Abstract
The recently developed International Consensus (IC) classification of hematologic neoplasms is primarily based on input from clinical advisory committees composed of pathologists, hematologists, oncologists, and genomic scientists. Morphology continues to represent a fundamental element in the definition of hematologic neoplasms. Acknowledging that the abnormal morphology is a result of dysregulated hematopoiesis driven by somatic gene mutations or altered expression, the IC classification considers genomic features more extensively. Defining nosologic entities based on underlying molecular mechanism(s) of disease is fundamental for enabling the development of precision treatments. Because translational and clinical research continuously advance the field, the classification of hematologic neoplasms will need to be regularly refined and updated; the basic question is what mechanism should be used for this purpose. Scientific hematopathology societies, in collaboration with hematology societies, should be primarily responsible for establishing a standing International Working Group, which would in turn collaborate with the World Health Organization (WHO)/International Agency for Research on Cancer (IARC) to realize and disseminate the classification. The current classification, with its strong morphology component, represents a basis for refinement. Through data sharing, the creation of large comprehensive patient data sets will allow the use of methods of inference, including statistical analyses and machine learning models, aimed at further identifying distinct disease subgroups. A collaborative clinico-pathologic review process will provide a mechanism for updating pathologic and genomic criteria within a clinical context. An interactive Web-based portal would make the classification more immediately available to the scientific community, while providing accessory features that enable the practical application of diagnostic, prognostic, and predictive information.
Introduction
Human disease must be defined and named before it can be studied, diagnosed, and treated.1 The International Statistical Classification of Diseases and Related Health Problems (ICD) of the World Health Organization (WHO) is the most widely used categorization, the most recent version being ICD-11.2 The ICD was originally conceived as a system of diagnostic codes for classifying human diseases, with the main purpose of enabling systematic recording, analysis, interpretation, and comparison of mortality and morbidity data collected in different countries or regions of the world, and it represents “the bedrock for health statistics.”2
The International Agency for Research on Cancer (IARC) was created in France in 1965 as a specialized cancer agency of the WHO. The objective of IARC is to promote international collaboration in cancer research; IARC publishes handbooks, textbooks, and manuals for cancer prevention and treatment.3 Among these publications, the series on the classification of tumors, or the WHO Blue Books, began in 1967.4 These were initially tumor atlases that included figures documenting the main tumor types but with minimal text. They were compiled by a few designated experts and largely lacked references to the published literature.
Historic review of the development of the WHO classification of hematologic neoplasms
After the Revised European-American Lymphoma (REAL) Classification of Lymphoid Neoplasms was published in 19945 and validated in an international study,6 IARC invited representatives of the major hematopathology societies, the Society for Hematopathology (SH) and the European Association for Haematopathology (EAH), to coordinate the preparation of the classification of neoplastic diseases of the hematopoietic and lymphoid tissues. A new vision was adopted that used a multidisciplinary approach to tumor classification which incorporated histology, genetics, clinical features, epidemiology, and etiology. The organizers recognized the importance of involving clinicians in the classification process. The first Clinical Advisory Committee (CAC) meeting was held in 1997, and the resulting classification was published in 1999,7 before the Blue Book-3rd Edition was published.8 The Blue Books became comprehensive monographs that integrated key information about each disease entity and tumor type; this effort was the first true worldwide consensus classification of hematologic neoplasms.
Subsequent revisions were published in 2008 and 2017 after CAC meetings in each case.9,10 The current WHO classification (revised 4th edition) was last updated in 2016 and fully published 1 year later. It was also reported in 2 articles Blood that subsequently became the most viewed, downloaded, and cited articles in this journal.11,12 This clearly illustrates the acceptance and relevance of the 2016 WHO classification of hematologic neoplasms within the scientific community.
Development of an independent classification of hematologic neoplasms
The current WHO classification was developed by a group of pathologists who were advised by clinicians and scientists within the CAC process. Pathologists served as editors of the last WHO Blue Book, whereas clinicians and scientists were co-authors of specific chapters9,10; the role of IARC essentially consisted of editing, publishing, and distributing the handbook in 2017.
In 2020, the IARC adopted a new strategy for preparing the 5th edition of the classification of hematologic neoplasms. The agency created an ad hoc editorial board consisting of standing and expert members that had little continuity with those involved in previous efforts. Importantly, this process no longer involved the hematopathology societies and did not include a CAC process. Because there were concerns regarding the lack of a collaborative process with the new IARC model, an international group of pathologists and clinicians independently proceeded with a CAC effort by organizing the International Consensus Conference on the Classification of Myeloid and Lymphoid Neoplasms, which occurred in Chicago in September 2021.13 This led to the development of the International Consensus (IC) classification of myeloid and lymphoid neoplasms, which reflects an international consensus on disease entities, terminology, and diagnostic criteria as developed through broad representation of individuals involved in the diagnosis and treatment of patients with hematologic neoplasms. The IC classifications of myeloid neoplasms, acute leukemia, and mature lymphoid neoplasms are described in 2 Special Reports in Blood.14,15
This proposed classification represents a major revision of previous classifications and is a significant step forward, further recognizing molecular heterogeneity as an integral component. In this perspective article, we aim to highlight a few distinctive features. Acknowledging the dynamic nature of the classification of hematologic neoplasms, we endeavor to envision how future classifications should evolve.
Increasing impact of genomics on the classification of hematologic neoplasms
Like all tumors, hematologic neoplasms have been primarily classified according to the tissue of origin and histologic features. Morphologic evaluation was the sole modality used for decades and was the framework upon which the classification of myeloid and lymphoid neoplasms was built. Various techniques of immunophenotyping have supplemented conventional morphology over time, and evaluation of immunologic phenotype has become instrumental for defining subtypes of hematologic malignancies.
Morphology will undoubtedly continue to represent a fundamental approach to the diagnosis of hematologic neoplasms and a robust basis for future classification. However, abnormal cellular morphology is a result of dysregulated blood cell production, differentiation, or survival that is driven by gene mutations or altered expression. The so-called driver mutations are most prominent and are typically somatic, although it is also recognized that inherited genetic abnormalities may serve as predisposing or cooperative factors.10 Not surprisingly, genomic profiling has become an increasingly crucial tool in the diagnostic workup of hematologic neoplasms. Cytogenetics, which has long been used to evaluate chromosome number and structure, is an essential component of current clinical guidelines and prognostic systems for hematologic neoplasms.16 The beginning of the genomics era, however, has been marked by the introduction of massively parallel DNA sequencing methods, commonly known as next-generation sequencing.17 Gene panel sequencing is currently the most common method used in clinical settings, but whole-genome sequencing is likely to become a routine diagnostic test in the future. In fact, whole-genome sequencing represents a potential replacement for both conventional cytogenetic and current sequencing approaches because it provides rapid and accurate comprehensive genomic profiling.18
A previous review article in this journal has exhaustively summarized how genomic analysis has influenced the diagnosis and clinical management of patients affected by diverse forms of hematologic neoplasms.19Figure 1 schematically illustrates how genomic profiling can improve the classification of hematologic neoplasms and their clinical management; in the following paragraphs, we will briefly discuss a few examples regarding the IC classification.
The potential impact of genomic profiling on the classification and clinical management of hematologic neoplasms.
The potential impact of genomic profiling on the classification and clinical management of hematologic neoplasms.
First, the detection of a driver mutation allows for establishing the neoplastic nature of disease when morphology is not diagnostic. Clonal cytopenia of undetermined significance (CCUS) is found in patients with unexplained cytopenia and insufficient criteria for a diagnosis of myelodysplastic syndrome (MDS) because of the lack of overt dysplasia or excess blasts.20 CCUS can be diagnosed only through molecular profiling, with the detection of a somatic mutation at a variant allele frequency (VAF) of at least 2% in 1 or more genes that are recurrently mutated in myeloid neoplasms. CCUS is now included in the IC classification of myeloid neoplasms along with other premalignant clonal cytopenias.15 A recent study of the evolution of myeloproliferative neoplasms (MPNs) indicates that somatic driver mutations may occur years before clinical manifestations, which suggests that genomic profiling may allow early diagnosis when morphology is not yet diagnostic.21 Similarly, the IC classification of mature lymphoid neoplasms now recognizes 2 subtypes of immunoglobulin M (IgM) monoclonal gammopathy of undetermined significance (MGUS), plasma cell type or not otherwise specified (NOS), based on genomic findings such as cytogenetic abnormalities typical of multiple myeloma (MM) or MYD88 mutation, which correlate with the risk of progression to either myeloma or lymphoplasmacytic lymphoma (LPL). In addition, primary cold agglutinin disease (which lacks MYD88 mutation but displays recurrent trisomies of chromosomes 3, 12, and 18 and recurrent mutations of KMT2D and CARD11) has been recognized as a new diagnostic category, distinct from LPL or IgM MGUS.14
Second, the use of massively parallel DNA sequencing has allowed the identification of a plethora of conditions with a germ line genetic predisposition for the development of hematologic neoplasms.22 Malignancies that derive from germ line predisposition may have distinct clinical features, including prognosis, as is the case with DDX41-mutated myeloid neoplasms.23 In the IC classification of myeloid neoplasms and acute leukemias, the section on hematologic neoplasms with germ line predisposition has now expanded considerably and includes acute lymphoblastic leukemia (ALL) with germ line predisposition linked to PAX5 or IKZF1 mutation, consistent with the emerging evidence that germ line predisposition to lymphoid neoplasms is more common than was previously thought.15
Third, genomic profiling can allow the categorization of morphologically defined neoplasms into distinct genomic subgroups. The study of the driver landscape of acute myeloid leukemia (AML) has revealed molecular entities with different outcomes, which informs disease classification and prognostic stratification.24 In the new IC classification, new AML subtypes include AML with mutated TP53 and AML with myelodysplasia-related gene mutations. Within MDS, isolated SF3B1 mutation defines a condition with a relatively good prognosis that may respond to luspatercept with the resolution of a transfusion requirement.25 To reflect this, MDS with mutated SF3B1 is now a distinct nosologic entity in the classification of MDS, replacing the previous morphologically defined entity MDS with ring sideroblasts.15 By contrast, MDS with TP53 multi-hit state is associated with a high risk of leukemic transformation and death.26 The IC classification of MDS includes MDS with mutated TP53 whose genomic features are TP53 multi-hit state or TP53 mutation and complex karyotype.15 MM is a genetically heterogeneous disease with 2 main groups defined by cytogenetics: those with recurrent IGH translocations with various partner genes and those that lack IGH translocations, which can be associated with prognosis and response to treatment.27 Therefore, MM has been formally divided within the IC classification of mature lymphoid neoplasms into mutually exclusive diagnostic groups termed MM, NOS and MM with recurrent genetic abnormalities, including MM with CCND family translocations, MM with MAF family translocation, MM with NSD2 translocation, and MM with hyperdiploidy.14 In addition, large B-cell lymphoma with IRF4 rearrangement, which is most common in children, has been upgraded to a definite entity, which can also occur in adults. Large B-cell lymphoma with 11q aberration has been entered as a provisional entity and replaces the previous Burkitt-like lymphoma with 11q aberration based on molecular profiling, which suggests that it is more similar to diffuse large B-cell lymphoma (DLBCL). Finally, the category of high-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangement has been refined, such that dual rearrangement of MYC and BCL6 is now separate and provisional until discrete biological foundation is confirmed, whereas on the basis of genomic profiling studies, high-grade B-cell lymphoma with rearrangement of MYC and BCL2 (with or without BCL6 rearrangement) has been demonstrated to be distinct from germinal center DLBCL, NOS.
Fourth, the detection of somatic mutations may provide biomarkers for disease monitoring. For instance, in patients with MDS after allogeneic hematopoietic stem-cell transplantation, sequencing-based monitoring of measurable residual disease has been found to have prognostic significance.28 Circulating tumor DNA represents the fraction of cell-free DNA released by tumor cells into body fluids such as plasma. In addition to its potential use as a liquid biopsy for genotyping and subtype classification, it allows for the measurement of tumor burden and may provide a reliable tool for dynamic monitoring of response to treatment.29 Circulating tumor DNA has shown promise as an early response predictor for DLBCL, although this requires further validation.30
Fifth, genomic profiling may lay the foundations for personalized treatments. In a study of patients with MPNs, genomic characterization allowed the identification of distinct genomic subgroups, which provides a classification based on driver mutations.31 Subsequently, the combined use of clinical and genomic data enabled the creation of prognostic models capable of generating personally tailored predictions of clinical outcomes.31 Genomic profiling has led to further dissection of DLBCL, with recently proposed discrete recognizable entities associated with variable prognoses.32,33 The algorithm for the diagnostic work-up of large B-cell lymphoma within the IC classification of mature lymphoid neoplasms integrates morphology, immunophenotyping, clinical features, and genomic analyses. Within DLBCL, NOS, the cell-of-origin designation has been retained. Although novel subgroups identified by next-generation sequencing have been acknowledged, further validation will be required before they are incorporated in future iterations.14
Future steps in classification development
Thus far, the mechanism by which the classification of hematologic neoplasms has been developed has relied on interactions within groups of experts in the field that have been intermittently convened. Individual experts have provided their input, and 1 or more group leaders have coordinated the working groups’ efforts. Ad hoc reviews of the literature have been performed, and consensus decision making was used to generate final recommendations.
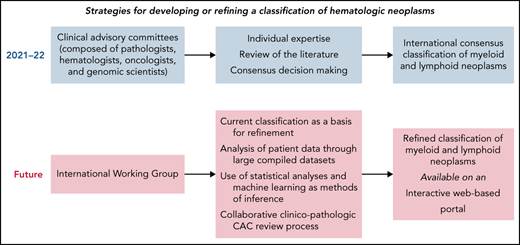
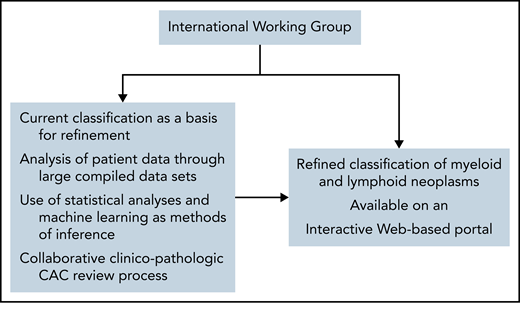
These procedures have historically been appropriate, but moving forward, additional mechanisms will be required to harness the potential of emerging data. Currently, an abundance of detailed clinical and biological data are being collected on individual patients within the context of translational and clinical research and even more frequently within the context of clinical care.34 These include clinical variables stored within electronic health records and the high-throughput data collected through omics analyses. All these data can be useful for defining novel nosologic entities, but interpretation will require collating information on large patient cohorts, which may be aided by using automated procedures. To accomplish this, we propose a systematic shift as illustrated in Figure 2 and discussed below.
A potential strategy for refining the classification of hematologic neoplasms to improve patient care through the implementation of precision treatments. The different processes could be coordinated by an International Working Group (IWG), which would oversee the classification. The current International Consensus classification, with its strong morphology component, will represent a basis for refinement. Access to patient data would be important for allowing the use of various methods of inference aimed at identifying distinct disease subgroups. A collaborative clinico-pathologic CAC review process will provide a mechanism to update pathologic and genomic criteria within clinical context. An interactive Web-based portal, also accessible through a mobile app, would make the classification more immediately available to the scientific community.
A potential strategy for refining the classification of hematologic neoplasms to improve patient care through the implementation of precision treatments. The different processes could be coordinated by an International Working Group (IWG), which would oversee the classification. The current International Consensus classification, with its strong morphology component, will represent a basis for refinement. Access to patient data would be important for allowing the use of various methods of inference aimed at identifying distinct disease subgroups. A collaborative clinico-pathologic CAC review process will provide a mechanism to update pathologic and genomic criteria within clinical context. An interactive Web-based portal, also accessible through a mobile app, would make the classification more immediately available to the scientific community.
Establishment of an International Working Group (IWG) for future revisions to the classification of hematologic neoplasms
Future revisions of the classification of hematologic neoplasms should be increasingly aimed at refining prognostication and informing personalized precision therapy. This will require ongoing close collaboration among pathologists, hematologists, oncologists, genomics scientists, bioinformaticians, and clinical trialists. To promote this collaboration and to ensure it is a continuous process, a formalized IWG should be established.
Scientific hematopathology societies, in collaboration with hematology societies, are well placed to be primarily responsible for establishing the IWG, which would, in turn, collaborate with the WHO and IARC to realize and disseminate the classification. Worldwide representation in this IWG is paramount to ensure that future proposals have applicability in regions where resources may not be equitable. The classification of hematologic neoplasms should be strongly based on data sources generated from laboratories, hospitals, and research institutions and must be interpreted by the pathologists, clinicians, and scientists who contribute to them. The IWG participants should be selected on the basis of merit and should be assigned a limited term; individual terms would be staggered to ensure a strong element of continuity.
Toward an increasingly mechanistic classification of hematologic neoplasms
The IC classification of hematologic neoplasms is based on a combination of clinical, morphologic, and genomic data.14,15 For instance, MDS with del(5q) is defined by the following features: cytopenia, no excess blasts, no Auer rods, and presence of del(5q) alone or with 1 additional abnormality except −7 or del(7q).15 Patients with MDS with del(5q) are likely to respond to lenalidomide with correction of anemia and cytogenetic remission, but those who harbor recurrent variants of TP53 or RUNX1 quickly become resistant to treatment and may progress to AML through a selective pressure mechanism that leads to expansion of lenalidomide-resistant TP53- or RUNX1-mutant cells.20,35,36 Therefore, distinguishing between MDS del(5q) without or with concomitant TP53 or RUNX1 mutations is clinically relevant. One could argue that the absence or presence of these mutations should be considered a prognostic or predictive factor rather than a classifier. However, the boundary between classification and prognosis is becoming more and more subtle with the implementation of precision medicine approaches. As we prepare for routine diagnostic whole-genome sequencing,18 we expect that a future pathology report may contain a conclusion something like this: “MDS del(5q); concomitant somatic mutation in TP53 (VAF 2%).” A note on potential clinical outcomes might also be included.
A mechanistic classification of hematologic neoplasms would be potentially advantageous for therapeutic decision making. In a recent study, the assessment of telomere maintenance mechanisms and RAS or p53 pathway mutations enabled a mechanistic classification of clinical phenotypes in neuroblastoma with the identification of 3 subgroups with distinct clinical outcomes.37 Patients belonging to these subgroups are potential candidates for substantially different treatments, which range from active surveillance with deferred treatment to enrollment in an experimental clinical trial.
Developing a mechanistic classification of hematologic neoplasms requires access to patient data to be analyzed with various methods of inference for identifying distinct disease subgroups. Large comprehensive data sets are needed for reaching firm conclusions on precision treatments and could include both registry information and data generated within prospective clinical trials. Data sharing provides an incredible opportunity for collating original data and strengthening research, and various models and approaches are currently available.38 Of note, Swarm Learning has recently been proposed as a method for a decentralized and confidential analysis of patient data.39
Different methods of inference can be used for developing a genomic classification of hematologic neoplasms, ranging from traditional statistical analyses to machine learning. Papaemmanuil et al24 combined driver mutations in cancer genes with cytogenetic and clinical data and then used Bayesian processes to establish classification rules that partitioned patients into distinct subgroups of AML.
Machine learning uses artificial intelligence (AI) technology. It consists of analyzing data with mathematical models to train and enhance the performance of an intelligent agent, that is, a computer system instructed to operate through AI. Deep learning, an evolution of machine learning, has been shown to exceed human abilities in the classification of images.40 In a recent study published in Blood,41 the use of deep neural networks allowed highly accurate differentiation of bone marrow cell morphologies. To refine the classification of hematologic neoplasms, both supervised and unsupervised machine learning models can be used to analyze patient data.42 Unsupervised learning may enable the identification of novel disease subgroups when analyzing large data sets of patients with multiple variables, including clinical, morphologic, and omics data.
Accessing a mechanistic classification of hematologic neoplasms in the digital era
The WHO Blue Books have been sitting on the desktops of pathologists and clinicians for decades and will always remain a testament to an important effort toward standardizing the categorization and nomenclature of hematologic neoplasms. However, the rapid rate of scientific progress requires a mechanism that can accommodate real-time updates so that major advances can be quickly disseminated. Digital platforms have revolutionized our lives and are increasingly relied on in medicine to access the most up-to-date information. Thus, a universal digital portal (accessed through an internet browser or mobile app) will likely be instrumental for the routine diagnostic workup and management of hematologic neoplasms in the future.
The IWG would optimally create a Web-based portal, which could be readily accessed via computer or mobile device worldwide. This portal would include the most current version of the classification (that is, disease names and their defining diagnostic criteria), along with several accessory features that enable practical application of diagnostic, prognostic, and predictive information, as illustrated in Figure 3. Although the latter features could be updated frequently in real-time, based on periodic review by the IWG, it is anticipated that the actual classification would only be updated every several years through a rigorous CAC process to ensure a systematic process informed by lessons learned from the previous classification.
Hypothetical web-based portal on the classification of hematologic neoplasms.
Web-based computational methods that help physicians implement the classification for diagnostic purposes in clinical practice may be available through the portal. In addition, the portal may include tools for personalized prognostic models, such as the one for AML prepared by the Sanger Institute43 or the recently developed Molecular International Prognostic Scoring System (IPSS-M) for MDS.44 Biomarker-driven, predictive tools may also be incorporated to guide personalized treatment. Finally, an interactive feature allowing ongoing exchange between the IWG and the scientific community will be of fundamental importance to solicit feedback and enable refinement on a continual basis.
Conclusions
The IC classification of myeloid and lymphoid neoplasms represents a significant step forward, further integrating genomic data to designate discrete entities that may be selectively treated. We emphasize the need for novel approaches to further enhance the classification of hematologic neoplasms in the future. Although our vision illustrated in Figure 2 may seem to be excessively ambitious, a few studies on subsets of hematologic neoplasms have already shown that with collective action, this is feasible.24,31,43,44
Acknowledgments
The authors acknowledge their many colleagues in the IC classification of hematologic neoplasms meetings for their constructive comments concerning this article.
Authorship
Contribution: M.C. and L.H.S. conceived this Perspective and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mario Cazzola, Fondazione Istituto di Ricovero e Cura a Carattere Scientifico Policlinico San Matteo, Viale Golgi 19, 27100 Pavia, Italy; e-mail: mario.cazzola@unipv.it; and Laurie H. Sehn, BC Cancer Centre for Lymphoid Cancer, 600 W 10th Ave, Vancouver, BC, Canada V5Z 4E6; e-mail: lsehn@bccancer.bc.ca.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal