Abstract
Myeloid neoplasms and acute leukemias derive from the clonal expansion of hematopoietic cells driven by somatic gene mutations. Although assessment of morphology plays a crucial role in the diagnostic evaluation of patients with these malignancies, genomic characterization has become increasingly important for accurate diagnosis, risk assessment, and therapeutic decision making. Conventional cytogenetics, a comprehensive and unbiased method for assessing chromosomal abnormalities, has been the mainstay of genomic testing over the past several decades and remains relevant today. However, more recent advances in sequencing technology have increased our ability to detect somatic mutations through the use of targeted gene panels, whole-exome sequencing, whole-genome sequencing, and whole-transcriptome sequencing or RNA sequencing. In patients with myeloid neoplasms, whole-genome sequencing represents a potential replacement for both conventional cytogenetic and sequencing approaches, providing rapid and accurate comprehensive genomic profiling. DNA sequencing methods are used not only for detecting somatically acquired gene mutations but also for identifying germline gene mutations associated with inherited predisposition to hematologic neoplasms. The 2022 International Consensus Classification of myeloid neoplasms and acute leukemias makes extensive use of genomic data. The aim of this report is to help physicians and laboratorians implement genomic testing for diagnosis, risk stratification, and clinical decision making and illustrates the potential of genomic profiling for enabling personalized medicine in patients with hematologic neoplasms.
Introduction
Genomic characterization is essential for the management of myeloid neoplasms and acute leukemia, providing critical information for diagnosis, risk assessment, therapeutic decisions, residual disease monitoring, progression, and treatment resistance (Figure 1). Chromosome banding analysis complemented by a variety of molecular studies are a central facet of evaluation, with new genomic techniques increasingly being used to improve characterization as described herein and in Table 1. This article is meant to be a practical guide for the application of genomic methods in the clinical evaluation of myeloid neoplasms and acute leukemia.
How molecular profiling can inform clinical decision making in MDS. IPSS-M, Molecular International Prognostic Scoring System; MDS, myelodysplastic syndrome; MRD, minimal/measurable residual disease. Professional illustration by Patrick Lane, ScEYEnce Studios.
How molecular profiling can inform clinical decision making in MDS. IPSS-M, Molecular International Prognostic Scoring System; MDS, myelodysplastic syndrome; MRD, minimal/measurable residual disease. Professional illustration by Patrick Lane, ScEYEnce Studios.
General advantages, limitations, and clinical applications of comprehensive genomic methods
| Technique . | CG . | FISH . | CMA . | OGM . | Targeted . | Exome . | WGS . | RNA-seq . |
|---|---|---|---|---|---|---|---|---|
| Viable cells | Yes | No | No | No | No | No | No | No |
| Resolution | ∼5 Mb | 100-200 kb | 20-100 kb | 5-50 kb | 1 bp | 1 bp | 1 bp | 1 bp |
| Coverage | Genome | Targeted | Genome | Genome | Targeted | Exome | Genome | Genome, Targeted |
| Alterations | CNV, SV | CNV, SV | CNV, LOH | CNV, SV | ← SNV, Indel, CNV, SV, LOH → | Gene expression, SV | ||
| Sensitivity (VAF) | 5%-10% | 1%-5% | 30% | 5% | 2% | 5%-10% | 10% | 5% |
| TAT (days)∗ | 2-21 | 1-3 | 3-14 | 4-7 | 5-14 | 5-14 | 3-14 | 5-14 |
| Cost∗ | $ | $ | $$ | $$$ | $$-$$$ | $$$ | $$$$ | $$-$$$ |
| Worldwide use∗ | High | High | Low | Low | Medium | Low | Low | Low |
| Used in | ||||||||
| MDS and MDS/MPN | D, FU | D, FU | D, R | D, R | D, MRD† | D | D, ND | |
| MPN | D | D | D | D | D, MRD† | D | D | ND |
| AML | D, R | D | D | D, R | D, MRD† | D | D | D |
| ALL | D, R | D | D, R | D, R | D | D | D | D |
| Technique . | CG . | FISH . | CMA . | OGM . | Targeted . | Exome . | WGS . | RNA-seq . |
|---|---|---|---|---|---|---|---|---|
| Viable cells | Yes | No | No | No | No | No | No | No |
| Resolution | ∼5 Mb | 100-200 kb | 20-100 kb | 5-50 kb | 1 bp | 1 bp | 1 bp | 1 bp |
| Coverage | Genome | Targeted | Genome | Genome | Targeted | Exome | Genome | Genome, Targeted |
| Alterations | CNV, SV | CNV, SV | CNV, LOH | CNV, SV | ← SNV, Indel, CNV, SV, LOH → | Gene expression, SV | ||
| Sensitivity (VAF) | 5%-10% | 1%-5% | 30% | 5% | 2% | 5%-10% | 10% | 5% |
| TAT (days)∗ | 2-21 | 1-3 | 3-14 | 4-7 | 5-14 | 5-14 | 3-14 | 5-14 |
| Cost∗ | $ | $ | $$ | $$$ | $$-$$$ | $$$ | $$$$ | $$-$$$ |
| Worldwide use∗ | High | High | Low | Low | Medium | Low | Low | Low |
| Used in | ||||||||
| MDS and MDS/MPN | D, FU | D, FU | D, R | D, R | D, MRD† | D | D, ND | |
| MPN | D | D | D | D | D, MRD† | D | D | ND |
| AML | D, R | D | D | D, R | D, MRD† | D | D | D |
| ALL | D, R | D | D, R | D, R | D | D | D | D |
ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; CG, cytogenetics; CMA, chromosomal microarray; CNV, copy number variations; D, diagnosis; ES, exome sequencing; FISH, fluorescence in situ hybridization; FU, follow-up; Indel, small insertion/deletions; LOH, loss-of-heterozygosity; MPN, myeloproliferative neoplasm; ND, not done; OGM, optical genome mapping; R, relapse; SNV, single nucleotide variant; SV, structural variant; TAT, turnaround time.
TATs, cost, and use approximated. Actual TATs, cost, and use vary significantly by region and laboratory.
When used in conjunction with high coverage sequencing and error correction methods for increased sensitivity/specificity for low-abundance mutations.
Conventional methods
Chromosome banding analysis
Karyotyping remains the most widely used and unbiased method for assessing chromosomal abnormalities including numerical (amplifications and losses) and structural (translocations, deletions, and inversions) abnormalities.1-3 The main limitations are the requirement for live culturable cells, low resolution (5-10 Mb), and low sensitivity (abnormalities present in 5%-10% of cells or an analytical sensitivity of ∼10−1). Turnaround times are generally between 2 and 21 days and may vary considerably between laboratories.
FISH
Fluorescence in situ hybridization (FISH) is often used to complement chromosome banding analysis and can be performed on both cultured dividing cells (metaphase) and fixed or nondividing cells (interphase). FISH probes can only identify genomic events at specific targeted regions but is more sensitive than cytogenetics (abnormalities in 1%-5% of cells or an analytical sensitivity of ∼10−2) and can detect cytogenetically cryptic abnormalities.4,5 Turnaround times are generally 1 to 3 days.
CMAs
Chromosomal microarrays (CMAs) are typically used to identify small, unbalanced abnormalities or cryptic copy number alterations (CNAs) but do not detect balanced rearrangements and, unlike karyotype, cannot distinguish changes occurring in separate clones. In addition, CMAs including single nucleotide polymorphism (SNP) probes (SNP arrays) can detect LOH and facilitate the determination of chromosomal ploidy.6 Unlike FISH, CMAs are unbiased and can detect abnormalities genome wide. CMAs are run from tumor DNA without requiring live cells and can detect small abnormalities (20-100 kb) present in 20% to 30% of tumor cells (or an analytical sensitivity of >10−1). Turnaround times are generally between 3 and 14 days. Although not array-based, multiplex ligation-dependent probe amplification (MLPA) can also be used to detect specific CNAs (including single exon events) through the use of multiple sequence-specific probes spanning a specific region, which are then amplified to determine DNA copy state.7
OGM
Optical genome mapping (OGM) methods are an unbiased approach that use genome-wide high-resolution enzymatic restriction digests of high-molecular-weight genomic DNA to identify structural variants such as translocations, inversions, and CNAs.8 Although not widely used in the clinical laboratory today, turnaround times are typically between 4 and 7 days with maximum sensitivity of ∼5%.9
PCR
Polymerase chain reaction (PCR) is a technique based on the enzymatic replication of DNA (or complementary DNA [cDNA] from reverse-transcribed RNA) and can generate tens of billions of copies of a particular small DNA or cDNA fragment (the sequence of interest), allowing gene mutations to be detected by various methods. Most clinical PCR applications use allele-specific PCR with primers for a specific mutation that only produce a PCR product when the mutation is present. Real-time, quantitative PCR (qPCR) can rapidly quantify specific fragments containing a sequence alteration and is used to detect specific single gene mutations or gene rearrangements. Digital PCR (dPCR) technologies enable absolute quantification through partitioning the reaction into thousands of independent PCRs to achieve high levels of sensitivity (1 mutation in 10 000 normal cells or a sensitivity of 10−4). qPCR and dPCR methods are generally suited to detect specific recurrent genetic alterations such as single gene mutations (eg, JAK2 p.V617F, KIT p.D816V) and distinct fusions (eg, BCR::ABL1) for diagnosis and disease monitoring. Turnaround times are generally 2 to 5 days.
Sanger sequencing
Sanger sequencing detects small gene-level DNA variation from PCR-amplified DNA fragments (<1 kb). Individual DNA bases are detected by electrophoresis due to the random incorporation of fluorescently labeled chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication. Sanger sequencing is generally used to detect gene mutations confined to single exons (eg, CEBPA, CALR) and has a relatively low analytic sensitivity of ∼20% (>10−1) but can be turned around more rapidly than next-generation sequencing (NGS)–based methods.
NGS-based methods
General concepts
Building upon the previously described conventional genomic methods, NGS, massive parallel sequencing, or high throughput sequencing uses millions or billions of parallel sequencing reactions to identify genomic abnormalities. NGS is highly scalable and can be coupled with enrichment technologies to interrogate a small subset of key genes (targeted gene panels), up to thousands of genes or genomic regions (whole-exome sequencing [WES]), or can be used without enrichment to detect full genome (whole genome sequencing [WGS]) or transcriptome-wide (whole-transcriptome sequencing [WTS] or RNA sequencing [RNA-seq]) genomic abnormalities. Depending on the design of the assay, NGS can be used to study the full range of genomic variation, including SNVs, small indels, structural changes (ie, CNAs), gene fusions or chromosomal translocations, gene expression, and DNA methylation, and may be used for initial diagnosis as well as monitoring.
The abundance of a detected variant is generally represented as a variant allele frequency (VAF, Figure 2A), which represents the ratio of sequencing reads that contain a variant at a given position in the genome divided by the total number of reads at that position. VAFs are considered a semiquantitative measure because the exact methods used to calculate VAFs, VAF precision, and VAF accuracy will differ slightly between laboratories. In addition, chromosomal aneuploidy, LOH, or gene amplification/deletion can skew both inherited and somatic variant VAFs either higher or lower. The methods used to separate somatic variants (those present in cancer cells) from germline variants (polymorphisms or pathogenic alterations in germline DNA) may also differ between laboratories. Although the gold standard for establishing somatic status of a variant involves sequencing both tumor and paired non-neoplastic tissue DNA, this approach is expensive and impractical for most clinical testing. Instead, many laboratories often infer variants with VAFs near 50% (in regions without CNAs) as germline; the use of population databases (1000 Genomes,10 gnomAD,11 dbSNP,12 etc) to filter out known polymorphisms is highly suggested to separate germline variants from somatic mutations. The exact methods used to filter polymorphisms varies widely between laboratories13 and may give rise to both false-positive somatic calls, especially in genes with low probability of loss intolerance (pLI)14 scores such as TET2,15 and false-negative calls, often resulting from the presence of clonal hematopoiesis of indeterminate potential (CHIP) variants (ie, DNMT3A p.R882) coded as polymorphisms in "normal" population databases.16,17 The limit of detection for standard NGS assays is generally determined by the sequencing coverage depth and typically ranges from 2% to 5% VAF (Figure 2B); reliable detection of variants below this level typically requires error-correction methods (described in further sections).
Key concepts in sequencing-based diagnostics. (A) VAF represents the ratio of sequencing reads that contain a variant divided by the total number of reads at that position. Because most somatic mutations are heterozygous, doubling the VAF generally indicates the fraction of cells with the mutation (except when the mutation occurs in a region of copy number alteration). (B) Coverage represents the number of sequencing reads (red and blue indicating forward and reverse reads, respectively) that span a particular region. Approximate coverage levels for different sequencing approaches are compared. Higher coverage (or more independent observations) generally yields more sensitive sequencing. Shown on the right is the coverage depth required to detect mutations at various VAFs. Binomial sampling probability for detection of variants with VAFs of 50% (typical inherited variants; black), 2% (general sensitivity for targeted panels; red), and 0.1% (MRD assays; blue) assuming each variant must be seen at least twice. (C) DNA-sequencing methods. In WGS, libraries are created by ligating sequencing adapters (gray and orange) to the 3′ and 5′ ends of short genomic DNA fragments called “inserts.” Gene panels or exome sequencing enriches DNA of interest form a library using antisense capture probes (green) labeled with biotin, which are then hybridized to DNA inserts from the sequencing and then physically enriched using streptavidin-coated magnetic beads. (D) High-sensitivity sequencing for MRD detection requires error correction to reliably identify mutations below the intrinsic error rate of the sequencer and to account for PCR errors. Error-corrected deep sequencing reduces false-positive calls for low VAF variants by tagging individual DNA molecules with unique molecular identifiers (UMIs). In this example a “true” mutation “T” is present in a single DNA molecule that labeled with a UMI (green). Library amplification and sequencing will result in duplicate DNA molecules each labeled with the same UMI. Randomly accumulated sequencing and PCR errors (orange) will be present in only a subset of reads with a particular UMI (green, purple, red). During sequencing analysis, variants present on only a subset of reads from a particular “read family” with the same UMI will be discarded as errors; true mutations present in the original DNA molecule will be detected in all reads within a read family with the same UMI. UMI methods can be further improved by tracking both DNA strands using “duplex sequencing,” which can yield sensitivities of 10−6.31 Professional illustration by Patrick Lane, ScEYEnce Studios.
Key concepts in sequencing-based diagnostics. (A) VAF represents the ratio of sequencing reads that contain a variant divided by the total number of reads at that position. Because most somatic mutations are heterozygous, doubling the VAF generally indicates the fraction of cells with the mutation (except when the mutation occurs in a region of copy number alteration). (B) Coverage represents the number of sequencing reads (red and blue indicating forward and reverse reads, respectively) that span a particular region. Approximate coverage levels for different sequencing approaches are compared. Higher coverage (or more independent observations) generally yields more sensitive sequencing. Shown on the right is the coverage depth required to detect mutations at various VAFs. Binomial sampling probability for detection of variants with VAFs of 50% (typical inherited variants; black), 2% (general sensitivity for targeted panels; red), and 0.1% (MRD assays; blue) assuming each variant must be seen at least twice. (C) DNA-sequencing methods. In WGS, libraries are created by ligating sequencing adapters (gray and orange) to the 3′ and 5′ ends of short genomic DNA fragments called “inserts.” Gene panels or exome sequencing enriches DNA of interest form a library using antisense capture probes (green) labeled with biotin, which are then hybridized to DNA inserts from the sequencing and then physically enriched using streptavidin-coated magnetic beads. (D) High-sensitivity sequencing for MRD detection requires error correction to reliably identify mutations below the intrinsic error rate of the sequencer and to account for PCR errors. Error-corrected deep sequencing reduces false-positive calls for low VAF variants by tagging individual DNA molecules with unique molecular identifiers (UMIs). In this example a “true” mutation “T” is present in a single DNA molecule that labeled with a UMI (green). Library amplification and sequencing will result in duplicate DNA molecules each labeled with the same UMI. Randomly accumulated sequencing and PCR errors (orange) will be present in only a subset of reads with a particular UMI (green, purple, red). During sequencing analysis, variants present on only a subset of reads from a particular “read family” with the same UMI will be discarded as errors; true mutations present in the original DNA molecule will be detected in all reads within a read family with the same UMI. UMI methods can be further improved by tracking both DNA strands using “duplex sequencing,” which can yield sensitivities of 10−6.31 Professional illustration by Patrick Lane, ScEYEnce Studios.
In addition to interlaboratory differences in VAFs and assignment of somatic status to variants, variant classification and annotation, including assigning variants to different "tiers" based on pathogenicity or clinical significance, may differ between laboratories. In general, it is advised that interpretations follow professional guidelines such as the Association for Molecular Pathology, the American Society for Clinical Oncology, and the College of American Pathologists guidelines,13 or disease-specific National Comprehensive Cancer Network (NCCN) guidelines, or World Health Organization guidelines, where possible. Variant annotations should also be considered in the context of the patient’s disease, and genomic data should be interpreted in conjunction with blood and bone marrow (BM) morphology, flow cytometry, and relevant clinical data.
Targeted gene panels
Driver mutations (recurrent somatic mutations known to be involved in disease pathogenesis) in specific myeloid neoplasms and acute leukemias tend to occur in a core group of 20 to 50 genes and are ideally suited to detection by small gene panels.18,19 Targeted panels, used most frequently in clinical laboratories, direct sequencing to specific genes or genetic regions that have defined clinical relevance and dictate clinical management.20,21 Key genes that should be included in sequencing panels for different diagnostic entities are summarized in Table 2. Enrichment strategies (Figure 2C) include the capture of genetic regions of interest though hybridization (hybrid capture using DNA or RNA probes) or PCR amplification (amplicon enrichment). This provides critical benefits over broad exome (WES) and genome (WGS) sequencing by increasing sensitivity in clinically relevant regions and by decreasing sequencing cost. Depending on the design, targeted panels may be able to detect CNAs and chromosomal translocations in addition to SNVs and indels. It should be noted that the detection of larger insertions including FLT3 internal tandem duplications (ITDs) and KMT2A (MLL) partial tandem duplications (PTDs) generally require specialized informatics approaches and may not be detected by all panels.22,23 Targeted sequencing methods may also be applied to RNA via capture or PCR amplification of cDNA, techniques used primarily to detect recurrent chromosomal translocations in hematologic malignancies. Turnaround times for targeted gene panels are generally 5 to 14 days.
Gene mutations in myeloid neoplasms and leukemia indicated for clinical testing
| Indication . | Single gene mutations . | Structural variants∗ . |
|---|---|---|
| MDS, MDS/MPN, cytopenia | ASXL1, BCOR, BCORL1, CBL, CEBPA, CSF3R, DDX41, DMNT3A, ETV6, ETNK1, EZH2, FLT3-ITD, FLT3-TKD, GATA2, GNB1, IDH1, IDH2, JAK2, KIT, KRAS, KMT2A-PTD, NF1, NPM1, NRAS, PHF6, PPM1D, PRPF8, PTPN11, RAD21, RUNX1, SAMD9†, SAMD9L†, SETBP1, SF3B1, SRSF2, STAG2, TET2, TP53, U2AF1, UBA1, WT1, ZRSR2 | |
| MPN and mastocytosis‡ | ASXL1, CALR, CBL, CSF3R, DNMT3A, EZH2, IDH1, IDH2 JAK2§, KIT, KRAS, MPL, NRAS, PTPN11, RUNX1, SETBP1, SF3B1, SH2B3, SRSF2, TET2, U2AF1, ZRSR2 | BCR::ABL1§ |
| Eosinophilia | ASXL1, CBL, DNMT3A, EZH2, KRAS, NRAS, RUNX1, SF3B1, SRSF2, STAT5B, TET2, U2AF1 | BCR::ABL1§, FGFR1::R, FLT3::R, JAK2::R, PDGFRA::R, PDGFRB::R |
| AML | Genes required for diagnosis and risk stratification: ASXL1, BCOR, CEBPA, DDX41, EZH2, FLT3-ITD§, FLT3-TKD§, IDH1§, IDH2§, NPM1, RUNX1, SF3B1, SRSF2, STAG2, TP53, U2AF1, ZRSR2 Additional genes recommended to test for at diagnosis and for use in disease monitoring: ANKRD26, BCORL1, BRAF, CBL, CSF3R, DNMT3A, ETV6, GATA2, JAK2, KIT, KRAS, NRAS, NF1, PHF6, PPM1D, PTPN11, RAD21, SETBP1, TET2, WT1 | BCR::ABL1§, CBFB::MYH11, DEK::NUP214 MECOM::R, KMT2A::R, NUP98::R, RUNX1::RUNX1T1, PML::RARA§ |
| B-ALL | CREBBP, CRLF2, FLT3, IDH1, IDH2, IKZF1, IL7R, JAK1, JAK2, JAK3, KMT2D, KRAS, NF1, NRAS, PAX5, PTPN11, SETD2, SH2B3, TP53 | ABL1::R§, ABL2::R, CRLF2::R, CSF1R::R, DUX4::R, EPOR::R, ETV6::R, JAK2::R, KMT2A::R, MEF2D::R, NUTM1::R, PAX5::R, PDGFRA::R, PDGFRB::R, TCF3::R, ZNF384::R |
| T-ALL | DNMT3A, ETV6, EZH2, FBXW7, FLT3, IDH1, IDH2, IL7R, JAK1, JAK3, KRAS, MSH2, NOTCH1, NRAS, PHF6, PTEN, U2AF1, WT1 | BCL11B::R, LMO2::R, MYB::R, NUP::ABL1, NUP214::R, STIL::R, TAL::R, TLX1::R, TLX3::R |
| Indication . | Single gene mutations . | Structural variants∗ . |
|---|---|---|
| MDS, MDS/MPN, cytopenia | ASXL1, BCOR, BCORL1, CBL, CEBPA, CSF3R, DDX41, DMNT3A, ETV6, ETNK1, EZH2, FLT3-ITD, FLT3-TKD, GATA2, GNB1, IDH1, IDH2, JAK2, KIT, KRAS, KMT2A-PTD, NF1, NPM1, NRAS, PHF6, PPM1D, PRPF8, PTPN11, RAD21, RUNX1, SAMD9†, SAMD9L†, SETBP1, SF3B1, SRSF2, STAG2, TET2, TP53, U2AF1, UBA1, WT1, ZRSR2 | |
| MPN and mastocytosis‡ | ASXL1, CALR, CBL, CSF3R, DNMT3A, EZH2, IDH1, IDH2 JAK2§, KIT, KRAS, MPL, NRAS, PTPN11, RUNX1, SETBP1, SF3B1, SH2B3, SRSF2, TET2, U2AF1, ZRSR2 | BCR::ABL1§ |
| Eosinophilia | ASXL1, CBL, DNMT3A, EZH2, KRAS, NRAS, RUNX1, SF3B1, SRSF2, STAT5B, TET2, U2AF1 | BCR::ABL1§, FGFR1::R, FLT3::R, JAK2::R, PDGFRA::R, PDGFRB::R |
| AML | Genes required for diagnosis and risk stratification: ASXL1, BCOR, CEBPA, DDX41, EZH2, FLT3-ITD§, FLT3-TKD§, IDH1§, IDH2§, NPM1, RUNX1, SF3B1, SRSF2, STAG2, TP53, U2AF1, ZRSR2 Additional genes recommended to test for at diagnosis and for use in disease monitoring: ANKRD26, BCORL1, BRAF, CBL, CSF3R, DNMT3A, ETV6, GATA2, JAK2, KIT, KRAS, NRAS, NF1, PHF6, PPM1D, PTPN11, RAD21, SETBP1, TET2, WT1 | BCR::ABL1§, CBFB::MYH11, DEK::NUP214 MECOM::R, KMT2A::R, NUP98::R, RUNX1::RUNX1T1, PML::RARA§ |
| B-ALL | CREBBP, CRLF2, FLT3, IDH1, IDH2, IKZF1, IL7R, JAK1, JAK2, JAK3, KMT2D, KRAS, NF1, NRAS, PAX5, PTPN11, SETD2, SH2B3, TP53 | ABL1::R§, ABL2::R, CRLF2::R, CSF1R::R, DUX4::R, EPOR::R, ETV6::R, JAK2::R, KMT2A::R, MEF2D::R, NUTM1::R, PAX5::R, PDGFRA::R, PDGFRB::R, TCF3::R, ZNF384::R |
| T-ALL | DNMT3A, ETV6, EZH2, FBXW7, FLT3, IDH1, IDH2, IL7R, JAK1, JAK3, KRAS, MSH2, NOTCH1, NRAS, PHF6, PTEN, U2AF1, WT1 | BCL11B::R, LMO2::R, MYB::R, NUP::ABL1, NUP214::R, STIL::R, TAL::R, TLX1::R, TLX3::R |
Conventional karyotype should be performed on all cases at diagnosis. Specific FISH, RT-PCR, or gene fusion NGS assays (targeted DNA/RNA or WGS) may be included depending on clinical context and results of other clinical studies.
Pediatric patients.
Mast cell disease with suspicion of associated hematologic neoplasm.
Food and Drug Administration–approved targeted therapy.
Genomic sequencing (WES and WGS) and transcriptomic sequencing (WTS or RNA-seq)
Broad genomic sequencing assays allow for the detection of genomic alterations anywhere in the coding genome (WES, interrogating 1%-2% of the whole genome) or entire genome (WGS).24,25 WGS and WES entail higher sequencing costs and more extensive data analysis pipelines, and do not typically achieve the same level of coverage depth as targeted gene panels, resulting in lower analytic sensitivity. Most WES applications are primarily limited to the research setting. In comparison, WGS, which can detect a full range of genomic alterations including CNAs and chromosomal rearrangements, has shown promise as a clinical application, especially in cases with unsuccessful conventional cytogenetics.26 Transcriptome-wide sequencing (WTS or RNA-seq) detects both chromosomal rearrangements and changes in messenger RNA (mRNA) and microRNA (miRNA) expression and is primarily limited to research and discovery.27 In ALL, WTS has led to the identification of unique B-cell ALL (B-ALL) subtypes and development of targeted panels for clinical use.28 Decreasing cost, increasing wider availability, and evidence for clinical utility will likely foster the integration of genomic sequencing technologies into routine clinical testing.
Molecular MRD methods
qPCR and FISH
The oldest and most established method for monitoring MRD in hematopoietic neoplasms rely on the detection of previously identified translocations (PML::RARA, BCR::ABL1, RUNX1::RUNX1T1) or recurrent insertions/deletion (NPM1 exon 11 mutation [ENST00000296930]; the same mutation may also be annotated in exon 12 depending on the transcript used by the laboratory). Recurrent translocations may be detected either by FISH or more sensitive qPCR from RNA, with sensitivities of 10−2 and 10−6, respectively.29 In AML, high sensitivity monitoring for highly recurrent NPM1 exon 11 gene mutation can be accomplished by qPCR with a sensitivity of ∼10−3 or lower.30
High-sensitivity sequencing for somatic variants
The sensitivity and specificity of NGS can be improved with UMIs to tag individual DNA templates on single or dual strands of the target and can increase the analytic detection sensitivity to up to 10−6.31-33 For detecting low levels of molecular disease after treatment in AML, a sensitivity of at least 10−3 is recommended.34 UMIs are used to computationally “collapse” DNA sequence information into “consensus” reads, allowing removal of PCR or sequencing errors absent in identically tagged templates (Figure 2D).35-38 UMI-based sequencing can be coupled with DNA enrichment to create generalized MRD panels or to monitor previously detected mutations in a patient-specific manner.
T-cell receptor (TR) and immunoglobulin NGS-based MRD approaches
NGS of the hypervariable regions of immunoglobulin (IGH, IGK, or IGL) and/or TR (TRB, TRG) can be used to measure MRD in B- or T-cell lymphoblastic leukemias. Although IGH/TR rearrangements are reasonably specific for identifying a patient’s neoplastic clone, importantly, these sequences may rarely occur as part of the normal immune repertoire at <10−4, and conversely, clonal sequences may continue to change through ongoing variable diversity joining recombination, potentially resulting in false-negative calls. Thus, caution should be exercised in interpreting very low levels of an IG/TR clone that is identical to the patient’s ALL clone.39-42 It is also recommended that laboratories follow more than 1 IG/TR clone (when possible) to reduce the chance of false-negative MRD errors.
Appropriate genomic testing depends on the clinical scenario or diagnostic disease category. The sections that follow provide recommendations for genomic testing in specific clinical contexts and myeloid neoplasm subgroups, for diagnosis, classification, prognosis, and disease monitoring after therapy. Suggested genes to be tested in specific diagnostic entities are summarized in Table 2.
Myeloid neoplasms and inherited/germline disorders
With the advent of NGS, individuals are increasingly recognized as having potentially deleterious germline variants that predispose to hematologic neoplasms, especially myeloid neoplasms (Table 3).43-49 Most of these are inherited, but some can occur de novo and are newly acquired in that individual’s germline, and as such, can be inherited by that individual’s descendants. Current indications for germline genetic testing include patients with ≥2 cancers, 1 of which is a hematologic neoplasm,50 and those with a hematologic neoplasm and a positive family history. Although, historically, germline genetic testing has been performed mainly on patients with myeloid neoplasms who received the diagnosis under the age of 40 to -50 years, it is now recognized that young age at diagnosis or positive family history are not required to justify genetic testing.51,52 Thus, germline predisposition risk should be considered for all patients diagnosed with a myeloid neoplasm regardless of age because some germline predisposition alleles, such as those in DDX41, present at older ages.43,48,53-55 In patients with mutations detected on sequencing panels that could represent pathogenic germline variants (eg, CEBPA, DDX41, GATA2, RUNX1, or TP53 mutations, among others) and occurring at ∼50% VAF, germline predisposition testing should be considered particularly if mutations persist in remission.56 Genetic counselors and health care providers should be familiar with testing options, including optimal sample types (eg, cultured skin fibroblasts to ensure exclusion of somatic mutations present in hematopoietic cells) and other types of tissues accepted by some laboratories (including hair follicles or skin biopsies washed to remove blood), as well as available testing platforms.57 Challenges to clinical testing for these disorders include the lack of training for most clinicians regarding these conditions, the rapid increase in genes under consideration, the high proportion of variants of uncertain significance (VUSs) in less well-studied genes, the need to distinguish germline from somatic mutations, and a lack of standardization in the field regarding which patients and which genes should be tested.57
Clinical considerations for germline predisposition testing
| Clinical considerations regarding germline predisposition testing . | ||
|---|---|---|
| WHO? | Individual with ≥2 cancers, 1 of which is an HM | |
| OR | ||
| Individual with a history of an HN AND | ||
| A relative within 2 generations diagnosed with an HN, OR | ||
| A relative within 2 generations diagnosed with a solid tumor at age ≤50, OR | ||
| A relative within 2 generations diagnosed with another hematopoietic abnormality | ||
| OR | ||
| Individual whose tumor-based molecular profiling identified a deleterious variant with a VAF consistent with germline status∗ | ||
| OR | ||
| HM diagnosis at a much younger age than is typical | ||
| IDEAL AGE for testing? | Individuals of all ages should be considered for germline predisposition testing because some gene variants drive myeloid malignancies even at advanced ages (e.g., DDX41) | |
| WHAT SAMPLE? | Ideal: | Gold standard cultured skin fibroblasts (some clinical laboratories also accept BM-derived mesenchymal stromal cells) |
| Possible: | Skin biopsy, with washout of PB | |
| Hair follicles (may not yield sufficient DNA for comprehensive testing) | ||
| Buccal swab (may have low-level PB contamination) | ||
| Not recommended: | Saliva (highly contaminated with PB) | |
| Fingernails (may be contaminated with monocytes) | ||
| WHAT TEST?† | WES augmented with spike-in probes for noncoding regions known to contain predisposition loci followed by analysis of gene groups | |
| WGS (if available), with a virtual panel of appropriate genes, including noncoding regions and copy number variation studies | ||
| Panel-based NGS | ||
| COMPLEMENTARY testing | Telomere flow–FISH can identify individuals with short-telomere syndromes, although interpretation can be confounded by active disease and/or treatment | |
| Diepoxybutane and mitomycin C analyses identify excessive chromosome breakage and assist in the diagnosis of FA | ||
| HOW can you tell if a variant is germline? | Variant is present in DNA derived from a preferred tissue source (see above) at a VAF consistent with germline status∗ OR | |
| Variant is present in the index patient plus one other relative at a VAF consistent with germline∗ | ||
| WHEN? | At HN diagnosis | |
| At recognition of a potential germline allele from tumor or other screening, including somatic variants suggestive of an underlying germline variant (eg, R525H-encoding variant in DDX41) | ||
| before HSCT using a relative as a donor | ||
| WHY? | Plan surveillance for other cancers or organ dysfunction | |
| Plan HSCT using a related donor | ||
| Allow pre-implantation genetic testing | ||
| Cascade testing throughout the family | ||
| Clinical considerations regarding germline predisposition testing . | ||
|---|---|---|
| WHO? | Individual with ≥2 cancers, 1 of which is an HM | |
| OR | ||
| Individual with a history of an HN AND | ||
| A relative within 2 generations diagnosed with an HN, OR | ||
| A relative within 2 generations diagnosed with a solid tumor at age ≤50, OR | ||
| A relative within 2 generations diagnosed with another hematopoietic abnormality | ||
| OR | ||
| Individual whose tumor-based molecular profiling identified a deleterious variant with a VAF consistent with germline status∗ | ||
| OR | ||
| HM diagnosis at a much younger age than is typical | ||
| IDEAL AGE for testing? | Individuals of all ages should be considered for germline predisposition testing because some gene variants drive myeloid malignancies even at advanced ages (e.g., DDX41) | |
| WHAT SAMPLE? | Ideal: | Gold standard cultured skin fibroblasts (some clinical laboratories also accept BM-derived mesenchymal stromal cells) |
| Possible: | Skin biopsy, with washout of PB | |
| Hair follicles (may not yield sufficient DNA for comprehensive testing) | ||
| Buccal swab (may have low-level PB contamination) | ||
| Not recommended: | Saliva (highly contaminated with PB) | |
| Fingernails (may be contaminated with monocytes) | ||
| WHAT TEST?† | WES augmented with spike-in probes for noncoding regions known to contain predisposition loci followed by analysis of gene groups | |
| WGS (if available), with a virtual panel of appropriate genes, including noncoding regions and copy number variation studies | ||
| Panel-based NGS | ||
| COMPLEMENTARY testing | Telomere flow–FISH can identify individuals with short-telomere syndromes, although interpretation can be confounded by active disease and/or treatment | |
| Diepoxybutane and mitomycin C analyses identify excessive chromosome breakage and assist in the diagnosis of FA | ||
| HOW can you tell if a variant is germline? | Variant is present in DNA derived from a preferred tissue source (see above) at a VAF consistent with germline status∗ OR | |
| Variant is present in the index patient plus one other relative at a VAF consistent with germline∗ | ||
| WHEN? | At HN diagnosis | |
| At recognition of a potential germline allele from tumor or other screening, including somatic variants suggestive of an underlying germline variant (eg, R525H-encoding variant in DDX41) | ||
| before HSCT using a relative as a donor | ||
| WHY? | Plan surveillance for other cancers or organ dysfunction | |
| Plan HSCT using a related donor | ||
| Allow pre-implantation genetic testing | ||
| Cascade testing throughout the family | ||
HN, hematopoietic neoplasm.
Generally considered to be a VAF between 30% to 60% when tested on an appropriate sample type.
Genes curated as those in which deleterious variants confer risk for hematopoietic malignancies are increasing in number. Resources that delineate up to date genes to consider for testing include: https://dnatesting.uchicago.edu/ and https://panelapp.genomicsengland.co.uk/panels/59/. Several biases regarding testing need to be kept in mind and considered. There may be ascertainment bias in some publications, with gene variants described in a cancer cohort but not in a control, noncancer cohort, resulting in a study that lacks a comparison of an observed variant frequency vs an expected variant frequency. Confounding factors, such as age, prior genotoxic therapies, and other familial factors, may contribute to cancer risk along with that conferred by the germline variant. Pathologic classifications of myeloid malignancies, including myelodysplastic syndromes, clonal cytopenias, and clonal hematopoiesis, shift over time and may complicate interpretations of individual and family histories.
Germline variants are categorized into 5 tiers according to the American College of Medical Genetics and Genomics/AMP as pathogenic, likely pathogenic, VUS, likely benign, and benign58; but only pathogenic and likely pathogenic variants are considered disease causing and followed clinically. Germline variant classifications can be found in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar). Gene-specific guidelines are available for RUNX1 and are under development for other myeloid predisposition genes.59 Importantly, as additional information regarding gene/allele function and additional patient cases (both unrelated cases and segregation data from known families) accrue, gene variant classification can change over time, and VUSs can be reclassified as likely pathogenic or pathogenic, complicating individual and family counseling. Recognition of hereditary myeloid neoplasms may alter patient management, especially regarding the consideration of allogeneic hematopoietic stem cell transplant (allo-HSCT) using a related donor, as well as health surveillance strategies for the patient and relatives who share the deleterious variant.60-63 Testing for germline risk alleles should be performed as early as possible during clinical management to facilitate treatment plans that may include allo-HSCT.
Certain germline disorders may be associated with additional clinical features, such as those associated with quantitative and qualitative platelet defects: ANKRD26, ETV6, and RUNX1; and those variably associated with additional organ dysfunction, for example: GATA2 with immunodeficiency; Shwachman-Diamond syndrome with exocrine pancreatic insufficiency and skeletal dysplasia; Fanconi anemia (FA) with congenital anomalies, squamous cell carcinomas, and liver tumors; and dyskeratosis congenita with pulmonary fibrosis, liver cirrhosis, and vascular anomalies; among others. Some genes, such as CEBPA, confer germline risk only to myeloid neoplasms, whereas other genes may confer risk to a variety of hematologic neoplasms and solid tumors. Additional testing can be a helpful complement to germline genetic testing. For example, telomere flow FISH can identify patients with short telomere syndromes, ∼30% of whom will not have a gene variant identified on panel testing.64 FA chromosomal breakage studies by diepoxybutane/mitomycin-C analysis are useful because of the challenges of FA genetic testing, including common VUSs in FA genes, deletions, distinguishing cis vs trans arrangements of FA gene mutations, treatment effects, and somatic mosaicism, which may mask the diagnosis. Of note, the tumor spectrum associated with each disorder may expand over time as longer follow-up of additional individuals and families becomes available. In addition, germline predisposition to lymphoid malignancies is emerging in importance and often overlaps with myeloid neoplasm risk genes. Future work will reveal a more comprehensive list of hematologic neoplasm predisposition genes and will influence how broadly to offer predisposition testing among patients with established myeloid neoplasms as well as those with sustained cytopenias.
Patients with cytopenia with suspicion of MDS
A key diagnostic challenge for hematologists is determining whether persistent cytopenia reflects MDS or other causes. Increasingly, gene-panel sequencing is being used in this population to aid diagnosis. The absence of clonal driver mutations in a patient with unexplained cytopenia, referred to as idiopathic cytopenia of undetermined significance (ICUS), has high negative predictive value (NPV) for MDS.65 Conversely, the presence of mutations in this scenario suggests either clonal cytopenia of undetermined significance (CCUS) or MDS, necessitating further workup in most cases. Mutant hematopoietic cell clones may also be found in individuals with normal blood counts and no evidence of hematologic neoplasm, referred to as CHIP or age-related clonal hematopoiesis.66 Although CHIP is associated with increased risk of developing hematologic67-70 and cardiovascular disease,71,72 and all-cause mortality,70,73 testing in patients who are noncytopenic is not currently recommended due to lack of evidence that intervention is indicated. In cases where CHIP is identified incidentally, patients may be seen by clinicians who can provide reassurance that the clinical course is generally benign, with suggestions for lifestyle modifications to lower cardiovascular disease risk.74 For those with CHIP or CCUS with higher risk features (discussed in further sections), more frequent surveillance of blood counts may be warranted to assess for early signs of malignant transformation, as discussed in further sections. Features of CHIP, ICUS, CCUS, and MDS are summarized in Table 4.
Features of CHIP, ICUS, CCUS, and MDS
| . | Cytopenia/dysplasia . | VAF cutoff . | Commonly mutated driver genes . | Higher risk features . |
|---|---|---|---|---|
| CHIP | No/Minimal (<10%) to none | >2% | DNMT3A, TET2, ASXL1, PPM1D, JAK2, ZBTB33, ZNF318, TP53, CBL, GNB1, SF3B1, SRSF2, loss of Y chromosome | Mutations in TP53, ASXL1, JAK2, SF3B1, SRSF2, U2AF1, or IDH1/IDH2; >1 driver mutations; VAF > 10% |
| ICUS | Yes/Minimal (<10%) to none | None | None | None |
| CCUS | Yes/Minimal (<10%) to none | >2% | TET2, DNMT3A, ASXL1, SRSF2, ZRSR2, SF3B1, U2AF1, IDH1/2, RUNX1, EZH2, JAK2, CBL, KRAS, CUX1, TP53 | Spliceosome gene mutations DNMT3A, ASXL1, TET2 in comutational patterns (RUNX1, EZH2, CBL, BCOR, CUX1, TP53, or IDH1/IDH2 most specific), >1 driver mutation; VAF > 10% |
| MDS | Yes/Yes | None | See text | See text |
| . | Cytopenia/dysplasia . | VAF cutoff . | Commonly mutated driver genes . | Higher risk features . |
|---|---|---|---|---|
| CHIP | No/Minimal (<10%) to none | >2% | DNMT3A, TET2, ASXL1, PPM1D, JAK2, ZBTB33, ZNF318, TP53, CBL, GNB1, SF3B1, SRSF2, loss of Y chromosome | Mutations in TP53, ASXL1, JAK2, SF3B1, SRSF2, U2AF1, or IDH1/IDH2; >1 driver mutations; VAF > 10% |
| ICUS | Yes/Minimal (<10%) to none | None | None | None |
| CCUS | Yes/Minimal (<10%) to none | >2% | TET2, DNMT3A, ASXL1, SRSF2, ZRSR2, SF3B1, U2AF1, IDH1/2, RUNX1, EZH2, JAK2, CBL, KRAS, CUX1, TP53 | Spliceosome gene mutations DNMT3A, ASXL1, TET2 in comutational patterns (RUNX1, EZH2, CBL, BCOR, CUX1, TP53, or IDH1/IDH2 most specific), >1 driver mutation; VAF > 10% |
| MDS | Yes/Yes | None | See text | See text |
The decision to perform gene-panel sequencing in patients with cytopenia is dependent on the pretest probability of a myeloid neoplasm, such as MDS, which is in turn informed by the clinical context and additional laboratory testing. Initial workup should include examination of a peripheral blood (PB) smear and lineage-appropriate studies, including nutritional deficiencies (iron, B12, folate, copper), or toxicity evaluation for hemolysis, renal failure, liver disease, splenomegaly or monoclonal gammopathy, and reconciliation with medications. Cytopenia(s) identified below certain thresholds (hemoglobin < 10 g/dL, platelets < 100 000/μL, absolute neutrophil count < 18 000/μL) increases the pretest probability of MDS75; however, MDS is not confined to these defined thresholds and gene-panel sequencing may be considered even in patients with mild cytopenia depending on clinical suspicion of MDS. Although at least 6 months of unexplained cytopenia has been suggested to establish its chronicity for some MDS subtypes,75 gene-panel sequencing might be considered earlier, particularly in those with severe cytopenia or where clinical suspicion is high. Clinical factors that positively influence the pretest probability of MDS include (1) a history of prior chemotherapy or radiation, particularly with delayed count recovery on therapy and (2) family history of a hematologic neoplasm, in which case germline testing is also indicated.
There is high concordance in mutation detection between PB and BM, and testing from PB can be used to predict the likelihood of a myeloid neoplasm in the BM.76,77 The high NPV of 95% from targeted sequencing of PB in unselected patients with cytopenia suggests that gene panel sequencing can identify those with a very low risk of a myeloid neoplasm who may not require an invasive and costly BM assessment.65 However, BM sampling is the only means for assessing BM morphology and is required to diagnose and distinguish between MDS, other clonal cytopenias (such as CCUS), and unexpected alternative malignancies, which may be missed with PB-only screening. In addition, BM sampling ensures accurate disease classification, enables conventional cytogenetics, and provides key information for treatment decisions or clinical trial eligibility. The detection of somatic driver mutations (especially high-risk ones, see further sections) in the PB warrants a BM biopsy for further evaluation, although it is not necessary to repeat gene panel sequencing on the concurrent BM sample.76-78
In the absence of a morphologic diagnosis of MDS, the presence of a driver mutation in a patient with cytopenia (ie, CCUS) is strongly predictive of a subsequent myeloid neoplasm. There are typical high-risk patterns of mutations, which further increase this likelihood, including the number of detected driver mutations and a higher VAF. A 10% VAF threshold is predictive of progression79; however, higher thresholds (eg, 20%) may be more applicable in unselected populations of patients with cytopenia in whom other etiologies for the cytopenia have not already been excluded.65 Mutations in spliceosome genes and/or comutation patterns of epigenetic genes DNMT3A, TET2, and ASXL1 are highly predictive of progression to a myeloid malignancy with cooccurring mutations in RUNX1, EZH2, CBL, BCOR, CUX1, TP53, or IDH1/IDH2 being most specific for progression to a myeloid neoplasm with MDS.79,80 Although (with the exception of multihit TP53 mutation) these high-risk mutational patterns are not currently part of the diagnostic criteria for MDS, studies have demonstrated that high-risk CCUS has comparable clinical outcomes to low-risk MDS.79 In contrast, solitary DNMT3A mutations, even in the context of cytopenias, may have an indolent course mimicking CHIP.80 Multihit TP53 variants (>1 mutation or mutation plus loss of the alternate allele) are associated with genomic instability and high-risk disease and are considered to be diagnostic of a myeloid neoplasm with mutated TP53 in the current International Consensus Classification.81,82 In patients who have undergone chemotherapy or radiation therapy, the identification of pathogenic variants in TP53, PPM1D, and CHEK2 indicate high risk for developing a therapy-related myeloid neoplasm.83
MDS and MDS/MPNs
The NCCN recommends incorporation of somatic gene mutation testing in PB or BM for patients with MDS, given the correlation of somatic mutations with disease risk in MDS and the potential to use targeted therapies such as IDH inhibitors.77,84 Although somatic gene mutations do not replace morphologic assessment, they are relevant to MDS categorization. The ICC guidelines recognize the mutation-defined entities: MDS with mutated SF3B1, MDS and MDS/AML with mutated TP53, and MDS/MPNs with thrombocytosis and SF3B1 mutation.82 In addition, specific somatic mutations represent exclusion criteria (eg, BCR::ABL1 rearrangement) for these and other entities, further supporting the need for genetic testing for accurate class assignment. NGS also significantly aids risk assessment and clinical decision making in patients with intermediate-risk MDS, according to IPSS-Revised, who are potential candidates for allo-HSCT.85
In the MDS/MPN entity, chronic myelomonocytic leukemia (CMML) clonal driver mutations can be detected in >90% of cases, with the combination of TET2 (especially biallelic variants) and SRSF2 being highly specific for a myelomonocytic phenotype.86 Consequently, genomic profiling can provide supportive evidence for a diagnosis of CMML, and focused gene panel testing is recommended.87 In patients presenting with monocytosis, the absence of a clonal driver mutation in the PB has a very high NPV for CMML, whereas a demonstrable mutation is not only strongly predictive of a neoplastic diagnosis in the BM but also significantly affects overall survival even in those without a confirmed morphologic diagnosis (clonal monocytosis of undetermined significance).88
The integration of somatic mutations into prognostic scoring systems provides more accurate risk stratification of individual patients.89,90 Within the International Working Group for Prognosis in MDS, a clinical-molecular IPSS model (IPSS-M) has been developed recently and validated for MDS (https://mds-risk-model.com).91 In the MDS/MPN entity CMML, somatic mutations are integrated in clinical/molecular prognostic systems resulting in improved risk stratification.92 This includes the analysis of ASXL1, NRAS, RUNX1, and SETBP1 with sequencing of these genes being strongly recommended in patients eligible for transplantion.87 Analysis of a minimum of 20 genes is recommended for patients with CMML who are being considered for active treatment.87 Somatic mutations may also predict response and/or outcome after selected treatments in MDS and MDS/MPN, for example, TP53 mutations are consistently associated with shorter survival after allo-SCT,93,94 and somatic mutations in TP53 also predict increased response to hypomethylating agents (HMAs).95 As with other disease-related variables, somatic mutations may require reassessment to update individual risk in case of significant clinical changes or before disease-modifying treatments. It must be noted that although combining genomic profiling with hematologic and cytogenetic variables improves risk assessment, prospective real-world data and clinical trials are required to translate this improved stratification into evidence-based recommendations for clinical decision making.
In patients undergoing disease-modifying therapies, genomic profiling is potentially instrumental to measure response and MRD in MDS and CMML. Persistent disease-associated mutations after allo-SCT are associated with a higher risk of progression.96 Molecular monitoring of MRD has also been applied after HMA therapy97; however, additional studies are warranted to confirm its clinical value. In patients with CMML, response to HMA has been associated with changes in DNA methylation, without decrease in mutation allele burden, arguing for a predominantly epigenetic effect.98 However, to date, somatic mutation analysis has not been incorporated in consensus-based response measurement in MDS or MDS/MPN, but evidence of molecular clonal evolution (ie, the acquisition of new pathogenic mutations and/or cytogenetic aberrations) has been proposed as a criterion for disease progression in adult MDS/MPN.99
Finally, compounds targeting proteins or signaling pathways disrupted by recurrently mutated genes have been tested in recent clinical trials in MDS and MDS/MPN, and the number of compounds entering clinical investigation will likely increase in the future.100 Although, at present, few agents besides lenalidomide for MDS with del(5q) have been licensed for clinical use in MDS or MDS/MPN, genomic profiling is instrumental to giving patients access to these targeted therapies within the context of clinical trials.
Inclusion of probes for copy number detection or use of SNP array–based karyotyping is highly recommended to capture chromosomal abnormalities, especially copy-neutral LOH.26,93 Detection of copy-neutral LOH is particularly important to capture multihit TP53 lesions. In fact, whereas most of the multihit TP53 lesions can be detected on the basis of VAF >50% or the presence of deletion 17p, del(17p), as many as 6% of these lesions display <50% VAF, thus potentially escaping detection based on these criteria.81 The use of FISH panels to detect recurrent cytogenetic abnormalities (−5, del(5q), −7, etc) is not required in the setting of adequate metaphase cytogenetic studies, and most studies have not increased diagnostic yield.101-103
MPNs, mast cell neoplasms, and eosinophilic neoplasms
Classical MPNs
Screening for mutations in the known driver genes JAK2, CALR, and MPL is mandatory for establishing MPN diagnosis.75,104 Panel-based NGS may not be required to establish the initial diagnosis but is recommended in patients who are driver mutation negative to identify uncommon somatic variants (eg, indels in JAK2 exon 12 in PV, noncanonical somatic mutations in JAK2 and MPL)105 as well as rare germline variants in JAK2/MPL that cause hereditary thrombocytosis or erythrocytosis and mimic MPNs.106 It is recommended that JAK2 p.V617F VAF be obtained in DNA from PB samples or purified granulocytes. Because the advent of disease-modifying agents (eg, interferon)107 requires serial measurements to inform treatment, quantitative mutation abundance should be reported. An increasing VAF from baseline has been associated with disease progression to post–polycythemia vera/essential thrombocythemia (PV/ET) myelofibrosis.108,109 NGS for testing for variants in other myeloid neoplasm–associated genes also has prognostic value: ASXL1, EZH2, SRSF2, IDH1, IDH2, and U2AF1, are considered “high molecular risk” mutations in primary myelofibrosis110 and are included in current risk classification schemes,111,112 whereas the clinical value of additional mutations in ET and PV is still uncertain. Although at present there is no general consensus on how extensive the search for additional mutations by NGS should be,113 inclusion of TP53, NRAS/KRAS, and RUNX1 may be worthwhile because these mutations are likely to have an impact on the outcome of and/or resistance to treatment.114,115 Similarly, CSF3R mutation status should be evaluated because truncating mutations occurring outside of the proximal membrane region have shown sensitivity to dasatinib.116,117 Persistence of MPN-associated variants 3 to 6 months after allo-HSCT associates with an increased risk of relapse; the use of highly sensitive JAK2 p.V617F PCR tests (sensitivity of at least 0.01%) may be particularly useful in this scenario.118 Post–allo-HSCT monitoring of variants after confirmation of molecular remission is not routinely performed but may be useful in cases of suspected relapse (eg, falling donor chimerism).
Chronic neutrophilic leukemia
Variants in CSF3R, which encodes the granulocyte colony-stimulating factor receptor, are found in 60% to 80% of individuals with this rare MPN.116,119,120 Identification of the most common activating mutation, p.T618I, has therapeutic significance because responses to ruxolitinib have been reported.116,121 Although CSF3R variants are most prevalent in chronic neutrophilic leukemia, they are not specific to this diagnosis and can also be seen in other myeloid disorders, particularly in atypical chronic myeloid leukemia (aCML) and CMML.
Systemic mastocytosis (SM)
In SM, the KIT p.D816V mutation is identified in >90% of patients at diagnosis.122,123 A BM sample should be used for greater sensitivity, however PB positivity for KIT mutation indicates multilineage involvement and suggests an associated hematologic neoplasm and establishes a B-finding for diagnosis of smoldering SM when present with a VAF ≥ 10%.124,125 High-sensitivity assays such as allele-specific PCR (from RNA or DNA) or dPCR are recommended over standard NGS for the identification of KIT p.D816V. The VAF (abundance) of KIT p.D816V should ideally be reported, which is more easily calculated from DNA rather than RNA. With the advent of KIT inhibitors (midostaurin, avapritinib),126,127 measurement of changes of KIT p.D816V VAF in PB, if positive at baseline, might have important value for monitoring and prognostication, although this is still exploratory.127,128 In KIT D816V–negative cases, sequencing rarely detects other KIT mutations at position 816 (eg, p.D816H/N/Y) or 822 (p.N822K) in exon 17 or in the extracellular domains.125 In advanced SM, prognosis is adversely affected by additional somatic mutations, for example, in SRSF2, ASXL1, or RUNX1 (so-called S/A/R mutations), identified by NGS.129 In patients with chemotherapy- or tyrosine kinase inhibitor (TKI)–resistant/refractory disease, an NGS myeloid panel might identify the emergence or expansion of clones with new (eg, KRAS/NRAS or TP53) or preexisting mutations, but at present, it is not mandatory for patient management. Cytogenetics may reveal prognostically negative abnormalities (eg, −5, −7, complex karyotype) and should be performed at diagnosis and at progression or relapse.130,131
Neoplasms with eosinophilia
Eosinophilia is most often reactive, and efforts should be made to identify secondary reactive etiologies before embarking on costly and unnecessary molecular tests.132 The 2 most frequent molecular abnormalities to search for, concurrently or stepwise, are FIP1L1::PDGFRA (which can be detected by reverse transcriptase PCR [RT-PCR], FISH, or NGS assays) and KIT D816V (which points to SM as the underlying cause for the eosinophilia).133 Cytogenetic analysis on a BM aspirate identifies reciprocal translocations indicating rearrangement of the TKs PDGFRA (4q12), PDGFRB (5q31-33), FGFR1 (8p11), FLT3 (13q12), and JAK2 (9p24), associated with myeloid/lymphoid neoplasms with eosinophilia (MLN-eos) and TK gene fusions. Other cytogenetic abnormalities, for example, deletions, monosomies, or complex karyotype, as well as presence of somatic mutations may help to classify eosinophilia as clonal. FISH analysis is used primarily to identify the specific TK gene rearrangement; however, the specific fusion may also be confirmed by RT-PCR or DNA/RNA NGS fusion detection and NGS methods may be particularly useful in identifying patients with cryptic fusions. The monitoring of PDGFRA/B rearrangement by FISH or fusion transcripts by RT-PCR is commonly used in patients treated with TKIs, although there is no standardization equivalent to the BCR::ABL1 international scale for CML. Some patients treated with TKIs have achieved undetectable FIP1L1::PDGFRA fusion transcripts and have been able to discontinue treatment and remain in molecular remission.134
Apart from MLN-eos with TK fusion genes, MPN-unclassified with eosinophilia and chronic eosinophilic leukemia are rarely associated with mutations in STAT5B (p.N642H),135JAK2 (p.V617F, ex13InsDel),136 and JAK1 (p.R629_S632delinsSA).137 Additional mutations in ETNK1, RUNX1, ASXL1, or EZH2 may be prognostically informative. T-cell clonality (detected by PCR or NGS) is found in both reactive and clonal eosinophilia and may point to the lymphocytic hypereosinophilic syndrome variant.
CML
CML is a model of molecularly based diagnosis and monitoring because all patients have the causal BCR::ABL1 fusion, which can be detected by FISH or RT-PCR in PB samples. The BCR::ABL1 transcript type should be characterized at diagnosis. For the ∼97% of patients with the common e13a2/e14a2 types, BCR::ABL1 transcript levels measured on an international reporting scale determine treatment response and guide therapeutic decisions.138,139 A rise during treatment can signal drug resistance and should trigger BCR::ABL1 kinase domain mutation analysis because these mutations are the major known resistance mechanism. Emerging data suggest that mutations in other cancer-related genes are implicated in drug resistance.140 These include RUNX1 and ASXL1 mutations, and IKZF1 deletions. The NCCN suggests myeloid mutation panel testing for patients diagnosed with accelerated or blast phase or to identify BCR::ABL1-independent resistance mutations in patients without kinase domain mutations.139 However, BCR::ABL1 mutations mostly cooccur with other mutated genes,141,142 and myeloid mutation panels will not detect all relevant variants associated with lymphoid blast phase.
AML
The diagnostic workup of AML includes annotation of cytogenetic and molecular aberrations in the setting of morphologic assessment to confirm the diagnosis; in particular, the blast count defining AML is lower in the setting of some recurrent genetic abnormalities as described in the ICC (supplemental Table 1 available on the Blood website). As knowledge of genomic abnormalities is paramount to AML treatment decisions, we recommend reporting results of mutations associated with diagnostic classification within 5 days when possible; in most cases, elaboration of a therapeutic plan can safely await these results.143
Complete genomic evaluation including cytogenetics, NGS panel, FLT3-ITD testing, and FISH/CMA (if needed to confirm cytogenetic findings or to provide more rapid results, in the case of FISH compared with metaphase cytogenetics)144 should be performed to identify genomic abnormalities that define specific AML subtypes, as well as for abnormalities within the 2022 European Leukemia Network (ELN) risk classification to determine prognosis in patients treated with standard intensive chemotherapy and to inform consolidation treatment choice, especially pertaining to the role of allo-HSCT in first remission. Important updates in the 2022 ELN risk classification145 include the categorization of all FLT3-ITD mutations within the intermediate-risk group regardless of FLT3-ITD allelic ratio or NPM1 comutation, and the addition of AML with MDS-related mutations in the adverse-risk group. Favorable-risk disease associated with CEBPA mutations has been revised to specify bZIP in-frame mutations of CEBPA, regardless of monoallelic or biallelic status.146,147
With more than a dozen genes incorporated within current AML classification and risk stratification systems, the use of gene-panel testing provides the most cost-effective testing approach. Due to limitations with most NGS-based assays, FLT3-ITD and FLT3-TKD determination is often performed separately by PCR and capillary electrophoresis. Rapid annotation of additional mutations of therapeutic relevance, such as IDH1, IDH2, FLT3-ITD, and FLT3-TKD, is necessary to determine best treatment approaches given the availability of targeted mutant-specific inhibitors. Immunohistochemistry can rapidly detect abnormal cytoplasmic expression of mutant NPM1 in formalin-fixed paraffin-embedded samples or cell blocks, providing utility in situations of myeloid sarcomas, NPM1 mutations outside exon 11, or in resource-limited settings where molecular techniques are not available.148,149 A subset of hotspot mutations in IDH1 and IDH2 can also be rapidly evaluated by immunohistochemistry, and p53 protein accumulation or null-pattern expression correlates with the presence of TP53 mutations in most cases of AML.150-155TP53 mutation present at a VAF > 10% now defines the new category of AML with mutated TP53. For other class-defining or risk-defining mutations, the VAF cutoff has not been established.
Conventional karyotyping at diagnosis can be aided by rapid testing for gene fusions (either by qPCR, FISH, or NGS-based fusion panels), for example, PML::RARA, RUNX1::RUNX1T1, and CBFB::MYH11. “Myeloid FISH panels” to test for common MDS-associated chromosomal aberrations associated with adverse risk can be useful, particularly in settings where metaphase cytogenetics are not available.144 Any clonal karyotype or FISH positivity present above the validated laboratory threshold should be considered a positive result. In the setting of nonevaluable cytogenetics, CMA can also be a useful adjunct to identify unbalanced abnormalities as well as cryptic CNAs. More recently, WGS has been proposed as a single comprehensive assay for the evaluation of AML.26
Once in remission, monitoring of MRD by molecular methods (qPCR, dPCR, NGS) and multiparameter flow cytometry (MFC) allows ongoing refinement of relapse risk estimations, providing the opportunity to identify impending relapse and possibly allow for early intervention or modified treatment approaches, such as consideration of allo-HSCT in patients with favorable risk who retain detectable MRD by qPCR after completion of planned consolidation therapy. The importance of MRD in AML was confirmed in a meta-analysis of >80 publications with >10 000 patients; the estimated 5-year overall survival was 68% vs 34% in patients in AML remission with MRD− vs MRD+ status.156 Although proven interventions to eradicate MRD are currently lacking, the detection of persistent MRD after completion of consolidation, or MRD “relapse,” correlates with inferior outcomes including increased risk of relapse and decreased overall survival. Current guidelines recommend MRD assessments after 2 cycles of standard therapy, at the end of treatment, and then, evaluation every 3 months (if BM) or every 4 to 6 weeks (if PB) for 24 months.34 Recommended time points for MRD assessment in patients receiving less-intensive treatment regimens are not yet established.
MRD monitoring by molecular methods and MFC may provide complementary data.157 If an RT-qPCR assay (eg, NPM1, core-binding factor [CBF] fusion) is available, this is considered the preferred method for MRD evaluation and should be performed in the diagnostic sample to allow for estimation of the kinetics of response during treatment.158 Both PB and BM may be used for MRD evaluation, but detection sensitivity in PB may be lower.
Outside of RT-PCR and MFC, NGS is an alternative method for MRD assessment, which can provide useful information about emerging mutations not present at AML diagnosis (Figure 3). However, NGS MRD analysis may be performed in conjunction with FC, as some AMLs will be more amenable to detection by one method or the other, depending on the mutations present and phenotype, respectively.157 Note that the sensitivity of most routine NGS panels is ∼2% VAF; however, the ELN recommends error-corrected NGS with a minimum sensitivity of ∼0.1% VAF.34 Although driver alterations such as NPM1 and CBF-fusions (RUNX1::RUNX1T1 and CBFB::MYH11) are typically present in the founding clone and retained at relapse, mutations of signaling genes (ie, FLT3-ITD, NRAS/KRAS) are often subclonal and may vary in their presence over time and during treatment, with low NPV. This is especially true in patients with FLT3-mutated AML after receipt of FLT3-directed treatments, as ∼40% of patients can relapse with FLT3–wild type clones.159 Caution must be taken in the interpretation of residual epigenetic mutations, including “DTA” (DNMT3A, TET2, ASXL1), which represent preleukemic clones and are not predictive of relapse.156,160 Residual SRSF2 and IDH1/2 mutations may be similarly noninformative for MRD assessment.161
Sequencing-based tumor burden monitoring in myeloid neoplasms. (A) In this AML example, sequencing identifies NPM1 and DNMT3A mutations at diagnosis with the NPM1 mutation representing the founding clone (green: based on higher mutation VAF) and DNMT3A representing a subclone (gray: based on lower mutation VAF). Estimated sensitivity to detect mutations for different sequencing approaches is shown below. As the patient enters remission, the NPM1-mutated clone is partially cleared, becoming undetectable by panel-based sequencing and WGS but remains detectable by high-sensitivity MRD sequencing. In this example, the DNMT3A-mutated clone remains without significant change in VAF, consistent with persistent clonal hematopoiesis. Finally, the patient relapses with the same NPM1-mutated founding clone plus a newly acquired NRAS mutation. (B) A comparison of sequencing methods for MRD monitoring. Although WGS offers the greatest sequencing breadth and is capable of detecting structural variants such as CNAs and chromosomal translocations, standard coverage is generally only ∼60×, limiting detection to mutations with VAFs > 10%. Targeted sequencing is generally limited to 50 to 500 genes, providing minimum sequencing breadth but can achieve high coverages (1000×) at a lower cost than WGS and provides adequate sensitivity (2% VAF) for initial diagnostic evaluation. MRD panels are similar to targeted panels but use much higher sequencing depths and use UMIs to achieve sensitivities of ∼0.1% VAF allowing for MRD monitoring. MRD panels are generally easy to implement but may be of limited clinical utility for patients with few mutations covered by the panel. In patient-specific MRD sequencing, mutations are identified at diagnosis using broad methods such as exome sequencing or WGS, ensuring an adequate number of mutations to track. These mutations are then individually targeted using custom probes at subsequent time points. By focusing sequencing on known mutations, patient-specific MRD can achieve extremely high detection sensitivities for nearly all patients. Patient-specific MRD, however, can be logistically challenging and expensive to implement because it requires custom probe designs and validation for every patient. Patient-specific methods also cannot detect newly acquired mutations that were not targeted by probes at diagnosis. Professional illustration by Patrick Lane, ScEYEnce Studios.
Sequencing-based tumor burden monitoring in myeloid neoplasms. (A) In this AML example, sequencing identifies NPM1 and DNMT3A mutations at diagnosis with the NPM1 mutation representing the founding clone (green: based on higher mutation VAF) and DNMT3A representing a subclone (gray: based on lower mutation VAF). Estimated sensitivity to detect mutations for different sequencing approaches is shown below. As the patient enters remission, the NPM1-mutated clone is partially cleared, becoming undetectable by panel-based sequencing and WGS but remains detectable by high-sensitivity MRD sequencing. In this example, the DNMT3A-mutated clone remains without significant change in VAF, consistent with persistent clonal hematopoiesis. Finally, the patient relapses with the same NPM1-mutated founding clone plus a newly acquired NRAS mutation. (B) A comparison of sequencing methods for MRD monitoring. Although WGS offers the greatest sequencing breadth and is capable of detecting structural variants such as CNAs and chromosomal translocations, standard coverage is generally only ∼60×, limiting detection to mutations with VAFs > 10%. Targeted sequencing is generally limited to 50 to 500 genes, providing minimum sequencing breadth but can achieve high coverages (1000×) at a lower cost than WGS and provides adequate sensitivity (2% VAF) for initial diagnostic evaluation. MRD panels are similar to targeted panels but use much higher sequencing depths and use UMIs to achieve sensitivities of ∼0.1% VAF allowing for MRD monitoring. MRD panels are generally easy to implement but may be of limited clinical utility for patients with few mutations covered by the panel. In patient-specific MRD sequencing, mutations are identified at diagnosis using broad methods such as exome sequencing or WGS, ensuring an adequate number of mutations to track. These mutations are then individually targeted using custom probes at subsequent time points. By focusing sequencing on known mutations, patient-specific MRD can achieve extremely high detection sensitivities for nearly all patients. Patient-specific MRD, however, can be logistically challenging and expensive to implement because it requires custom probe designs and validation for every patient. Patient-specific methods also cannot detect newly acquired mutations that were not targeted by probes at diagnosis. Professional illustration by Patrick Lane, ScEYEnce Studios.
ALL
Genomic studies have led to the identification of new ALL entities28,162,163 of prognostic and therapeutic significance,164,165 even in the context of MRD-based risk-adapted therapy. These optimally require sequencing-based approaches to identify all genomic features of clinical importance. However, the choice of diagnostic approach depends in part on how genomic information will be used to guide management and on the availability of genomic and conventional diagnostic assays in individual laboratories.
Routine diagnostic approaches
Chromosome banding analysis and FISH are widely used for identification of aneuploidy (hyperdiploidy and hypodiploidy) and subtype-defining chromosomal alterations (eg, BCR::ABL1, ETV6::RUNX1, KMT2A::AFF1, TCF3::PBX1, iAMP21, etc), many of which are used for risk assignment and treatment stratification. FISH assays may be used for rapid identification of translocations and gene fusions (Figure 4), including those in BCR::ABL1-like B-ALL for which targeted therapies are currently available (eg, ABL-family kinase genes, JAK2, CRLF2, and NTRK3). However, these assays do not detect all clinically relevant alterations, for example, focal insertions of EPOR into immunoglobulin loci166 and sequence mutations (eg, JAK1/JAK2/JAK3) and deletions (eg, SH2B3) that also drive kinase signaling.167 PCR assays can identify subtypes defined by gene fusions and point mutations (eg, PAX5 p.P80R and IKZF1 p.N159Y). Quantitative RT-PCR may be used to identify the gene expression profile of BCR::ABL1-like ALL,168 but subsequent testing (eg, FISH, targeted or transcriptome sequencing) is required to identify the driver kinase-activating alterations. Quantitative RT-PCR can also be used to identify deregulated gene expression characteristic of recently identified entities (DUX4, EPOR, NUTM1, and CDX2/UBTF),163,169 but alteration-specific confirmatory diagnostic approaches are desirable.
Identification of distinct subtypes of ALL through gene expression profiling. (A) Representative break-apart FISH for KMT2A::AFF1 fusion. The upper panel shows a cell with DNA FISH for KMT2A 5′ and 3′ showing 1 intact allele and 1 disrupted allele. The lower panel shows a second hybridization added on top of the first with AFF1 3′, which confirms disruption of KMT2A and fusion to AFF1 3′. (B) Illustration showing overexpression of CRLF2 and detection by flow cytometry. The image was created in Biorender (https://biorender.com/). (C) Schematic representation of NUTM1 rearrangements with multiple fusion partners and multiple breakpoints detected by WTS and visualized in ProteinPaint (https://proteinpaint.stjude.org/). Ex, exon. The approach in parenthesis (WGS) is alternative to WTS. (D) Integrative Genomics Viewer visualization of BCL11B Enhancer Tandem Amplification (BETA), observed in 20% of BCL11B-activated lineage ambiguous leukemia.162 (E) t-distributed stochastic neighbor embedding (t-SNE) representation from WTS data of B-ALL subtypes highlighted in different colors. Each dot represents a sample (N = 2004). Image is from Kimura et al.191
Identification of distinct subtypes of ALL through gene expression profiling. (A) Representative break-apart FISH for KMT2A::AFF1 fusion. The upper panel shows a cell with DNA FISH for KMT2A 5′ and 3′ showing 1 intact allele and 1 disrupted allele. The lower panel shows a second hybridization added on top of the first with AFF1 3′, which confirms disruption of KMT2A and fusion to AFF1 3′. (B) Illustration showing overexpression of CRLF2 and detection by flow cytometry. The image was created in Biorender (https://biorender.com/). (C) Schematic representation of NUTM1 rearrangements with multiple fusion partners and multiple breakpoints detected by WTS and visualized in ProteinPaint (https://proteinpaint.stjude.org/). Ex, exon. The approach in parenthesis (WGS) is alternative to WTS. (D) Integrative Genomics Viewer visualization of BCL11B Enhancer Tandem Amplification (BETA), observed in 20% of BCL11B-activated lineage ambiguous leukemia.162 (E) t-distributed stochastic neighbor embedding (t-SNE) representation from WTS data of B-ALL subtypes highlighted in different colors. Each dot represents a sample (N = 2004). Image is from Kimura et al.191
Several entities may benefit from flow cytometry analysis of subtype-defining antigen expression patterns, such as CD371 in DUX4-rearranged ALL and surface expression of TSLPR (encoded by CRLF2), a sensitive and specific indicator of CRLF2-rearrangement (Figure 4). A major advantage of flow cytometry–based evaluation is rapid turnaround time within 1 to 2 days.
Capture-based sequencing approaches
The diverse genomic landscape of some subtypes, in particular BCR::ABL1-like B-ALL, can make diagnosis challenging. Capture-based approaches170 or amplicon-based sequencing can detect most common chimeric fusions in B-ALL simultaneously and are well suited to identify the wide spectrum of rearrangements in BCR::ABL1-like B-ALL; however they may fail to detect complex rearrangements, like EPOR in BCR::ABL1-like B-ALL, and some fusions are difficult to capture if breakpoints involve regions in the introns, which may not be feasible to comprehensively tile. DNA-based NGS panels are commonly used to detect common secondary mutations, such as mutations in Jak and Ras pathway signaling genes and mutations associated with relapsed ALL (eg, TP53, CREBBP, and NT5C2).
MLPA assays are widely used to identify focal DNA CNAs in single genes and the “IKZF1plus” composite genotype171 (defined as IKZF1 deletions cooccurring with deletions in CDKN2A/B, PAX5, or in the pseudoautosomal region 1, PAR1, in the absence of ERG deletion), which has been associated with high-risk features. However, MLPA cannot be used to identify all relevant alterations: PAR1 deletions accompany P2RY8::CRLF2 but not IGH::CRLF2 rearrangements, and ERG is deleted in only ∼50% of DUX4-rearranged ALL. Thus, PAR1 and ERG deletions identify only a subset of CRLF2- and DUX4-rearranged ALL, respectively.
Transcriptomic and genomic sequencing
In contrast to targeted approaches, genome-wide sequencing can identify the full spectrum of alterations in a single approach, virtually diagnosing all different entities.
Transcriptome sequencing provides comprehensive characterization of fusion transcript chimeras (Figure 4), mutant allele expression (Figure 4), gene expression profiling, and ploidy169,172 to identify subgroups and several phenocopies (eg, BCR::ABL1-like ALL, ETV6-RUNX1-like, KMT2A-like, and ZNF384/362-like).169,173 The availability of a reference dataset of leukemia transcriptomes, such as the St Jude Cloud (https://www.stjude.cloud),174 allows analysis and classification of individual samples against a reference data set, without the need for an extensive local cohort. Limitations of WTS include poor sensitivity to detect rearrangements that involve complex/repetitive regions (eg, involving antigen receptor loci and DUX4)175,176 or those that do not generate a chimeric transcript, and limited sensitivity for sequence variants that are not expressed or result in nonsense-mediated decay; however, these alterations can be detected by WGS.
T-ALL subgroups are mostly defined by deregulation of T-lineage transcription factors. These are highly diverse in terms of the genes involved and the genomic drivers of deregulation, challenging to identify comprehensively, and are not consistently associated with outcome, thus are not typically identified in current diagnostic workflows.28,162 WGS can detect the diverse genomic alterations that, more commonly in T-ALL than B-ALL, involve intergenic regions (eg, TLX3, T-cell receptor gene loci) that deregulate oncogenes, and noncoding sequence mutations that generate neo-enhancers (eg, TAL1 and LMO1/2).28,162 One entity of clinical relevance is early T-cell precursor ALL (ETP ALL), a high-risk subset of early T-lineage and stem cell leukemias most commonly identified by immunophenotyping (CD7+ and typically cytoplasmic CD3+; CD2+; CD1a−; CD8−; myeloperoxidase negative but positive for at least 1 stem cell/myeloid marker).177 ETP ALL is genetically diverse, but one-third of ETP ALL and T/myeloid mixed phenotype acute leukemia cases have structural variants deregulating BCL11B, which may be detected by WGS (Figure 4) or, for the majority, by FISH to detect disruption of the BCL11B locus.178
Molecular quantitation of MRD
Early MRD monitoring at the end of induction and consolidation phases of therapy has important prognostic and, subsequently, therapeutic implications.179-181 Conventional approaches include MFC and allele-specific PCR for IG/TR gene rearrangements.180,182 To be clinically relevant, MRD analysis needs to be accurate and sensitive (at least ≤10−4). Recently, high-throughput NGS of IG/TR rearrangements,182 which can reach a sensitivity of 10−6, is becoming more widely used.
General conclusions and future directions
Myeloid neoplasms and acute leukemias are characterized by a complex coexistence of multiple clones that evolve over time.183 Historically, these have been studied in bulk samples, precluding a more direct understanding of the clinicopathologic effect of such clonal complexity. Recent studies of clonal architecture at a single-cell level offer unique insights into the interaction of clones, suggesting that the presence of distinct clones may potentially affect the growth and fitness of the others.184-186 At present the use of single-cell sequencing remains confined to a research setting; however, in the future such assays could help predict disease progression and may guide therapeutic strategies to intercept clonal evolution and allow for individual targeting of clones in multiclonal disease. The field will continue to be shaped by advanced methods such as proteomics and cellular indexing of transcriptomes and epitopes sequencing, which can identify potential therapeutic targets and characterize simultaneous gene and protein expression at the single-cell level,187,188 and computation artificial intelligence approaches that can identify new relationships in complex data sets.189,190 In the nearer term, it is expected that the continued decline in sequencing costs will drive further adoption of more frequent panel-based and MRD testing for disease monitoring and broader genomic methods such as WGS for comprehensive genomic evaluation.
It is important to note that these recommendations reflect current practice, based on current treatments and disease classification. As the classification of myeloid neoplasms and their treatments evolve and as genomic testing methods continue to advance, the recommendations for testing will inevitably change and require updating.
Acknowledgments
E.J.D. thanks Jill Guess for managing meetings and emails and David Spencer at Washington University for his discussions of diagnostic genomic methods.
L.M. was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC), Milan, Italy (Investigator Grant #20125; AIRC 5x1000 project #21267); Cancer Research UK, FC AECC and AIRC under the International Accelerator Award Program (project #C355/A26819 and #22796). S.B. was supported by the National Health and Medical Research Council of Australia: APP1117718.
Authorship
Contribution: E.J.D., A.B., R.P.H., C.D.D., L.A.G., I.I., S.J., L.M., A.M.V., K.P.P., D.A.A., M.E.A., R.B., N.B., M.J.B., S.B., A.L.B., C.A.C., H.D., B.F., G.G.-M., T.H., E.H.-L., A.S.K., J.M.K., R.K., M.L.-C.L., S.L., C.G.M., S.O., A.O., E.P., A.R., D.M.R., M.S., A.S., R.C.S., F.S., R.M.S., A.T., M.J.W., D.W., B.L.E., and M.C. were responsible for writing sections of the manuscript and overall review of the content.
Conflict-of-interest disclosure: E.J.D. is a consultant for Cofactor Genomics, Genescopy LLC, and Vertex, and has received honoraria from Blueprint bio and AbbVie. C.D. research support (to institution): AbbVie, Astex, BMS, Beigene, Cleave, Foghorn, ImmuneOnc, Loxo, Servier Consultant with honoraria: Abbvie, BMS, Foghorn, GSK, Jazz, Novartis, Notable Labs, Servier, Takeda. I.I. received honoraria from Amgen and Mission Bio. S.J. received consulting fees from Novartis, Roche Genentech, AVRO Bio, and Foresite Labs; speaking fees from GSK; is an equity holder and on the scientific advisory board of Bitterroot Bio; and is a founder, equity holder, and scientific advisory board member of TenSixteen Bio. A.M.V. received an advisory board and lecture fee from Novartis, AbbVie, Incyte, Blueprint, BMS, and GSK. K.P.P. has served as consultant for Novartis and Astellas and received speaker fees from Astella. M.E.A. received honoraria from Janssen Global Services, Bristol-Myers Squibb, AstraZeneca, Roche, Biocartis, Invivoscribe, Physician Educational Resources, Peerview Institute for Medical Education, Clinical Care Options, and RMEI Medical Education. R.B. is employed by and owns stocks in Aptose Biosciences; ad-hoc advisory board for BMS; DMC chair for Gilead and Epizyme; and received research funding from Takeda. M.B. serves on advisory boards for Amgen and Blueprint Medicines. S.B. is a member of the advisory board of Qiagen, Novartis, and Cepheid; received honoraria from Qiagen, Novartis, Bristol-Myers Squibb, and Cepheid; and received research support from Novartis and Cepheid. C.A.C. serves on the advisory board for Novartis and AOP Orphan and received research funding from BMS. H.D. consults with honoraria for AbbVie, Agios, Amgen, Astellas, AstraZeneca, Berlin-Chemie, BMS, Celgene, GEMoaB, Gilead, Janssen, Jazz, Novartis, Servier, and Syndax and received clinical research funding (to institution) from AbbVie, Agios, Amgen, Astellas, Bristol Myers Squibb, Celgene, Jazz Pharmaceuticals, Kronos Bio, and Novartis. R.K. is a speaker bureau and advisory board member of AbbVie; received a research grant from and serves on advisory board of BMS; is a speaker bureau and advisory board member of CTI biopharma; consults for Geron; is a speaker bureau and advisory board member of Jazz; is an advisory board member of Novartis and Taiho; and is a speaker bureau and advisory board member of PharmaEssentia and Servio. S.L. has received advisory fees from AbbVie, Blue Print Medicine, Daiichi Sankyo, Guidepoint; is a consultant for Gerson Lehrman Group, Qual World, Guidepoint; has received honoraria from Path Education Partners, Pathology Learning Center, Peer View; has stock ownership in AbbVie; has research support from Astellas, Amgen. C.G.M. received research funding from Loxo Oncology, Pfizer, AbbVie; received honoraria from Amgen and Illumina; and holds stock in Amgen. E.P. is a founder, equity holders and hold fiduciary roles in Isabl Inc as well as has scientific advisor shares in ten sixteen bio. D.M.R. received honoraria from Novartis, BMS, and Keros. B.L.E. has received research funding from Celgene, Deerfield, Novartis, and Calico and consulting fees from GRAIL; and is a member of the scientific advisory board and shareholder for Neomorph Therapeutics, TenSixteen Bio, Skyhawk Therapeutics, and Exo Therapeutics. The remaining authors declare no competing financial interests.
Correspondence: Eric J. Duncavage, Department of Pathology and Immunology, 660 Euclid Ave #8118, Washington University in St. Louis, St. Louis, MO 63105; e-mail: duncavagee@wustl.edu.
References
Author notes
This review manuscript contains no original data.
The online version of this article contains a data supplement.

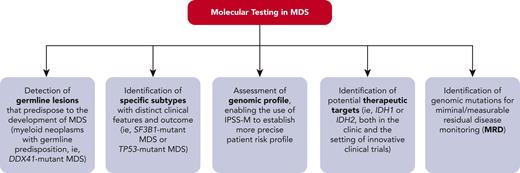
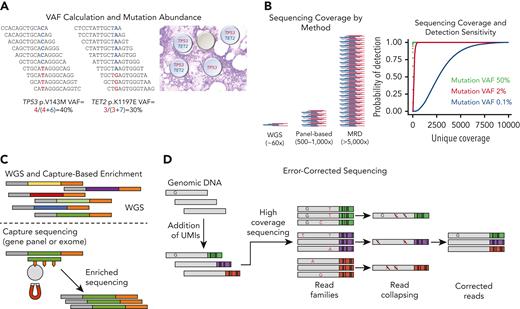
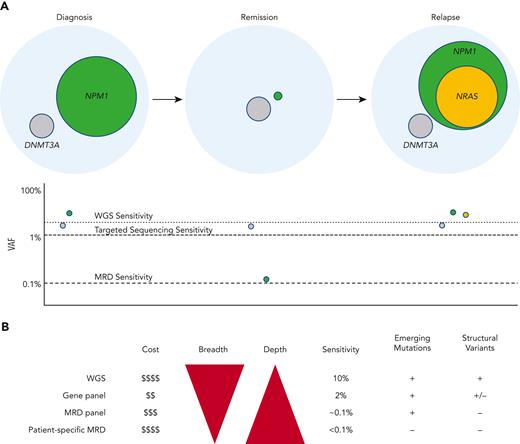
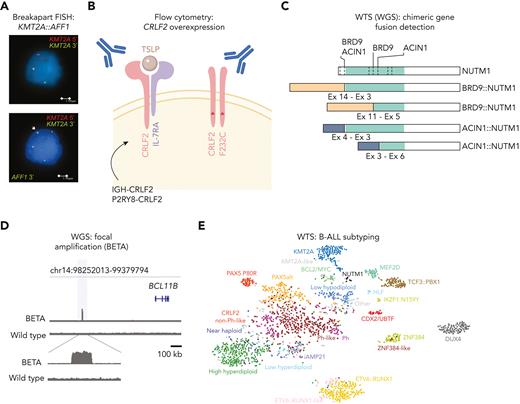
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal