the DZsigpos subgroup. Log-rank test was used to compare survival curves. ∗∗∗∗P < .0001, ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.
Proportion by age group and clinical outcomes of DLBCL patients in the study population according to GEP–defined molecular subgroups using the DLBCL90 assay. (A) COO subgroup distribution according to age group at diagnosis. (B) DZsig distribution according to age group at diagnosis. P values of categorical comparisons across age groups were derived from χ2 test. (C-D) FFP and OS according to COO classification in patients treated with curative-intent R-CHOP. (E-F) FFP and OS in patients treated with R-CHOP after removing patients with DZsig positive (DZsigpos) DLBCL from the GCB and unclassified COO (UNCLASS) subgroups, to form the DZsigpos subgroup. Log-rank test was used to compare survival curves. ∗∗∗∗P < .0001, ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.
To determine the impact of molecular signatures on real-world clinical outcomes, freedom from progression (FFP) and overall survival (OS) were examined according to COO in patients treated with R-CHOP. GCB-DLBCL had 14% and 12% higher 2-year FFP and OS compared with ABC-DLBCL, respectively (Figure 2C-D). A similar relationship was observed when all 804 patients with GEP data were analyzed, irrespective of treatment (supplemental Figure 6A-B).
Next, DZsigpos was incorporated as a fourth group which subsumed 90 GCB and 3 UNC tumors, constituting 18% of all GCB/UNC-DLBCL. Two ABC tumors that were DZsigpos, remained assigned to the ABC group based on the algorithm in supplemental Figure 1. Patients with DZsigind tumors had outcomes intermediate between DZsigpos and DZsigneg following R-CHOP (supplemental Figure 7). DZsigind and DZsigneg tumors remained in their respective COO groups in subsequent analyses. Comparisons of baseline clinical and pathologic characteristics among the molecular subgroups in patients treated with R-CHOP showed significant differences in stage and lactate dehydrogenase levels (Table 2). Of the 4 subgroups, DZsigpos had the worst outcomes with 2-year FFP and OS of 51% and 57%, respectively. After removing patients with DZsigpos tumors from the GCB-DLBCL group, the remaining patients with DZsigneg/ind GCB-DLBCL had excellent outcomes, with 2-year FFP and OS at 87% and 89%, respectively. A nonsignificant trend toward inferior outcomes of DZsigpos relative to ABC was seen (2-year FFP: 51% vs 67%, P = .07; 2-year OS: 57% vs 71%, P = .12) (Figure 1E-F). After adjusting for individual international prognostic index (IPI) risk factors by multivariable Cox regression, DZsigpos was significantly associated with inferior FFP (hazard ratio, 3.9; 95% confidence interval [CI], 2.5-6.0) and OS (hazard ratio, 2.7; 95% CI, 1.9-3.9) relative to GCB-DLBCL (supplemental Table 5). When restricting the analysis to 157 patients treated with R-CHOP with limited-stage disease, ABC-DLBCL was associated with inferior FFP and OS compared with GCB-DLBCL. Limited by a small number of patients with DZsigpos (n = 13), no significant difference in outcomes between DZsigpos and the other subgroups was found in limited-stage disease. In advanced-stage disease, patients with DZsigpos had significantly inferior outcomes compared with patients with GCB-DLBCL, in whom excellent outcomes were maintained with a 2-year OS of 90% (supplemental Figure 8).
Baseline clinical and pathological characteristics of patients treated with R-CHOP according to molecular subgroups defined by gene expression profile (DLBCL90)
| Characteristic of R-CHOP-treated patients . | DZsigpos . | GCB . | UNC . | ABC . | P . |
|---|---|---|---|---|---|
| Total (n) | 70 | 279 | 67 | 213 | |
| Age, median (range, y) | 64 (35-88) | 67 (22-92) | 64 (20-92) | 67 (31-93) | .18∗ |
| Age > 60 y (n, %) | 45 (64) | 181 (65) | 40 (60) | 143 (67) | .74 |
| Female sex (n, %) | 26 (37) | 122 (44) | 24 (36) | 102 (48) | .23 |
| Stage (n, %) | .01 | ||||
| I/II | 30 (43) | 151 (55) | 30 (46) | 85 (42) | |
| III/IV | 39 (57) | 120 (44) | 35 (54) | 119 (58) | |
| NA | 1 | 8 | 2 | 9 | |
| PS (n, %) | .29 | ||||
| 0-1 | 45 (65) | 190 (72) | 40 (65) | 128 (65) | |
| 2-4 | 24 (35) | 73 (28) | 22 (35) | 69 (35) | |
| NA | 1 | 16 | 5 | 16 | |
| LDH (n, %) | <.01 | ||||
| Normal | 24 (37) | 144 (61) | 37 (61) | 94 (53) | |
| Elevated | 40 (63) | 93 (39) | 24 (39) | 83 (47) | |
| NA | 6 | 42 | 6 | 36 | |
| Extranodal sites (n, %) | .47 | ||||
| 0-1 | 53 (77) | 221 (82) | 55 (85) | 158 (77) | |
| ≥ 2 | 16 (23) | 50 (18) | 10 (15) | 46 (23) | |
| NA | 1 | 12 | 2 | 9 | |
| B symptoms (n, %) | .34 | ||||
| No | 46 (67) | 197 (74) | 43 (66) | 136 (67) | |
| Yes | 23 (33) | 71 (26) | 22 (34) | 68 (33) | |
| NA | 1 | 7 | 2 | 9 | |
| Bulky disease ≥10 cm (n, %) | .34 | ||||
| No | 43 (67) | 186 (71) | 50 (79) | 146 (75) | |
| Yes | 21 (33) | 76 (29) | 13 (21) | 48 (24) | |
| NA | 6 | 17 | 4 | 19 | |
| IPI risk group (n, %) | .09 | ||||
| Low (0-1) | 20 (30) | 105 (44) | 22 (37) | 60 (33) | |
| Low-intermediate (2) | 16 (24) | 64 (27) | 16 (27) | 38 (21) | |
| High-intermediate (3) | 17 (26) | 42 (18) | 12 (20) | 42 (23) | |
| High (4, 5) | 13 (20) | 29 (12) | 10 (17) | 40 (22) | |
| Not calculable | 4 | 39 | 7 | 33 | |
| FISH-DH status, (n, %) | <.001 | ||||
| HGBCL-DH-BCL2 | 25 (38) | 9 (3) | 1 (2) | 1 (0.5) | |
| HGBCL-DH-BCL6 | 3 (5) | 1 (1) | 0 (0) | 4 (2) | |
| Atypical-DH† | 13 (20) | 57 (22) | 13 (20) | 42 (20) | |
| Non-DH | 25 (38) | 193 (74) | 51(78) | 159 (77) | |
| NA | 4 | 19 | 2 | 7 | |
| FISH total R, (n total/n data available, %)‡ | |||||
| Total MYC-R | 38/67 (57) | 15/262 (6) | 4/66 (6) | 16/207 (8) | <.001 |
| Total BCL2-R | 44/62 (71) | 81/221 (37) | 15/58 (26) | 3/178 (2) | <.001 |
| Total BCL6-R | 11/61 (18) | 43/213 (20) | 27/53 (51) | 55/167 (33) | <.001 |
| mRNA by digital GEP, mean log count (SD)‡ | |||||
| MYC | 11.9 (1.4) | 10.7 (0.81) | 10.8 (0.8) | 11.5 (0.9) | < .0001∗ |
| BCL2 | 12.4 (1.9) | 11.2 (1.6) | 11.7 (1.3) | 12.0 (1.3) | < .0001∗ |
| IHC, (n total/n data available, %)‡ | |||||
| MYC/BCL2 DPE | 32/58 (55) | 16/238 (7) | 12/57 (21) | 79/173 (46) | <.001 |
| MYC | 36/53 (68) | 37/215 (17) | 13/52 (25) | 85/165 (52) | <.001 |
| BCL2 | 53/66 (80) | 145/255 (57) | 52/64 (81) | 178/201 (89) | <.001 |
| BCL6 | 54/60 (90) | 216/235 (92) | 45/58 (78) | 108/165 (65) | <.001 |
| CD10 | 62/65 (95) | 161/243 (66) | 13/59 (22) | 9/179 (5) | <.001 |
| MUM1 | 2/56 (4) | 44/224 (20) | 28/53 (53) | 134/163 (82) | <.001 |
| GCB by Hans | 64/65 (99) | 207/232 (89) | 29/54 (54) | 23/159 (14) | <.001 |
| Characteristic of R-CHOP-treated patients . | DZsigpos . | GCB . | UNC . | ABC . | P . |
|---|---|---|---|---|---|
| Total (n) | 70 | 279 | 67 | 213 | |
| Age, median (range, y) | 64 (35-88) | 67 (22-92) | 64 (20-92) | 67 (31-93) | .18∗ |
| Age > 60 y (n, %) | 45 (64) | 181 (65) | 40 (60) | 143 (67) | .74 |
| Female sex (n, %) | 26 (37) | 122 (44) | 24 (36) | 102 (48) | .23 |
| Stage (n, %) | .01 | ||||
| I/II | 30 (43) | 151 (55) | 30 (46) | 85 (42) | |
| III/IV | 39 (57) | 120 (44) | 35 (54) | 119 (58) | |
| NA | 1 | 8 | 2 | 9 | |
| PS (n, %) | .29 | ||||
| 0-1 | 45 (65) | 190 (72) | 40 (65) | 128 (65) | |
| 2-4 | 24 (35) | 73 (28) | 22 (35) | 69 (35) | |
| NA | 1 | 16 | 5 | 16 | |
| LDH (n, %) | <.01 | ||||
| Normal | 24 (37) | 144 (61) | 37 (61) | 94 (53) | |
| Elevated | 40 (63) | 93 (39) | 24 (39) | 83 (47) | |
| NA | 6 | 42 | 6 | 36 | |
| Extranodal sites (n, %) | .47 | ||||
| 0-1 | 53 (77) | 221 (82) | 55 (85) | 158 (77) | |
| ≥ 2 | 16 (23) | 50 (18) | 10 (15) | 46 (23) | |
| NA | 1 | 12 | 2 | 9 | |
| B symptoms (n, %) | .34 | ||||
| No | 46 (67) | 197 (74) | 43 (66) | 136 (67) | |
| Yes | 23 (33) | 71 (26) | 22 (34) | 68 (33) | |
| NA | 1 | 7 | 2 | 9 | |
| Bulky disease ≥10 cm (n, %) | .34 | ||||
| No | 43 (67) | 186 (71) | 50 (79) | 146 (75) | |
| Yes | 21 (33) | 76 (29) | 13 (21) | 48 (24) | |
| NA | 6 | 17 | 4 | 19 | |
| IPI risk group (n, %) | .09 | ||||
| Low (0-1) | 20 (30) | 105 (44) | 22 (37) | 60 (33) | |
| Low-intermediate (2) | 16 (24) | 64 (27) | 16 (27) | 38 (21) | |
| High-intermediate (3) | 17 (26) | 42 (18) | 12 (20) | 42 (23) | |
| High (4, 5) | 13 (20) | 29 (12) | 10 (17) | 40 (22) | |
| Not calculable | 4 | 39 | 7 | 33 | |
| FISH-DH status, (n, %) | <.001 | ||||
| HGBCL-DH-BCL2 | 25 (38) | 9 (3) | 1 (2) | 1 (0.5) | |
| HGBCL-DH-BCL6 | 3 (5) | 1 (1) | 0 (0) | 4 (2) | |
| Atypical-DH† | 13 (20) | 57 (22) | 13 (20) | 42 (20) | |
| Non-DH | 25 (38) | 193 (74) | 51(78) | 159 (77) | |
| NA | 4 | 19 | 2 | 7 | |
| FISH total R, (n total/n data available, %)‡ | |||||
| Total MYC-R | 38/67 (57) | 15/262 (6) | 4/66 (6) | 16/207 (8) | <.001 |
| Total BCL2-R | 44/62 (71) | 81/221 (37) | 15/58 (26) | 3/178 (2) | <.001 |
| Total BCL6-R | 11/61 (18) | 43/213 (20) | 27/53 (51) | 55/167 (33) | <.001 |
| mRNA by digital GEP, mean log count (SD)‡ | |||||
| MYC | 11.9 (1.4) | 10.7 (0.81) | 10.8 (0.8) | 11.5 (0.9) | < .0001∗ |
| BCL2 | 12.4 (1.9) | 11.2 (1.6) | 11.7 (1.3) | 12.0 (1.3) | < .0001∗ |
| IHC, (n total/n data available, %)‡ | |||||
| MYC/BCL2 DPE | 32/58 (55) | 16/238 (7) | 12/57 (21) | 79/173 (46) | <.001 |
| MYC | 36/53 (68) | 37/215 (17) | 13/52 (25) | 85/165 (52) | <.001 |
| BCL2 | 53/66 (80) | 145/255 (57) | 52/64 (81) | 178/201 (89) | <.001 |
| BCL6 | 54/60 (90) | 216/235 (92) | 45/58 (78) | 108/165 (65) | <.001 |
| CD10 | 62/65 (95) | 161/243 (66) | 13/59 (22) | 9/179 (5) | <.001 |
| MUM1 | 2/56 (4) | 44/224 (20) | 28/53 (53) | 134/163 (82) | <.001 |
| GCB by Hans | 64/65 (99) | 207/232 (89) | 29/54 (54) | 23/159 (14) | <.001 |
P values derived from Fisher exact test unless otherwise stated. P < .05 have been highlighted in bold.
DH, double-hit; DPE, dual protein expressor; DZsigpos, DZsig positive; GCB, germinal B-cell-like; IPI, international prognostic index; LDH, lactate dehydrogenase; mRNA, messenger RNA; MYC-R, MYC-rearrangement; NA, not available; PS, performance status; R, rearrangement; SD, standard deviation; UNC, unclassified COO.
Multifactor analysis of variance.
Atypical-DH lymphoma as defined in supplemental Figure 11A.
Each row represents a unique variable compared separately.
Baseline characteristics and treatment outcomes among the 4 molecular subgroups were further assessed in all patients, irrespective of treatment. DZsigpos was associated with a higher proportion of patients with stage III/IV, elevated lactate dehydrogenase, and high-intermediate/high IPI scores (supplemental Table 6) as well as inferior FFP and OS relative to GCB-DLBCL (supplemental Figure 6C-D). Taken together, DZsigpos identifies a poor-prognosis DLBCL subgroup following R-CHOP, whereas excellent outcomes were observed with GCB-DLBCL lacking DZsig expression.
Real-world outcomes of FISH-defined subgroups
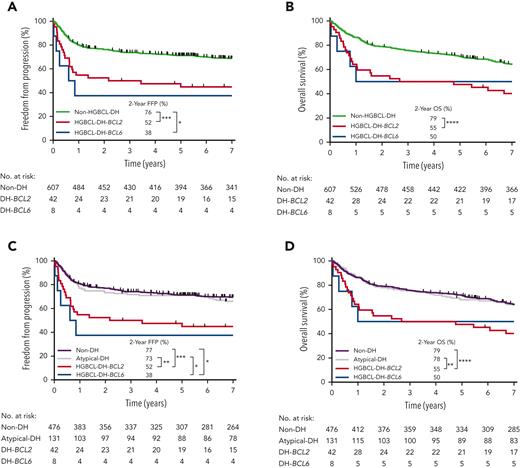
Given that all evaluable biopsies underwent unbiased FISH testing, we next assessed the clinical outcomes of FISH-defined subgroups. In total, 849 patients had biopsies evaluable for HGBCL-DH, 53 (6%) and 15 (2%) of which were HGBCL-DH-BCL2 and HGBCL-DH-BCL6, respectively. Of 657 patients treated with R-CHOP, 42 (6%) were HGBCL-DH-BCL2, 8 (1.2%) were HGBCL-DH-BCL6, and 607 (92%) lacked concurrent MYC and BCL2 and/or BCL6 rearrangement (non-HGBCL-DH) (Table 1). HGBCL-DH-BCL2 was associated with significantly inferior 2-year FFP (52%; P < .001) and OS (55%; P < .0001) compared with non-HGBCL-DH (Figure 3A-B). Analysis according to clinical stage showed that in advanced-stage disease, HGBCL-DH-BCL2 was associated with significantly inferior outcomes compared with non-HGBCL-DH. In limited-stage disease, HGBCL-DH-BCL2 was associated with a significantly inferior OS compared with non-HGBCL-DH but not FFP (supplemental Figure 9). Outcome estimates of HGBCL-DH-BCL6 and HGBCL-DH-BCL2 stratified by the MYC-rearrangement partner gene are limited by small patient numbers, the latter shown in supplemental Figure 10.
Clinical outcomes in patients treated with R-CHOP according to SVs in MYC, BCL2 and/or BCL6 detected by FISH. (A-B) FFP and OS according to FISH-defined double-hit lymphoma subgroups: HGBCL-DH-BCL2 and HGBCL-DH-BCL6, with the remaining majority of patients defined as non-HGBCL-DH. (C-D) FFP and OS after separating patients with atypical-DH lymphoma as defined in supplemental Figure 11A from the non-HGBCL-DH group into a distinct subgroup with the remaining majority of patients defined as non-double-hit (non-DH). ∗∗∗∗P < .0001, ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.
Clinical outcomes in patients treated with R-CHOP according to SVs in MYC, BCL2 and/or BCL6 detected by FISH. (A-B) FFP and OS according to FISH-defined double-hit lymphoma subgroups: HGBCL-DH-BCL2 and HGBCL-DH-BCL6, with the remaining majority of patients defined as non-HGBCL-DH. (C-D) FFP and OS after separating patients with atypical-DH lymphoma as defined in supplemental Figure 11A from the non-HGBCL-DH group into a distinct subgroup with the remaining majority of patients defined as non-double-hit (non-DH). ∗∗∗∗P < .0001, ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.
To assess the clinical significance of atypical-DH, patients with atypical-DH tumors (as defined in supplemental Figure 11A) were separated from the non-HGBCL-DH group into a distinct subgroup, with the remaining tumors defined as non–double-hit (non-DH). Among all R-CHOP–treated tumors with evaluable FISH data, 131 (20%) were atypical-DH and 476 (72%) were non-DH. Patients with atypical-DH tumors had outcomes equivalent to the non-DH group (Figure 3C-D) and further stratifying atypical-DH based on the presence of MYC/BCL2 or MYC/BCL6 SVs showed no difference in outcomes between these 2 groups (supplemental Figure 11B-C). A similar outcome association for each FISH-defined subgroup was observed when including all patients, irrespective of treatment (supplemental Figure 12).
Clinical and molecular characteristics of DZsigpos-DLBCL
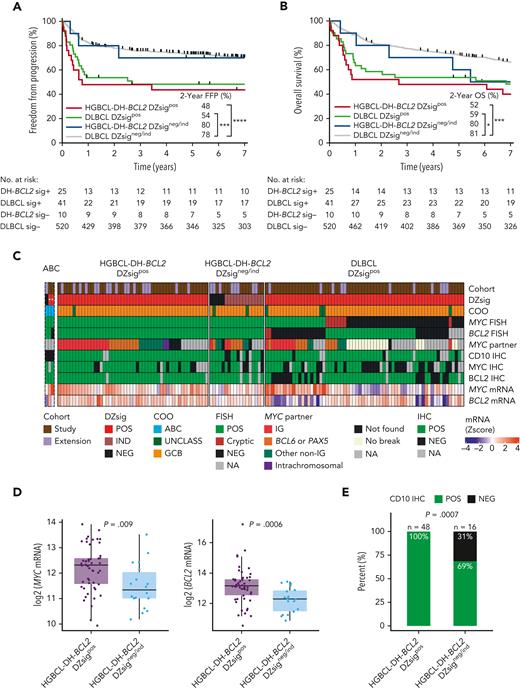
We next examined the clinical outcomes and molecular characteristics of DZsigpos tumors in relation to HGBCL-DH-BCL2. As shown in Figure 1A, 33 (38%) DZsigpos tumors were HGBCL-DH-BCL2 (HGBCL-DH-BCL2 DZsigpos) as analyzed by FISH and 55 (62%) lacked detectable MYC and/or BCL2 rearrangements (DLBCL DZsigpos). Notably, 10 of 44 (23%) HGBCL-DH-BCL2 tumors with available GEP data did not express DZsig (HGBCL-DH-BCL2 DZsigneg/ind). A FISH-cryptic rearrangement that would otherwise render a HGBCL-DH-BCL2 diagnosis was detected in 5 of 39 (13%) DLBCL DZsigpos with available sequencing data (supplemental Figure 13A). These tumors remained in their original FISH-defined categories for this analysis. Comparison of the baseline clinical characteristics between patients with HGBCL-DH-BCL2 DZsigpos and patients with DLBCL DZsigpos treated with R-CHOP revealed no significant differences (supplemental Table 7). Patients in both groups had similar clinical outcomes with overlapping FFP (P = .46) and OS curves (P = .36). In contrast, patients with HGBCL-DH-BCL2 DZsigneg/ind had favorable outcomes comparable to DLBCL DZsigneg/ind (Figure 4A-B). A similar relationship was found when all patients with available GEP and FISH results were analyzed, irrespective of treatment regimen (supplemental Figure 14).
The clinical and molecular characteristics of DZsig positive (DZsigpos) tumors in relation to HGBCL-DH-BCL2. (A-B) FFP and OS of patients treated with R-CHOP harboring DZsigpos tumors in the presence or absence of a FISH-based diagnosis of HGBCL-DH-BCL2 (HGBCL-DH-BCL2 DZsigpos vs DLBCL DZsigpos respectively), including the small subset of HGBCL-DH-BCL2 tumors that lack DZsig expression (HGBCL-DH-BCL2 DZsigneg/ind). DLBCL DZsigneg/ind are the remaining majority of patients with DLBCL, NOS. Patients harboring tumors with evaluable FISH and GEP data were included. Patients with HGBCL with MYC and BCL6 rearrangements (n = 8) were included in the DLBCL groups. Two patients with ABC tumors that are DZsigpos were excluded from the analysis. Log-rank test was used to compare survival curves. ∗∗∗∗P < .0001, ∗∗∗P < .001, ∗P < .05. (C) Molecular characterization of HGBCL-DH-BCL2-DZsigpos, DLBCL DZsigpos, and the HGBCL-DH-BCL2 DZsigneg/ind in a cohort combining the study population and extension cohort (1112 evaluable biopsies). Tumors were profiled by: (1) Digital GEP by the DLBCL90 assay for COO, DZsig, and relative MYC and BCL2 mRNA expression (z scores) displayed as a heat map in the bottom 2 rows. (2) FISH for MYC and BCL2 rearrangements. (3) Targeted capture or whole genome sequencing to identify MYC rearrangement partner in MYC rearranged tumors. (4) IHC for MYC, BCL2 and the GC marker CD10. “ABC” is a separate track of rare ABC tumors (n = 3) that were DZsigpos (marked with ∗, as not applicable in ABC) and/or HGBCL-DH-BCL2, which were excluded from subsequent analysis. (D) Box plots showing relative mRNA expression of MYC (left) and BCL2 (right) in HGBCL-DH-BCL2-DZsigpos tumors vs HGBCL-DH-BCL2 DZsigneg/ind shown as z scores from log2 normalized counts with means compared by t test. (E) Proportion of tumors positive for CD10 by IHC in HGBCL-DH-BCL2-DZsigpos vs HGBCL-DH-BCL2 DZsigneg/ind compared by Fisher exact test. IG, immunoglobulin; ind, indeterminate DZsig; mRNA, messenger RNA; UNCLASS, unclassified COO.
The clinical and molecular characteristics of DZsig positive (DZsigpos) tumors in relation to HGBCL-DH-BCL2. (A-B) FFP and OS of patients treated with R-CHOP harboring DZsigpos tumors in the presence or absence of a FISH-based diagnosis of HGBCL-DH-BCL2 (HGBCL-DH-BCL2 DZsigpos vs DLBCL DZsigpos respectively), including the small subset of HGBCL-DH-BCL2 tumors that lack DZsig expression (HGBCL-DH-BCL2 DZsigneg/ind). DLBCL DZsigneg/ind are the remaining majority of patients with DLBCL, NOS. Patients harboring tumors with evaluable FISH and GEP data were included. Patients with HGBCL with MYC and BCL6 rearrangements (n = 8) were included in the DLBCL groups. Two patients with ABC tumors that are DZsigpos were excluded from the analysis. Log-rank test was used to compare survival curves. ∗∗∗∗P < .0001, ∗∗∗P < .001, ∗P < .05. (C) Molecular characterization of HGBCL-DH-BCL2-DZsigpos, DLBCL DZsigpos, and the HGBCL-DH-BCL2 DZsigneg/ind in a cohort combining the study population and extension cohort (1112 evaluable biopsies). Tumors were profiled by: (1) Digital GEP by the DLBCL90 assay for COO, DZsig, and relative MYC and BCL2 mRNA expression (z scores) displayed as a heat map in the bottom 2 rows. (2) FISH for MYC and BCL2 rearrangements. (3) Targeted capture or whole genome sequencing to identify MYC rearrangement partner in MYC rearranged tumors. (4) IHC for MYC, BCL2 and the GC marker CD10. “ABC” is a separate track of rare ABC tumors (n = 3) that were DZsigpos (marked with ∗, as not applicable in ABC) and/or HGBCL-DH-BCL2, which were excluded from subsequent analysis. (D) Box plots showing relative mRNA expression of MYC (left) and BCL2 (right) in HGBCL-DH-BCL2-DZsigpos tumors vs HGBCL-DH-BCL2 DZsigneg/ind shown as z scores from log2 normalized counts with means compared by t test. (E) Proportion of tumors positive for CD10 by IHC in HGBCL-DH-BCL2-DZsigpos vs HGBCL-DH-BCL2 DZsigneg/ind compared by Fisher exact test. IG, immunoglobulin; ind, indeterminate DZsig; mRNA, messenger RNA; UNCLASS, unclassified COO.
Additional characterization of the 10 HGBCL-DH-BCL2 DZsigneg/ind tumors suggested different molecular features compared with HGBCL-DH-BCL2 DZsigpos (supplemental Figure 13). Given the rarity of HGBCL-DH-BCL2 DZsigneg/ind tumors, we extended the study cohort by including 308 additional tumors with evaluable FISH and DLBCL90 data from 2 additional DLBCL cohorts with unbiased FISH testing.6,25 This yielded 9 additional HGBCL-DH-BCL2 tumors that lacked DZsig expression, one of which was ABC by COO classification and therefore was excluded from the HGBCL-DH-BCL2 DZsigneg/ind subgroup (Figure 4C). Compared with HGBCL-DH-BCL2 DZsigpos, HGBCL-DH-BCL2 DZsigneg/ind tumors expressed lower levels of MYC (P = .005) and BCL2 (P = .003) messenger RNA and were more frequently CD10 negative by IHC (P = .0007) (Figure 4D-E).
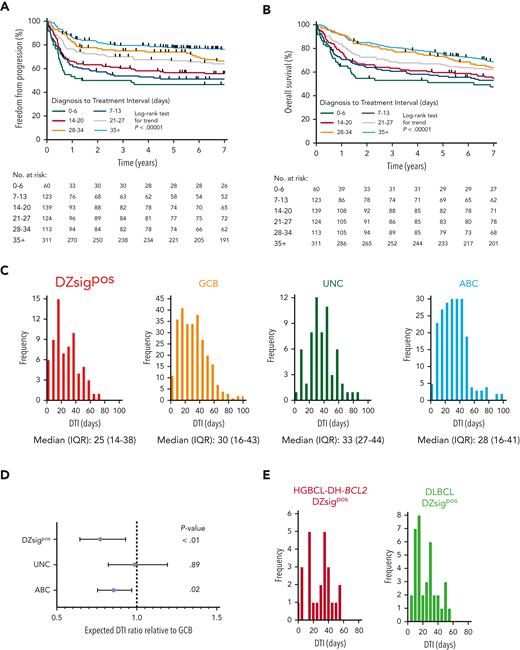
Poor-prognosis molecular subgroups are associated with shorter DTI
We confirmed the relationship between clinical outcomes and DTI in our study population using the same weekly intervals as defined by Maurer et al.28 DTI was strongly associated with FFP and OS in patients treated with R-CHOP (P < .00001, Figure 5A-B). Using patients with available GEP data (supplemental Figure 3), we analyzed the relationship between GEP–defined molecular subgroups in DLBCL and DTI. Analogous to the difference in clinical outcomes, patients with missing GEP data had significantly shorter DTI than patients with available GEP data (supplemental Figure 15). Frequency distributions of DTI for each molecular subgroup were generated (Figure 5C) and subgroups were compared using negative binomial regression. Relative to GCB-DLBCL, DZsigpos and ABC-DLBCL had significantly shorter DTI, with expected DTI ratios (95% CI) of 0.79 (0.65-0.96, P < .01) for DZsigpos and 0.85 (0.75-0.97, P = .02) for ABC (Figure 5D). There was no significant difference in DTI distribution between HGBCL-DH-BCL2 DZsigpos and DLBCL DZsigpos (Figure 5E).
Poor-prognosis molecular subgroups are associated with a shorter DTI in patients with DLBCL treated with R-CHOP. (A-B) FFP and OS in the study population according to DTI intervals previously defined by Maurer et al.28 (C) DTI frequency distribution according to molecular subgroup. Six outliers with DTI > 100 days are not shown (1 DZsigpos, 4 GCB, and 1 ABC). (D) Comparison of DTI ratios of molecular subgroups by negative binomial regression using the GCB subgroup as a reference. DTI ratios and 95% CI are shown. (E) Comparison of DTI distribution in patients with DLBCL with DZsig positive (DZsigpos) tumors based on the presence (HGBCL-DH-BCL2 DZsigpos, left panel) or absence (DLBCL DZsigpos, right panel) of a FISH–based diagnosis of HGBCL-DH-BCL2. DTI ratio (95% CI) of HGBCL-DH-BCL2 DZsigpos relative to DLBCL DZsigpos was 0.94 (0.67-1.34, P = .74) by negative binomial regression. IQR, interquartile range; UNC, unclassified COO.
Poor-prognosis molecular subgroups are associated with a shorter DTI in patients with DLBCL treated with R-CHOP. (A-B) FFP and OS in the study population according to DTI intervals previously defined by Maurer et al.28 (C) DTI frequency distribution according to molecular subgroup. Six outliers with DTI > 100 days are not shown (1 DZsigpos, 4 GCB, and 1 ABC). (D) Comparison of DTI ratios of molecular subgroups by negative binomial regression using the GCB subgroup as a reference. DTI ratios and 95% CI are shown. (E) Comparison of DTI distribution in patients with DLBCL with DZsig positive (DZsigpos) tumors based on the presence (HGBCL-DH-BCL2 DZsigpos, left panel) or absence (DLBCL DZsigpos, right panel) of a FISH–based diagnosis of HGBCL-DH-BCL2. DTI ratio (95% CI) of HGBCL-DH-BCL2 DZsigpos relative to DLBCL DZsigpos was 0.94 (0.67-1.34, P = .74) by negative binomial regression. IQR, interquartile range; UNC, unclassified COO.
We next expanded the cohort to examine the impact of molecular subgroups on DTI in all patients who received any immunochemotherapy, including palliative regimens. DTI remained both strongly predictive of outcomes and shorter in DZsigpos and ABC-DLBCL relative to GCB-DLBCL with DTI ratios of 0.73 (0.62-0.87, P < .001) and 0.85 (0.75-0.95, P < .01), respectively (supplemental Figure 16). Differences among subgroups were more pronounced compared with the former analysis of patients treated with R-CHOP only, possibly owing to the inclusion of patients treated with palliative immunochemotherapy who had a shorter DTI compared with patients treated with R-CHOP (median 21 vs 27 days, supplemental Figure 17). In conclusion, patients in molecular subgroups associated with poor outcomes had significantly shorter DTI.
Discussion
We profiled evaluable biopsies from all patients with DLBCL diagnosed in BC between 2005 and 2010 to establish the real-world outcomes associated with GEP and FISH-defined molecular subgroups and characterized their salient features. Several key findings emerged. First, DZsig defines a group of GC-origin, aggressive B-cell lymphomas with a common gene expression phenotype that extends beyond DLBCL and HGBCL-DH-BCL2. Second, DZsig identifies a subgroup of DLBCL tumors with poor prognosis, two-thirds of which are negative for HGBCL-DH-BCL2 by FISH. Further, HGBCL-DH-BCL2 tumors that lacked DZsig expression were associated with favorable outcomes. Third, poor-prognosis molecular subgroups were associated with shorter DTI. Fourth, atypical-DH conferred no prognostic effect. Fifth, the distribution of molecular subgroups within DLBCL was independent of age in this population.
HGBCL-DH-BCL2 tumors were previously shown to express the molecular high-grade (MHG) signature,32 which was derived from the molecular Burkitt signature.33,34 Here, we observed universal DZsig expression in BL, indicating that a common gene expression phenotype extends beyond HGBCL-DH-BCL2 to other aggressive B-cell lymphomas of GC origin. Previously, single-cell transcriptomic profiling of GC B cells showed that DZsigpos tumors share a gene expression pattern that overlaps with DZ GC B cells.35 Similarly, HGBCL-DH and MHG-positive tumors were enriched for the expression of a DZsig identified by digital spatial profiling of reactive lymph nodes.36 Altogether, these findings suggest that both DZsig and MHG converge to identify a dysregulated DZ phenotype shared by BL, HGBCL-DH-BCL2, and a subset of GCB-DLBCL, in favor of renaming the signature formerly known as DHITsig20,21,25 to DZsig.
We confirmed that DZsigpos identifies a poor-prognosis population following R-CHOP within GCB-DLBCL, consistent with the cohort used for the original description of DZsig, which partially overlaps with the current study cohort.20 In this larger, unselected population based cohort, patients with DZsigind tumors, in whom the DLBCL90 assay could not assign DZsig status with sufficient confidence, had relatively superior outcomes compared with DZsigpos. This contrasts with the original cohort, in which the outcomes of the DZsigpos and DZsigind subgroups were similar.20 Here we restricted the DZsig subgroup to DZsigpos only to minimize the risk of misclassification and to select out patients with the worst prognosis. Although one-third of patients with DZsigpos are HGBCL-DH-BCL2 by FISH, the majority would not be identified by techniques routinely used in pathology workflows. The outcomes of patients with DZsigpos with or without HGBCL-DH-BCL2 were equivalent. Intriguingly, we found a small group of HGBCL-DH-BCL2 that are not DZsigpos, generally lacking the typical features of this disease category and with the caveat of small numbers have outcomes that are similar to DLBCL, NOS following R-CHOP. The same finding was noted in patients with HGBCL-DH-BCL2 who lacked MHG expression.32 Given that many centers use intensified chemotherapy regimens for HGBCL-DH-BCL2, these observations raise the question of whether FISH is the optimal method to select patients for treatment intensification or that selection should be based on DZsig expression. In a representative patient sample selected from 2 phase II trials of young, high-risk patients with DLBCL morphology tumors treated with dose-dense immunochemotherapy, DZsigpos was not associated with inferior outcomes.37 Although confirmatory studies are needed, this suggests that chemotherapy intensification in appropriately selected patients may overcome the negative prognostic effect conferred by DZsigpos. Alternatively, patients with DZsigpos could be considered for trials that incorporate novel therapeutic approaches in the frontline treatment of patients with high-risk, large B-cell lymphoma.38 The use of DZsig would lead to broader, but more refined, patient selection for these strategies compared with FISH.
The difference in outcomes between GCB-DLBCL and ABC-DLBCL found in this cohort is significantly smaller than seen in previous studies conducted in the rituximab era,4,6 confirming that COO alone does not account for the heterogenous outcomes following R-CHOP and that further stratification is needed. Here we showed that further refining COO by removing patients with DZsigpos from GCB-DLBCL, leaves a patient group with excellent outcomes following R-CHOP, substantially superior to ABC-DLBCL. These excellent outcomes persisted even in advanced-stage disease, suggesting that R-CHOP is likely sufficient for GCB-DLBCL.
We established the association between DTI and outcomes in this cohort and extended these findings by showing that poor-prognosis molecular subgroups have shorter DTI in patients treated with curative and palliative intent.28 The shorter DTI seen in high-risk molecular subgroups likely contributes to their underrepresentation in DLBCL clinical trials because typical delays associated with trial enrollment select against these patients. In the Alliance/CALGB 50303 trial, only 1.2% of patients with DLBCL with evaluable FISH data were HGBCL-DH,39 compared with 8% in our population. In the GOYA trial, in which a Nanostring-based assay was used for COO assignment similar to our study, the proportion of patients with ABC was 26%,5 6% lower than in our cohort. This discrepant proportion of high-risk molecular subgroups between real-world and clinical trial populations further highlights the need to adjust trial design to facilitate the inclusion of high-risk patients who require urgent treatment initiation.
The lack of prognostic significance observed with atypical-DH provides the clinical correlation to our recent comprehensive molecular analysis of atypical-DH tumors.25 These findings support the current classification systems, whereby only rearrangements are considered in the HGBCL-DH definition.15,16 In contrast to atypical-DH, our data suggest that HGBCL-DH-BCL6 is associated with poor outcomes, with the caveat of small numbers. Further understanding of the biological underpinnings of the poor prognosis associated with HGBCL-DH-BCL6 despite its biological heterogeneity40 is required. Our study confirms the negative impact of MYC:IG partner on HGBCL-DH prognosis, albeit with small numbers compared with previous, larger studies.19,41
The COO age distribution in our population does not support an enrichment for ABC in elderly patients, as described in other DLBCL cohorts.9,42 Cohort differences such as the population registry-derived, treatment-agnostic nature of our cohort may explain these discrepant observations. Similarly, there was no enrichment for DZsigpos in the elderly. These findings argue against the overrepresentation of poor-prognosis molecular subgroups as a major contributing factor to the inferior outcomes in elderly patients with DLBCL. Future studies that incorporate genetic-based subgroups12 and global variation in COO distribution7,8 are required to further understand the relationship between age and DLBCL molecular subgroups.
Our study has some limitations inherent to retrospective population analyses. First, despite concerted efforts to work with minimal diagnostic material, adequate material was unavailable in some cases. Patients excluded because of insufficient remaining tissue or only bone/BM-derived biopsies had worse outcomes and shorter DTI compared with patients with evaluable biopsies. This indicates that, somewhat analogous to bias observed in clinical trials, patients may be excluded from real-world molecular testing owing to the impact of specific clinical scenarios on the nature of tumor sampling. Such scenarios include patients in need of immediate treatment precluding sizable biopsies and specific disease presentations (eg, disease restricted to bone/BM). Second, in some survival comparisons, the small sample sizes of rare molecular subgroups limited the power to detect statistically significant differences in this predefined population. Third, DTI analysis was limited by the inability to capture the precise timing of administration of systemic glucocorticoids or palliative radiation, although neither were considered in the original description.28
In conclusion, in this large, real-world population study of DLBCL, recognition of a DZsig refined the COO classification by distinguishing patients within GCB-DLBCL with clinically relevant differences in outcomes and diagnosis-to-treatment times. These findings may potentially improve patient selection for treatment intensification and clinical trial design in DLBCL.
Acknowledgments
This study was supported by the Canadian Cancer Society Research Institute (704848 and 705288), Genome Canada (4108 and 13124), Genome British Columbia (141LYM and 271LYM), the Canadian Institutes of Health Research (GPH-129347, GPI-155873 and 300738), the Terry Fox Research Institute (1061 and 1043), and the British Columbia Cancer Foundation. W.A. is supported by the Kuwait Ministry of Health, the Leukemia and Lymphoma Society of Canada/Canadian Institutes of Health Research Clinician Scientist Fellow award and the Michael Smith Health Research, British Columbia Research Trainee award. B.C. is supported by a Canada Graduate Scholarships Doctoral program award from the Canadian Institutes of Health Research and a Doctoral Fellowship from the University of British Columbia. D.W.S. is supported by a Michael Smith Foundation for Health Research Health Professional Investigator award (18646).
Authorship
Contribution: D.W.S., W.A., and B.C. conceived and designed the study; W.A., B.C., S.B.-N., L.K.H., M.B., B.M., L.C., T.M.-T., G.W.S., P.F., J.W.C., A.L., R.D.G., and D.W.S. performed the research and analyzed data; A.J., W.A., B.C., and L.K.H. performed statistical analysis; K.J.S., D.V., A.S.G., C.L.F., and J.M.C. participated in the collection and analysis of clinical data; R.D.M., A.J.M., and C.S. collected and analyzed molecular data; W.A., B.C., and D.W.S. wrote the manuscript; and all authors edited and approved the final version of the manuscript.
Conflict-of-interest disclosure: J.W.C.: consulting and honoraria from Bayer and BeiGene. K.J.S.: honoraria from SeaGen, BMS, Kyowa, Novartis, Merck, Janssen; research funding from BMS, Roche Research; Beigene steering committee member; DSMC for Regeneron. D.V.: honoraria and advisory boards from AbbVie, Janssen, Kite/Gilead, AstraZeneca, Roche, BeiGene, Sandoz Canada, Kyowa Kirin; research funding (to the institution) from AstraZeneca and Roche. A.S.G.: honoraria from AbbVie, AstraZeneca, Janssen and Sandoz; research funding from AbbVie, AstraZeneca and Janssen. C.L.F.: honoraria from BMS/Celgene, Sanofi, Incyte, AbbVie, Amgen and Janssen; consultancy for BMS/Celgene; research funding from Janssen. L.H.S.: consulting and honoraria from AbbVie, Acerta, Amgen, Apbiologix, AstraZeneca, Celgene, Chugai, Gilead, Incyte, Janssen, Kite, Karyopharm, Lundbeck, Merck, Morphosys, Roche/Genentech, Sandoz, Seattle Genetics, Servier, Takeda, Teva, TG Therapeutics and Verastem; research funding from Roche/Genentech and Teva. C.S.: consultancy for Seattle Genetics, Curis Inc, Roche, AbbVie, Juno Therapeutics and Bayer; and research support from Epizyme, Bristol Myers Squibb and Trillium Therapeutics Inc. D.W.S.: consultancy for AbbVie, AstraZeneca, Incyte, Janssen; research funds from Janssen and Roche/Genentech; named inventor on a patent describing the use of gene expression to subtype aggressive B-cell lymphomas, one of which is licensed to NanoString Technologies. The remaining authors declare no competing financial interests.
Correspondence: David W. Scott, British Columbia Cancer Research Centre, 675 West 10th Ave, Vancouver, BC V5Z 1L3, Canada; e-mail: dscott8@bccancer.bc.ca.
References
Author notes
∗W.A. and B.C. are first authors and contributed equally to this study.
Data are available on request from the corresponding author, David W. Scott (dscott8@bccancer.bc.ca).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal