Key Points
LEDGF/p75, an important cofactor required for MLL-rearranged leukemia, is not essential for steady-state hematopoiesis.
Loss of LEDGF/p75 blocks the development of MLL-rearranged leukemia supporting the MLL-LEDGF/p75 interaction as a new therapeutic target.
Abstract
Mixed lineage leukemia (MLL) represents a genetically distinct and aggressive subset of human acute leukemia carrying chromosomal translocations of the MLL gene. These translocations result in oncogenic fusions that mediate aberrant recruitment of the transcription machinery to MLL target genes. The N-terminus of MLL and MLL-fusions form a complex with lens epithelium-derived growth factor (LEDGF/p75; encoded by the PSIP1 gene) and MENIN. This complex contributes to the association of MLL and MLL-fusion multiprotein complexes with the chromatin. Several studies have shown that both MENIN and LEDGF/p75 are required for efficient MLL-fusion–mediated transformation and for the expression of downstream MLL-regulated genes such as HOXA9 and MEIS1. In light of developing a therapeutic strategy targeting this complex, understanding the function of LEDGF/p75 in normal hematopoiesis is crucial. We generated a conditional Psip1 knockout mouse model in the hematopoietic compartment and examined the effects of LEDGF/p75 depletion in postnatal hematopoiesis and the initiation of MLL leukemogenesis. Psip1 knockout mice were viable but showed several defects in hematopoiesis, reduced colony-forming activity in vitro, decreased expression of Hox genes in the hematopoietic stem cells, and decreased MLL occupancy at MLL target genes. Finally, in vitro and in vivo experiments showed that LEDGF/p75 is dispensable for steady-state hematopoiesis but essential for the initiation of MLL-mediated leukemia. These data corroborate the MLL-LEDGF/p75 interaction as novel target for the treatment of MLL-rearranged leukemia.
Introduction
The mixed lineage leukemia (MLL) gene is the mammalian homolog to Drosophila trithorax and is a master regulator of the clustered homeotic (HOX) genes.1-5 MLL is a frequent target of chromosomal rearrangements that are found in >70% of infant acute leukemia,6 ∼10% of acute myeloid leukemia (AML) cases in adults7 and secondary or therapy-related leukemias.8 More importantly, MLL-rearranged (MLL-r) leukemia is associated with poor clinical outcome.9 The most common rearrangements are balanced chromosomal translocations that fuse the N-terminus of the MLL gene in frame with the C terminus of >130 different partners, producing oncogenic MLL-fusion proteins (MLL-FPs).10,11
Although the exact molecular mechanism of most fusions remains unknown, MLL-FPs mediate aberrant recruitment of transcription machinery to MLL target genes, driving overexpression of HOX genes (eg, HOXA9) and other coregulators such as MEIS1 and PBX3.12-14 The extreme N-terminus of MLL forms a triple complex with MENIN and lens epithelium-derived growth factor/p75 (LEDGF/p75), a prerequisite for targeting the MLL oncogenic complex to target genes (Figure 1A-B).15-17 The existence of this complex is supported by structural studies in which MENIN acts as a molecular adaptor linking MLL with LEDGF/p75.18-20
Interaction of the MLL-fusion complex with LEDGF/p75. (A) Schematic representation of the MLL-fusion complex. The most common MLL chromosomal translocations result in expression of the N-terminus of MLL and the C-terminus of a fusion partner gene (MLL-fusion). The extreme N-terminus of MLL forms a triple complex with MENIN and LEDGF/p75. The latter recognizes H3K36 di- and trimethylated marks, whereby it tethers the MLL-fusion complex to its targets genes such as HOXA9, MEIS1, and PBX3. (B) Domain structure of wild-type MLL, MLL-fusion, LEDGF/p75, and LEDGF/p52. Wild-type MLL and MLL-fusions contain the MENIN binding domain (MBD) and the LEDGF/p75 binding domain (IBD-interacting motif [IBM]) at its extreme N-terminus. Further downstream, wild-type MLL contains 3 AT-hooks, 2 speckled nuclear localization signals (SNLs), a transcriptional repression domain (TRD), 4 PHD fingers, and a bromodomain. The transactivation domain (TAD) and SET domains are located at the MLL C-terminus. LEDGF/p75 harbors a PWWP (aa 1-93), a nuclear localization signal (NLS), 2 AT hook-like motifs (ATH), 3 charged regions (CRs), and the integrase binding domain (IBD) that binds to MLL. LEDGF/p52 shares similar structural domains but lacks the C-terminal IBD.
Interaction of the MLL-fusion complex with LEDGF/p75. (A) Schematic representation of the MLL-fusion complex. The most common MLL chromosomal translocations result in expression of the N-terminus of MLL and the C-terminus of a fusion partner gene (MLL-fusion). The extreme N-terminus of MLL forms a triple complex with MENIN and LEDGF/p75. The latter recognizes H3K36 di- and trimethylated marks, whereby it tethers the MLL-fusion complex to its targets genes such as HOXA9, MEIS1, and PBX3. (B) Domain structure of wild-type MLL, MLL-fusion, LEDGF/p75, and LEDGF/p52. Wild-type MLL and MLL-fusions contain the MENIN binding domain (MBD) and the LEDGF/p75 binding domain (IBD-interacting motif [IBM]) at its extreme N-terminus. Further downstream, wild-type MLL contains 3 AT-hooks, 2 speckled nuclear localization signals (SNLs), a transcriptional repression domain (TRD), 4 PHD fingers, and a bromodomain. The transactivation domain (TAD) and SET domains are located at the MLL C-terminus. LEDGF/p75 harbors a PWWP (aa 1-93), a nuclear localization signal (NLS), 2 AT hook-like motifs (ATH), 3 charged regions (CRs), and the integrase binding domain (IBD) that binds to MLL. LEDGF/p52 shares similar structural domains but lacks the C-terminal IBD.
LEDGF/p75 was first described as a transcriptional coactivator involved in stress response.21 It is an epigenetic reader of H3K36 di- and trimethylation marks via its Pro-Trp-Trp-Pro (PWWP) domain and as such preferentially associates with actively transcribed chromatin.22-25 The presence of the LEDGF/p75 PWWP domain was shown to be important for the association of the MLL-fusion complex with chromatin.16,26 In addition to its role in MLL-driven leukemia, LEDGF/p75 plays an essential role in the pathogenesis of HIV-1, where it functions as a molecular tether targeting HIV-1 integration into the body of active genes.27-31 This research resulted in the development of a new class of HIV inhibitors, LEDGINs, that block the LEDGF/p75-integrase interaction.32 In analogy to HIV-1 integrase, LEDGF/p75 binds the MLL-MENIN complex via its C-terminal integrase binding domain (IBD; Figure 1B).16,18 We and others demonstrated that Ledgf/p75 knockdown, as well as overexpression of the IBD, dramatically reduces clonogenic growth and decreases the expression of MLL target genes in vitro and in vivo.16,33 Additionally, we have shown that small peptides known to inhibit the LEDGF/p75–HIV-1 integrase interaction impair clonogenic growth of primary murine MLL-r cells.20 These results establish LEDGF/p75 as an important cofactor required for the leukemogenic function of MLL-FPs and suggest the MLL-LEDGF/p75 interface as a potential therapeutic target.
Before developing small molecules targeting this interaction, understanding the role of LEDGF/p75 in hematopoiesis is crucial. LEDGF/p75 and its splice variant LEDGF/p52, which lacks the IBD domain, are encoded by PSIP1 gene (PC4 and SFRS1-interacting protein 1, Figure 1B).34 For reasons as yet unknown, ubiquitous deletion of Psip1 is characterized by high perinatal lethality, limiting further analysis of adult hematopoiesis.35,36 Surviving knockout mice displayed homeotic skeletal transformations, suggesting a critical role in the control of Hox genes expression.
In this study, we established a conditional knockout mouse model to address the role of Psip1 in postnatal hematopoiesis and the initiation of MLL leukemogenesis. Mice with Psip1 deletion are viable but showed defects in hematopoiesis and displayed reduced colony-forming unit (CFU) activity and decreased Hox genes expression in the hematopoietic stem cells (HSCs). Although LEDGF/p75 is dispensable for the hematopoietic reconstitution, it is essential for the initiation of MLL-r leukemia in vitro and in vivo.
Materials and methods
Generation of Psip1 knockout mice
C57BL/6 Psip1-floxed mice (PsipFl/Fl) were engineered to harbor loxP sites flanking exon 3 of the Psip1 gene (supplemental Figure 1A-B, available on the Blood Web site).31 To knock out Psip1 in the hematopoietic system, PsipFl/Fl mice were crossed with Vav-iCre transgenic mice (a kind gift from Jody Haigh, Flemish Interuniversity Institute for Biotechnology, Ghent, Belgium). Polymerase chain reaction (PCR) genotyping confirmed Cre expression in PsipFl/Fl mice (supplemental Figure 1C). All primers are listed in supplemental Table 1. All animal experiments were approved by the Katholieke Universiteit Leuven ethical committee.
Animal experiments
To monitor peripheral blood counts at steady state, blood samples were collected from 8- to 12-week-old mice in EDTA-containing Microtainer tubes (BD Biosciences) and analyzed on Cell Dyn-3700 (Abbott Hematology). To extract lineage-depleted (lin−) progenitors, bone marrow (BM) cells were isolated from femurs and tibias of 8- to 10-week-old mice. After red blood cell lysis, lineage-negative cells were enriched (EasySep Mouse Hematopoietic Progenitor Cell Isolation Kit, STEMCELL Technologies) and cultured in RPMI-1640 (10% fetal calf serum, 50 µg/mL gentamicin), supplemented with murine interleukin-6 (10 ng/mL), murine interleukin-3 (6 ng/mL), and murine stem cell factor (100 ng/mL, PeproTech).
Transduction of primary cells
Lentiviral and murine stem cell virus (MSCV) vector productions were previously described.33 Lin− cells were transduced by spinoculation (90 minutes at 2500 rpm; 8 μg/mL polybrene) on 2 consecutive days. Cells were washed with phosphate-buffered saline (PBS) 24 hours after the second spinoculation and used as indicated for the different assays.
Flow cytometric analyses
To analyze the different hematopoietic compartments, single-cell suspensions were prepared from BM cells. Antibodies used in fluorescence-activated cell sorter (FACS) staining for hematopoietic stem/progenitor cells and B-cell precursors are listed in supplemental Tables 2 and 3, respectively. For flow cytometric analysis of CFU colonies, cells were washed twice with PBS and subsequently stained with phycoerythrin-conjugated anti-Sca-1 and allophycocyanin-conjugated anti-cKit or phycoerythrin-conjugated anit-Gr-1 and allophycocyanin-conjugated anti-Mac1.
Clonogenic growth in vitro
For myeloid colony formation assays, (transformed) lin− cells were plated in methylcellulose (M3534, STEMCELL Technologies). The number of colonies was scored after 6 to 7 days. For replating experiments, colonies were harvested and washed with PBS. Cells were counted in a Neubauer chamber using trypan blue for dead cell exclusion and plated in fresh medium for the subsequent round. For pre-B lymphoid CFU assays, lin− cells or spleen cells were plated in methylcellulose (M3231, STEMCELL Technologies) supplemented with murine interleukin-7 (10 ng/mL). May-Grünwald Giemsa staining was performed as described earlier.33
Further information about the experimental procedures is provided in supplemental Methods.
Results
Psip1 knockout mice have reduced peripheral blood cell counts
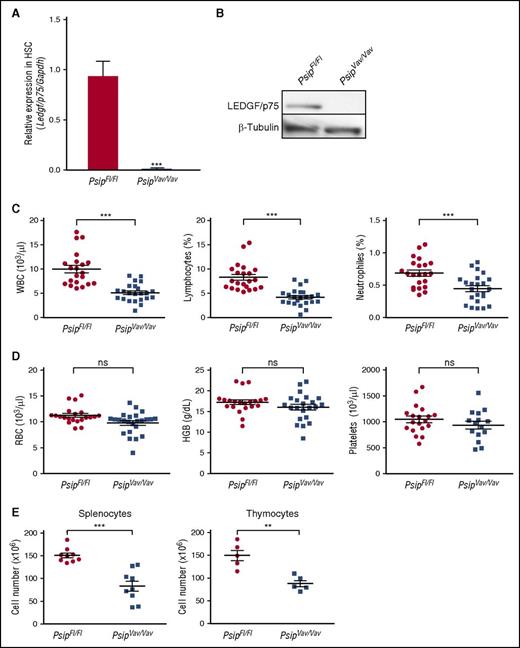
To study the role of LEDGF/p75 in normal hematopoiesis, we established a conditional Psip1 knockout mouse model in the hematopoietic compartment by crossing Psip1-floxed mice31 with Vav-iCre mice, which constitutively express Cre recombinase in HSC at early embryonic stages37 (supplemental Figure 1A-C). Complete depletion of LEDGF/p75 was confirmed by quantitative reverse transcription (qRT)-PCR in lin− cells and by western blot in whole BM cells using a LEDGF/p75-specific antibody (Figure 2A-B). Mice harboring homozygous deletion of Psip1 in the hematopoietic system (referred to as PsipFl/FlVav-Cre-positive mice or PsipVav/Vav) were viable, fertile, phenotypically normal, and born at the expected Mendelian frequencies compared with their PsipFl/FlVav-Cre-negative littermates (PsipFl/Fl) (data not shown).
Knockout of Psip1 reduces peripheral blood cell counts. (A-B) qRT-PCR of lineage-depleted BM cells (A) and western blot of whole BM cells (B) validating complete LEDGF/p75 depletion in Psip1 knockout (PsipVav/Vav) mice compared with the floxed control mice (PsipFl/Fl). Messenger RNA (mRNA) expression in (A) and protein expression in (B) were normalized to Gapdh and β-tubulin, respectively. (C-D) Peripheral blood cell counts measured in PsipFl/Fl and PsipVav/Vav mice (8-12 weeks of age). Mean values and standard error of the mean (SEM) are indicated. HGB, hemoglobin; RBC, red blood cells; ns, nonsignificant. (E) Total cell number extracted from spleens and thymuses of PsipFl/Fl and PsipVav/Vav mice (8-10 weeks of age). Mean values and SEM are indicated. P values (*) show significance (**P < .01, ***P < .001, Student t test).
Knockout of Psip1 reduces peripheral blood cell counts. (A-B) qRT-PCR of lineage-depleted BM cells (A) and western blot of whole BM cells (B) validating complete LEDGF/p75 depletion in Psip1 knockout (PsipVav/Vav) mice compared with the floxed control mice (PsipFl/Fl). Messenger RNA (mRNA) expression in (A) and protein expression in (B) were normalized to Gapdh and β-tubulin, respectively. (C-D) Peripheral blood cell counts measured in PsipFl/Fl and PsipVav/Vav mice (8-12 weeks of age). Mean values and standard error of the mean (SEM) are indicated. HGB, hemoglobin; RBC, red blood cells; ns, nonsignificant. (E) Total cell number extracted from spleens and thymuses of PsipFl/Fl and PsipVav/Vav mice (8-10 weeks of age). Mean values and SEM are indicated. P values (*) show significance (**P < .01, ***P < .001, Student t test).
Next, we determined whether knockout of Psip1 affects steady-state hematopoiesis. Peripheral blood cell counts from a pool of PsipFl/Fl and PsipVav/Vav mice were compared. Peripheral white blood cell (WBC) counts of PsipVav/Vav mice were ∼65% lower than PsipFl/Fl mice (Figure 2C). Significant reductions were also observed in the lymphocytes and neutrophils. In contrast, PsipVav/Vav mice had normal red blood cell, hemoglobin, and platelet counts (Figure 2D).
Furthermore, we checked the cellularity of the spleens and thymuses of PsipFl/Fl and PsipVav/Vav mice. Compared with the controls, the total numbers of splenocytes and thymocytes were significantly reduced by ∼55% and 60% in PsipVav/Vav, respectively (Figure 2E). Collectively, these results reveal that Psip1 inactivation affects the hematopoietic system at steady state.
In vivo ablation of Psip1 perturbs HSC compartments
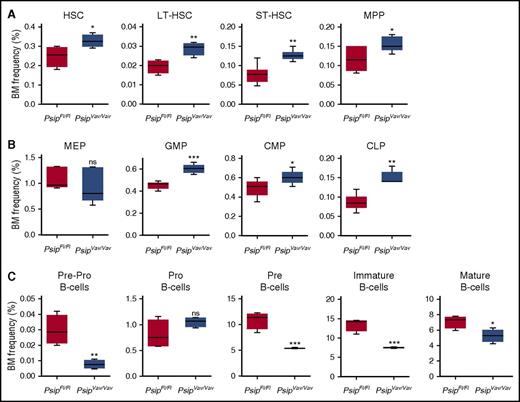
Next, we analyzed the effects of Psip1 ablation on different hematopoietic compartments by flow cytometric analysis of whole BM cells (supplemental Figure 2A-B). We first checked the frequency of HSC (Lineage−Sca-1+ckit+ [LSK]) progenitors in total BM. FACS analysis showed that the percentage of phenotypically defined HSC was significantly increased after Psip1 deletion (Figure 3A). Subsectioning according to CD34 and CD135 expression showed that subpopulations containing primitive HSCs (long-term HSC, short-term HSC and multipotent progenitors) were also increased in PsipVav/Vav cells (Figure 3A). Further analysis of different subsets of the more differentiated progenitors revealed that, although the percentage of the megakaryocyte-erythroid progenitors was comparable, the granulocyte-macrophage progenitors, and common myeloid progenitors were increased in Psip1-depleted cells (Figure 3B). Similarly, the frequency of the common lymphoid progenitors was increased in Psip1-excised cells (Figure 3B).
Psip1 excision perturbs the HSC compartments. (A) The proportion of BM corresponding to the HSC-containing LSK progenitors measured in PsipFl/Fl and PsipVav/Vav mice (4-7 animals per group, 8-10 weeks of age). Subsectioning according to CD34 and CD135 expression yielded phenotypic assessments of LT-HSC, ST-HSC, and MPP fractions. LT-HSC, long-term hematopoietic stem cells; MPP, multipotent progenitors; ST-HSC, short-term hematopoietic stem cells. (B) BM frequencies of the more differentiated progenitors gated in the LSK population, subsectioned based on CD16/32 and CD34 expression to compare common myeloid progenitors (CMPs), granulocyte-macrophage progenitors (GMPs), and megakaryocyte-erythroid progenitors (MEPs). The CLP fraction is gated based on CD127 expression. CLP, common lymphoid progenitors. (C) Percentage of BM cells at different steps in B-cell development. The frequencies of pre-pro-B, pro-B, pre-B, immature, and mature B cells in PsipFl/Fl and PsipVav/Vav mice are shown (4-7 animals per group). Mean values and SEM are shown. P values (*) show significance (*P < .05, **P < .01, ***P < .001, Student t test).
Psip1 excision perturbs the HSC compartments. (A) The proportion of BM corresponding to the HSC-containing LSK progenitors measured in PsipFl/Fl and PsipVav/Vav mice (4-7 animals per group, 8-10 weeks of age). Subsectioning according to CD34 and CD135 expression yielded phenotypic assessments of LT-HSC, ST-HSC, and MPP fractions. LT-HSC, long-term hematopoietic stem cells; MPP, multipotent progenitors; ST-HSC, short-term hematopoietic stem cells. (B) BM frequencies of the more differentiated progenitors gated in the LSK population, subsectioned based on CD16/32 and CD34 expression to compare common myeloid progenitors (CMPs), granulocyte-macrophage progenitors (GMPs), and megakaryocyte-erythroid progenitors (MEPs). The CLP fraction is gated based on CD127 expression. CLP, common lymphoid progenitors. (C) Percentage of BM cells at different steps in B-cell development. The frequencies of pre-pro-B, pro-B, pre-B, immature, and mature B cells in PsipFl/Fl and PsipVav/Vav mice are shown (4-7 animals per group). Mean values and SEM are shown. P values (*) show significance (*P < .05, **P < .01, ***P < .001, Student t test).
Our data indicate that Psip1 knockout results in lower peripheral WBC counts. We evaluated earlier steps in B-cell development. FACS analysis showed a significant decrease in the pre-pro-B-cell and pre-B-cell precursors in the BM of excised mice (Figure 3C). Consistent with the peripheral WBC counts, immature and mature B-cell populations were reduced in PsipVav/Vav mice (Figure 3C). Together, these findings suggest that loss of Psip1 affects the number of HSC and progenitor cells at steady state.
Absence of Psip1 affects colony formation potential of HSC
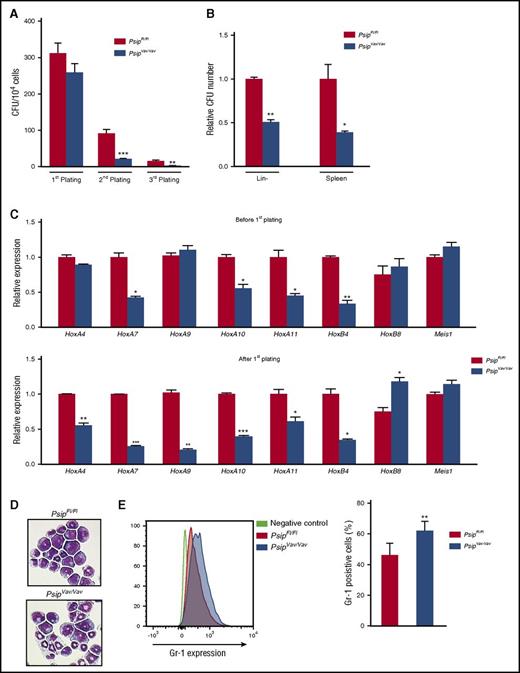
Although loss of Psip1 resulted in higher number of HSC and progenitor cells, it was not clear whether the functionality of the cells was affected. Accordingly, we sought to compare the colony-forming capacity of Psip1 wild-type and knockout cells using myeloid and pre-B lymphoid CFU assays. Lin− cells were harvested from PsipFl/Fl and PsipVav/Vav mice and serially plated in myeloid CFU culture. After 7 days (first plating), the number of colonies derived from PsipFl/Fl and PsipVav/Vav mice was not significantly different (Figure 4A). However, the total cell number harvested from the formed PsipVav/Vav colonies was reduced (8.9 × 106PsipFl/Fl cells vs 4.6 × 106PsipVav/Vav cells). In the second and third platings, the number of colonies of PsipVav/Vav mice was significantly reduced compared with the wild-type control (Figure 4A). Alternatively, we compared the colony-forming capacity of PsipFl/Fl and PsipVav/Vav cells in pre-B lymphoid CFU assays. The number of pre-B colonies in Psip1-depleted cells was reduced in lin− cells and spleen cells, compared with the controls (Figure 4B). To ensure that the decreased colony-forming potential in PsipVav/Vav cells was due to Psip1 deletion and not Cre expression, we compared the CFU potential of lin− cells harvested from C57BL/6 mice and Vav-iCre transgenic mice in the same background. Cre expression on its own did not affect the number of colonies formed upon replating, nor did it alter the total cell number harvested from these colonies (data not shown).
Psip1 knockout affects colony formation potential of HSC in vitro. (A) Number of colonies in 3 consecutive rounds of a myeloid CFU assay per 104 lin− BM cells harvested from PsipFl/Fl and PsipVav/Vav mice (10 weeks of age). (B) Pre-B CFU assays for PsipFl/Fl and PsipVav/Vav cells. Lin− cells and spleen cells were harvested and plated in methylcellulose for 7 to 10 days. The number of colonies formed was normalized to the control (PsipFl/Fl) cells. (C) qRT-PCR measuring expression levels (normalized to Gapdh) of different genes in lin− cells harvest from PsipFl/Fl or PsipVav/Vav cells (before first plating) and after 1 round in myeloid CFU assay (after first plating). (D) Comparison of cell morphology of PsipFl/Fl and PsipVav/Vav cells after 1 round in the CFU assay via May-Grünwald Giemsa staining. A representative picture is shown. (E) FACS analysis for Gr-1 expression in PsipFl/Fl and PsipVav/Vav cells (n = 8) harvested after 1 round in CFU assay. Mean and SEM values are indicated. Error bars (panels A, B, C, and E) represent standard deviation of triplicate measurements. Statistical differences were determined using Student t test; *P < .05, **P < .01, ***P < .001.
Psip1 knockout affects colony formation potential of HSC in vitro. (A) Number of colonies in 3 consecutive rounds of a myeloid CFU assay per 104 lin− BM cells harvested from PsipFl/Fl and PsipVav/Vav mice (10 weeks of age). (B) Pre-B CFU assays for PsipFl/Fl and PsipVav/Vav cells. Lin− cells and spleen cells were harvested and plated in methylcellulose for 7 to 10 days. The number of colonies formed was normalized to the control (PsipFl/Fl) cells. (C) qRT-PCR measuring expression levels (normalized to Gapdh) of different genes in lin− cells harvest from PsipFl/Fl or PsipVav/Vav cells (before first plating) and after 1 round in myeloid CFU assay (after first plating). (D) Comparison of cell morphology of PsipFl/Fl and PsipVav/Vav cells after 1 round in the CFU assay via May-Grünwald Giemsa staining. A representative picture is shown. (E) FACS analysis for Gr-1 expression in PsipFl/Fl and PsipVav/Vav cells (n = 8) harvested after 1 round in CFU assay. Mean and SEM values are indicated. Error bars (panels A, B, C, and E) represent standard deviation of triplicate measurements. Statistical differences were determined using Student t test; *P < .05, **P < .01, ***P < .001.
Previous studies showed that germ line deletion of Psip1 resulted in perinatal mortality and homeotic skeletal transformations similar to Hox-cluster mutant mice.35,36 Additionally, MLL, MENIN, and LEDGF/p75 have been shown to colocalize on HoxA7 and HoxA9 promoters, and knockdown of LEDGF/p75 resulted in decreased HOXA9 expression in MLL-FP transformed cells.16,33 These results suggest that Hox genes might be regulated by Psip1. Accordingly, we measured the expression of several HoxA and HoxB cluster genes as well as other cofactors such as Meis1 in lin− cells harvested directly from PsipFl/Fl and PsipVav/Vav mice and after plating in myeloid CFU assays. HoxA9 and HoxA4 expression were significantly reduced only after first plating (Figure 4C). In contrast, expression of other HoxA genes (HoxA7, HoxA10, and HoxA11) and HoxB4 was readily reduced at steady state. Although the expression of HoxB8 was increased after CFU plating in Psip1-depleted cells, Meis1 expression was comparable between PsipFl/Fl and PsipVav/Vav cells before and after plating. May-Grünwald Giemsa staining on the harvested colonies (after first plating) suggested a more differentiated morphology in Psip1-depleted cells compared with the control (Figure 4D). In agreement, FACS analysis of the harvested colonies showed reduced Sca-1 and cKit expression, suggesting increased differentiation in PsipVav/Vav cells (supplemental Figure 3A-B). Furthermore, PsipVav/Vav cells showed skewed differentiation toward the granulocytic lineage, as indicated by higher Gr-1 expression when compared with PsipFl/Fl cells (Figure 4E). Of note, Mac-1 expression was comparable in PsipFl/Fl and PsipVav/Vav cells (data not shown). Taken together, these results suggest that Psip1 depletion affects the colony formation capacity of HSCs and expression of Hox genes.
Specific loss of LEDGF/p75 and not LEDGF/p52 affects colony formation of HSCs
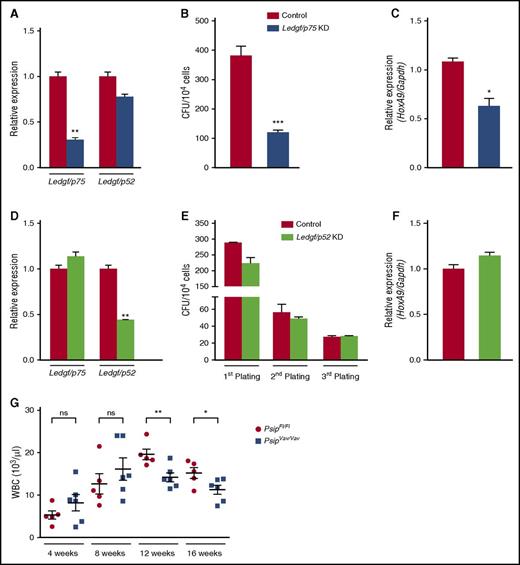
PSIP1 encodes LEDGF/p75 and the shorter LEDGF/p52 isoform. Because p52 lacks the IBD, it is unable to interact with MLL (Figure 1B).34 To analyze the specificity of the observed phenotype, we harvested lin− cells from C57BL/6J mice and transduced them with a lentiviral vector expressing a Ledgf/p75-specific microRNA (miRNA) or eGFP-miRNA as a negative control. We observed ∼40% knockdown for Ledgf/p75, whereas the p52 isoform was not significantly affected as measured by qRT-PCR (Figure 5A). After 7 days, the effects of Ledgf/p75 knockdown were readily observed by the reduced number of colonies (∼70% reduction) compared with the control (Figure 5B). Moreover, knockdown of Ledgf/p75 caused approximately a twofold reduction in HoxA9 expression (Figure 5C). In contrast, depletion of Ledgf/p52 did not affect colony-formation potential of lin− cells or HoxA9 expression, even after 3 rounds in culture (Figure 5D-F). Altogether, our results confirm that specific loss of LEDGF/p75 and not LEDGF/p52 affects the colony formation capacity of HSCs.
Specific knockdown of Ledgf/p75 affects CFU activity of HSC. (A) qRT-PCR measuring Ledgf/p75 and Ledgf/p52 mRNA expression levels in lin− cells harvested from 8-week-old C57BL/6J mice after transduction with lentiviral vector expressing a Ledgf/p75-specific miRNA (Ledgf/p75 KD) or eGFP-miRNA as a control. Expression levels were normalized to Gapdh. (B) CFU assay per 104 cells after Ledgf/p75 knockdown. (C) HoxA9 expression levels (normalized to Gapdh) as measured by qRT-PCR in Ledgf/p75 knockdown cells. (D) qRT-PCR measuring Ledgf/p75 and Ledgf/p52 expression levels and serial plating of a myeloid CFU assay (E) after Ledgf/p52 knockdown (KD). Expression levels in panel D were normalized to Gapdh. (F) qRT-PCR measuring HoxA9 expression levels (normalized to Gapdh) in Ledgf/p52 knockdown cells after first plating in the CFU assay (E). (G) Peripheral WBCs measured after BM transplantation in lethally irradiated recipients transplanted with a total of 1 × 106 lin− cells harvested from 8- to 10-week-old PsipFl/Fl and PsipVav/Vav mice. WBC was monitored 4, 8, 12, and 16 weeks posttransplantation. Mean values and SEM are indicated. Error bars (panels A-F) represent standard deviation of triplicate measurements. Differences in panels A-G were determined using Student t test; *P < .05, **P < .01, ***P < .001.
Specific knockdown of Ledgf/p75 affects CFU activity of HSC. (A) qRT-PCR measuring Ledgf/p75 and Ledgf/p52 mRNA expression levels in lin− cells harvested from 8-week-old C57BL/6J mice after transduction with lentiviral vector expressing a Ledgf/p75-specific miRNA (Ledgf/p75 KD) or eGFP-miRNA as a control. Expression levels were normalized to Gapdh. (B) CFU assay per 104 cells after Ledgf/p75 knockdown. (C) HoxA9 expression levels (normalized to Gapdh) as measured by qRT-PCR in Ledgf/p75 knockdown cells. (D) qRT-PCR measuring Ledgf/p75 and Ledgf/p52 expression levels and serial plating of a myeloid CFU assay (E) after Ledgf/p52 knockdown (KD). Expression levels in panel D were normalized to Gapdh. (F) qRT-PCR measuring HoxA9 expression levels (normalized to Gapdh) in Ledgf/p52 knockdown cells after first plating in the CFU assay (E). (G) Peripheral WBCs measured after BM transplantation in lethally irradiated recipients transplanted with a total of 1 × 106 lin− cells harvested from 8- to 10-week-old PsipFl/Fl and PsipVav/Vav mice. WBC was monitored 4, 8, 12, and 16 weeks posttransplantation. Mean values and SEM are indicated. Error bars (panels A-F) represent standard deviation of triplicate measurements. Differences in panels A-G were determined using Student t test; *P < .05, **P < .01, ***P < .001.
LEDGF/p75 is dispensable for hematopoietic reconstitution
Next, we evaluated the capacity of Psip1-depleted cells to reconstitute the hematopoietic system after BM transplantation (BMT). Lin− cells harvested from PsipFl/Fl and PsipVav/Vav mice were transplanted into lethally irradiated recipients. Subsequently, hematopoietic regeneration was monitored by measuring peripheral blood counts 4, 8, 12, and 16 weeks posttransplantation. Short-term reconstitution of PsipVav/Vav cells at 4 weeks did not differ from PsipFl/Fl cells, suggesting that LEDGF/p75 is not essential for HSC homing, engraftment, or reconstitution. Nonetheless, at 12 weeks posttransplantation, a mild but significant reduction in the number of peripheral WBC was observed (Figure 5G). This reduction persisted at 16 weeks posttransplantation, in agreement with the effects observed for LEDGF/p75 depletion on steady-state hematopoiesis (Figure 2C). Altogether, these data indicate that loss of LEDGF/p75 impairs, but does not abolish hematopoietic reconstitution.
Psip1 gene signature overlaps with that of MLL-FP target genes
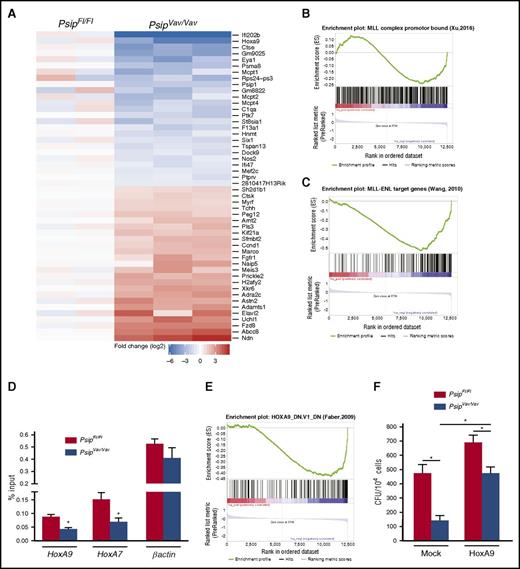
To better understand the observed defects in hematopoiesis, we performed gene expression profiling by RNA-sequencing (RNA-seq) on lin− cells harvested from PsipFl/Fl and PsipVav/Vav mice after 1 round in the myeloid CFU assay. We identified 44 downregulated (comprising several known MLL target genes) and 170 upregulated genes using the criteria of an absolute log2-fold change >1.5 and P value < .05 (Figure 6A; supplemental Table 4). Downregulation of known MLL target genes such as HoxA9 and Eya1 in PsipVav/Vav cells was validated by qRT-PCR (supplemental Figure 4A).38 It has been established that wild-type MLL and MLL-FP have partially overlapping target gene sets,39 suggesting that they can be recruited by different mechanisms. Gene set enrichment analysis (GSEA) revealed that the principal signature in the PsipVav/Vav samples was not significantly up- or downregulated when compared with a set of genes with wild-type MLL bound to the promoter regions (P = .21, supplemental Figure 4B).38 However, those genes with evidence for promoter-bound MLL complexes (overlapping MLL, WDR5, and H3K4me2 chromatin immunoprecipitation [ChIP]-seq peaks) are significantly enriched among the downregulated genes (P = .02, Figure 6B).38 A stronger enrichment was found when MLL-ENL target genes were tested against the PsipVav/Vav ranked list (P < .001, Figure 6C).38 Similar results were obtained with a set of MLL-AF9–bound genes (P < .001, supplemental Figure 4C).40 These data suggest that LEDGF/p75 recruits wild-type MLL to genes that are also targeted by MLL-FP.16,24 This is in line with conservation of the LEDGF/p75 interaction with MLL-FP, whereas other tethering mechanisms are lost or disrupted in these fusions. To verify this, we assessed MLL occupancy at Hox genes in lin− cells harvested from Psip1-depleted cells through ChIP-qPCR. In line with earlier reports,24,41 ChIP-qPCR analyses demonstrated that MLL occupancy at HoxA9 and HoxA7 promoters was decreased about twofold in PsipVav/Vav cells compared with the controls (Figure 6D).
Psip1 knockout gene expression signature overlaps with that of MLL-FP target genes. (A) Heat map of RNA-seq data showing the top differentially expressed genes in PsipFl/Fl and PsipVav/Vav cells harvested after 1 round in the CFU assay. GSEA showing that Psip1-regulated genes are enriched for genes with promoter regions bound by (B) MLL complex and (C) MLL-ENL fusion. (D) Quantitative ChIP assay for PsipFl/Fl and PsipVav/Vav cells using Mll antibody. The promoter regions amplified by qPCR are indicated below the respective panels. (E) GSEA showing the correlation between the principal signature in the PsipVav/Vav RNA-seq samples and genes downregulated in MOLM-14 cells (AML) upon knockdown of Hoxa9. (F) CFU assay per 104 cells harvested from PsipFl/Fl and PsipVav/Vav mice. Cells were transduced with pMSCV-HoxA9-pgk-neo or mock vector and plated for colony formation. Error bars indicate standard deviations of triplicate measurements. Differences were determined using Student t test; *P < .05.
Psip1 knockout gene expression signature overlaps with that of MLL-FP target genes. (A) Heat map of RNA-seq data showing the top differentially expressed genes in PsipFl/Fl and PsipVav/Vav cells harvested after 1 round in the CFU assay. GSEA showing that Psip1-regulated genes are enriched for genes with promoter regions bound by (B) MLL complex and (C) MLL-ENL fusion. (D) Quantitative ChIP assay for PsipFl/Fl and PsipVav/Vav cells using Mll antibody. The promoter regions amplified by qPCR are indicated below the respective panels. (E) GSEA showing the correlation between the principal signature in the PsipVav/Vav RNA-seq samples and genes downregulated in MOLM-14 cells (AML) upon knockdown of Hoxa9. (F) CFU assay per 104 cells harvested from PsipFl/Fl and PsipVav/Vav mice. Cells were transduced with pMSCV-HoxA9-pgk-neo or mock vector and plated for colony formation. Error bars indicate standard deviations of triplicate measurements. Differences were determined using Student t test; *P < .05.
HoxA9 overexpression rescues defective colony formation of PsipVav/Vav cells
One of the top hits revealed by GSEA was that of genes downregulated in MOLM-14 cells (AML) upon HOXA9 knockdown (q value = 0.001, Figure 6E).42 Constitutive HOXA9 expression is one of the main drivers of MLL-r leukemia.14 Moreover, overexpression of HoxA9 was shown to be sufficient to elicit cellular transformation.43 As such, we aimed to determine whether the defects observed in Psip1-depleted cells could be rescued by HoxA9 overexpression. Lin− cells harvested from PsipFl/Fl and PsipVav/Vav mice were transduced with a retroviral vector expressing HoxA9 or a mock control (supplemental Figure 4D) and plated for colony formation. Although Psip1 knockout led to ∼70% reduction in the number of colonies, overexpression of HoxA9 restored CFU activity to wild-type levels (Figure 6F). These data were corroborated in Ledgf/p75 knockdown cells (supplemental Figure 4E). In contrast to PsipVav/Vav cells, acute knockdown of Ledgf/p75 readily caused a significant reduction in HoxA9 expression associated with a fivefold reduction in colony formation (supplemental Figure 4F-G). This defect was rescued to wild-type levels upon HoxA9 overexpression. Collectively, these results suggest that LEDGF/p75 is important for proper HSC differentiation, likely through regulation of Hox genes expression.
Psip1 is essential for inducing MLL-rearranged leukemia
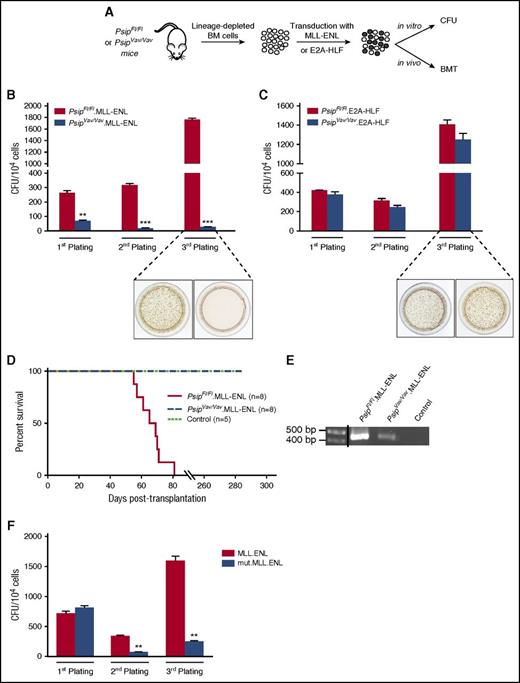
Earlier studies reported that LEDGF/p75 is important to maintain MLL-mediated transformation in human MLL-r cell lines and primary MLL-fusion transformed cells.16,33 The availability of Psip1 knockout cells allowed us to investigate whether LEDGF/p75 is important for the initiation of MLL-r leukemia (Figure 7A). Lin− cells harvested from PsipFl/Fl and PsipVav/Vav mice were transduced with an MLL-ENL fusion (PsipFl/Fl.MLL-ENL and PsipVav/Vav.MLL-ENL, respectively) and their transformation potential was compared in the CFU assay. Although the number of colonies in PsipFl/Fl.MLL-ENL cells increased after the third plating because of MLL-fusion–mediated transformation, Psip1 knockout cells proved refractory to transformation, as evident by the significant reduction in the number of colonies upon serial plating (Figure 7B). Similar data were obtained in MLL-AF9–transformed cells (supplemental Figure 5). In contrast, expression of an oncogenic E2A-HLF fusion (PsipFl/Fl.E2A-HLF and PsipVav/Vav.E2A-HLF), which is independent of HoxA9 for transformation,44 revealed comparable transformation capacity after serial plating in PsipFl/Fl and PsipVav/Vav cells (Figure 7C).
LEDGF/p75 is essential for MLL-rearranged transformation. (A) Schematic representation of the experimental setup. BM cells were harvested from 10-week-old PsipFl/Fl and PsipVav/Vav mice. After depletion of lineage-committed progenitors, cells were transduced with MLL-ENL or E2A-HLF fusions and assessed for leukemogenic activities in vitro in CFU assays and in vivo in BMT. CFU per 104PsipFl/Fl or PsipVav/Vav cells transduced with MSCV encoding MLL-ENL (B) and E2A-HLF (C) fusions. Representative images of tetrazolium-stained colonies after the third plating are shown. Error bars represent standard deviation of triplicate measurements. Differences were determined using Student t test; **P < .01; ***P < .001. (D) Kaplan-Meier survival curve for lethally irradiated recipients transplanted with PsipFl/Fl or PsipVav/Vav cells transduced with MLL-ENL (PsipFl/Fl.MLL-ENL and PsipVav/Vav.MLL-ENL, respectively) or control cells (PsipFl/Fl cells transduced with mock vector). Number of transplanted animals (n) per group is indicated. (E) PCR analysis of whole BM cells of moribund mice transplanted with PsipFl/Fl.MLL-ENL cells and whole BM cells of PsipVav/Vav.MLL-ENL or control animals (described in panel D) harvested at 180 days posttransplantation. Primers were designed to specifically detect the MLL-ENL fusion gene. A vertical line has been inserted to indicate a repositioned gel lane. (F) Replating CFU transformation assay for lin− cells transduced with MLL-ENL fusion or MLL-ENL mutated in the LEDGF/p75-binding domain (mut.MLL-ENL). Error bars represent standard deviation of triplicate measurements. Differences were determined using Student t test; **P < .01.
LEDGF/p75 is essential for MLL-rearranged transformation. (A) Schematic representation of the experimental setup. BM cells were harvested from 10-week-old PsipFl/Fl and PsipVav/Vav mice. After depletion of lineage-committed progenitors, cells were transduced with MLL-ENL or E2A-HLF fusions and assessed for leukemogenic activities in vitro in CFU assays and in vivo in BMT. CFU per 104PsipFl/Fl or PsipVav/Vav cells transduced with MSCV encoding MLL-ENL (B) and E2A-HLF (C) fusions. Representative images of tetrazolium-stained colonies after the third plating are shown. Error bars represent standard deviation of triplicate measurements. Differences were determined using Student t test; **P < .01; ***P < .001. (D) Kaplan-Meier survival curve for lethally irradiated recipients transplanted with PsipFl/Fl or PsipVav/Vav cells transduced with MLL-ENL (PsipFl/Fl.MLL-ENL and PsipVav/Vav.MLL-ENL, respectively) or control cells (PsipFl/Fl cells transduced with mock vector). Number of transplanted animals (n) per group is indicated. (E) PCR analysis of whole BM cells of moribund mice transplanted with PsipFl/Fl.MLL-ENL cells and whole BM cells of PsipVav/Vav.MLL-ENL or control animals (described in panel D) harvested at 180 days posttransplantation. Primers were designed to specifically detect the MLL-ENL fusion gene. A vertical line has been inserted to indicate a repositioned gel lane. (F) Replating CFU transformation assay for lin− cells transduced with MLL-ENL fusion or MLL-ENL mutated in the LEDGF/p75-binding domain (mut.MLL-ENL). Error bars represent standard deviation of triplicate measurements. Differences were determined using Student t test; **P < .01.
These data were corroborated in vivo by BMT. PsipFl/Fl.MLL-ENL and PsipVav/Vav MLL-ENL cells were transplanted into lethally irradiated mice. Whereas recipients transplanted with PsipFl/Fl.MLL-ENL cells succumbed to leukemia within 60 to 85 days after transplantation, animals transplanted with PsipVav/Vav.MLL-ENL or control cells (PsipFl/Fl cells transduced with pMSCV.Neo) did not show any sign of the disease, even at 285 days posttransplantation (Figure 7D). PCR analysis for MLL-ENL fusion in whole BM cells harvested from PsipFl/Fl.MLL-ENL moribund mice or PsipVav/Vav.MLL-ENL and control animals 180 days after BMT confirmed expression of the MLL-ENL fusion in PsipVav/Vav cells despite absence of disease progression (Figure 7E).
In accordance with previous studies,16 our data show that LEDGF/p75 can recruit both wild-type MLL and MLL-FPs to genes driving MLL-r leukemia. In contrast, it has been recently suggested that LEDGF/p75 is required to tether the wild-type complex, but not MLL-FP at target gene loci.24 To further investigate this, we introduced 2 mutations (F148A and F151A) in the consensus IBD-interacting motif45 of the MLL-ENL fusion (mut.MLL-ENL) that specifically abrogate the interaction with LEDGF/p75 and evaluated its oncogenic activity in vitro. In contrast to the wild-type MLL-ENL fusion, the mut.MLL-ENL was unable to transform myeloid progenitors (Figure 7F). Collectively, these data support a role for LEDGF/p75 in targeting both wild-type MLL and its oncogenic counterpart onto the chromatin and that the function of LEDGF/p75 in the MLL-FP complex is important to establish cellular transformation.
Discussion
Oncogenic fusion proteins resulting from chromosomal translocations of the MLL gene induce aggressive acute leukemias in children and adults, emphasizing the need for new therapeutic strategies. It is clear that epigenetic states play an important role in the proliferative capacity of transformed cells. Targeting epigenetic readers and writers has become an attractive anticancer strategy as evidenced by the identification and development of BET protein inhibitors.46 However, careful understanding of the proteins involved is necessary to avoid toxicity and to assess therapeutic windows. MLL forms a ternary complex with MENIN and the epigenetic reader LEDGF/p75. Several studies show that the latter tethers wild-type MLL and/or MLL-FP complexes to chromatin (Figure 1A).16,24,41 Several approaches have been taken to interfere with the function of MLL-fusion complexes, including DOT1L or BET protein inhibitors that target the fusion moiety of MLL-FPs.47-50 On the other hand, others directly target the MLL fragment of the fusion. In this regard, the development of MLL-MENIN interaction inhibitors proved the feasibility of targeting the MLL-MENIN-LEDGF/p75 complex.51,52 Furthermore, detailed structural characterization of the MLL-MENIN-LEDGF/p75 complex and strategies exploiting inhibitory peptides also support the MLL-LEDGF/p75 interaction as a potential new therapeutic target.19,20 However, because the role of LEDGF/p75 (more specifically, the MLL-LEDGF/p75 interaction) in hematopoiesis has not been studied, the specificity of potential MLL-LEDGF/p75 interaction inhibitors has not been addressed. This knowledge is required to fully appreciate the “druggability” of this interaction.
Because mice with systemic Psip1 deletion were shown to suffer from perinatal lethality,35,36 we investigated adult hematopoiesis by knocking out Psip1 in the hematopoietic system. These mice did not exhibit any evident phenotypic abnormalities, suggesting that Psip1 is not essential for HSC maintenance or survival in adult mice. Nonetheless, Psip1 knockout mice displayed specific hematopoietic defects including an approximate twofold decrease in peripheral WBC counts, perturbed numbers of HSCs and progenitor cells, and impaired colony-forming capacity upon serial plating.
Earlier gene targeting studies of Mll underscored its importance for HSC development during embryogenesis and adult hematopoiesis.53-55 Mll deletion resulted in hematopoietic defects and reconstitution failure, suggesting that LEDGF/p75 is not exclusively required for all physiological functions of MLL. Other mechanisms might exist that tether the MLL complex to its target genes, as has been suggested previously.56,57 We observed that expression changes in PsipVav/Vav samples are more pronounced at MLL-FP–bound genes than at MLL wild-type–bound genes. This result confirms that those genes targeted by the MLL-MENIN-LEDGF/p75 complex are also crucial for the development of MLL-r leukemia. In contrast to the p52 isoform, LEDGF/p75 contains the IBD domain, which specifically interacts with MLL and several other cellular proteins.45 However, among these interaction partners, only MLL is known to affect HOX expression. Although we did not observe decreased expression of HoxA9 in the lin− fraction in contrast to other members of the HoxA cluster, our RNA-seq data revealed downregulation of HoxA9 targets in Psip1-depleted cells after 1 week in culture and that the observed phenotypes could be rescued upon HoxA9 overexpression suggests that the HoxA9 axis is an important component of the observed differentiation defects and decreased CFU formation. Targeted disruption of HoxA9 was shown to display similar hematopoietic defects to Psip1 knockout mice such as lower peripheral lymphocytes, reduced spleen and thymus cellularity, defects in B-cell production, and impaired CFU capacity.58,59
Despite its dispensability for normal hematopoiesis, our data show that LEDGF/p75 is essential for the initiation of MLL-r leukemia. Although MLL-r leukemia is mainly induced by gain of function of the MLL-FPs, the remaining wild-type allele is typically expressed. Some studies suggested a functional role of the latter in cellular transformation.60,61 On the other hand, MLL-AF6 human leukemic cells (ML2 cells) do not express the wild-type MLL protein.38 Although all MLL-FPs retain the wild-type N-terminus, genome-wide analysis has shown that wild-type MLL and MLL-FPs have distinct chromatin-binding profiles.39 Whereas MLL-FPs exert their oncogenic activity through activation of a particular gene expression program, integrative analyses of MLL binding and expression profiling showed that MLL-FPs share only a small subset of target genes with the wild-type protein,38 suggesting different chromatin-tethering mechanisms. Earlier work claimed that LEDGF/p75 specifically associates with wild-type MLL and MLL-FP and colocalizes on target genes.16 In contrast, recent studies from the same group and others showed that knockdown of LEDGF/p75 led to increased MLL-FP recruitment.24,41 The authors suggested an independent role for LEDGF/p75 in the wild-type complex, which in turn competes with MLL-FP for chromatin-binding sites. Similar to Zhu et al, we observed a significant reduction of wild-type MLL binding to HoxA9 and HoxA7 promoter regions upon LEDGF/p75 depletion. Although our Psip1 knockout mice reveal that LEDGF/p75 is essential for MLL-r leukemia and that the absence of LEDGF/p75 affects Hox expression and hematopoiesis likely due to diminished MLL recruitment, it was unclear whether the leukemogenic defect is due to diminished tethering of wild-type MLL and/or MLL-FP to target genes. Based on recent structural data, we mutated the IBD-interacting motif in MLL-ENL, which is responsible for the interaction with LEDGF/p75.45 CFU assays revealed impaired transformation capacity in lin− cells. These data corroborate the function of LEDGF/p75 in the tethering the MLL-FP, as has been suggested earlier.16
In summary, we demonstrated that LEDGF/p75 is not essential for adult hematopoiesis, whereas MLL-r leukemia cannot be initiated in its absence. These data open new perspectives to develop small molecule inhibitors of the MLL-LEDGF/p75 interaction, validating a novel target with a sufficient therapeutic window to treat MLL leukemia and possibly other MLL-dependent diseases.62
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Martin Michiels and Sara T’Sas for the technical assistance and Thomas Milne for the technical advice.
This work was supported by grants from the Katholieke Universiteit Leuven Interdisciplinair onderzoeksprogramma (IDO) program (IDO/12/008-3E130241), the Flemish Research Foundation - Flanders (G.0595.13 and G065614N) (Z.D.), the Odysseus program (P.V.V.), the Flemish agency for Innovation by Science and Technology (SBO 140038 [ChromaTarget]), and the US National Institutes of Health, National Institute of Allergy and Infectious Diseases (AI039394) (A.E.). S.E.A. is a postdoctoral fellow supported by the IDO program. J.S. is supported by the Swiss National Science Foundation (31003A_149714), the Swiss Cancer League (KFS-3487-08-2014), and the Gertrude Von Meissner Foundation (Basel, Switzerland). T.P. and S.G. are postdoctoral fellows funded by the Belgian “Stand Up To Cancer” Foundation.
Authorship
Contribution: S.E.A. and J.D.R. designed the experiments and wrote the manuscript. S.E.A., J.D.R., T.P., S.G., S.J., and S.V.B. performed experiments and analyzed data. A.E. generated PsipFl/Fl mice. J.D. performed bioinformatics analyses. N.B. performed and analyzed May-Grünwald Giemsa stainings. J.S., F.C., P.V.V., Z.D., and J.D.R. supervised the project. All authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing interests.
The current affiliation for J.D. is The Francis Crick Institute, London, United Kingdom.
Correspondence: Jan De Rijck, Laboratory for Molecular Virology and Gene Therapy, Kapucijnenvoer 33, 3000 Leuven, Belgium; e-mail: jan.derijck@kuleuven.be; and Zeger Debyser, Laboratory for Molecular Virology and Gene Therapy, Kapucijnenvoer 33, 3000 Leuven, Belgium; e-mail: zeger.debyser@kuleuven.be.
References
Author notes
Z.D. and J.D.R. shared senior authorship.

![Figure 1. Interaction of the MLL-fusion complex with LEDGF/p75. (A) Schematic representation of the MLL-fusion complex. The most common MLL chromosomal translocations result in expression of the N-terminus of MLL and the C-terminus of a fusion partner gene (MLL-fusion). The extreme N-terminus of MLL forms a triple complex with MENIN and LEDGF/p75. The latter recognizes H3K36 di- and trimethylated marks, whereby it tethers the MLL-fusion complex to its targets genes such as HOXA9, MEIS1, and PBX3. (B) Domain structure of wild-type MLL, MLL-fusion, LEDGF/p75, and LEDGF/p52. Wild-type MLL and MLL-fusions contain the MENIN binding domain (MBD) and the LEDGF/p75 binding domain (IBD-interacting motif [IBM]) at its extreme N-terminus. Further downstream, wild-type MLL contains 3 AT-hooks, 2 speckled nuclear localization signals (SNLs), a transcriptional repression domain (TRD), 4 PHD fingers, and a bromodomain. The transactivation domain (TAD) and SET domains are located at the MLL C-terminus. LEDGF/p75 harbors a PWWP (aa 1-93), a nuclear localization signal (NLS), 2 AT hook-like motifs (ATH), 3 charged regions (CRs), and the integrase binding domain (IBD) that binds to MLL. LEDGF/p52 shares similar structural domains but lacks the C-terminal IBD.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/1/10.1182_blood-2017-05-786962/4/m_blood786962f1.jpeg?Expires=1769088431&Signature=XMItKjo1QlaNVf1pDzFxmSgp8n87nrNJ7HRdYYAuNtUEGpxVkNzMgDxVg-~rdf027SHJs5y8olnviDYPibwrv2-DhWrbCW8RZb8QqTS7~3KH6XLvl1xcDws8fpju6PV0shH03HlHPjgfxHvOPe5GZUQpTR4odObDQpDJnw8QzbsVS-VkLQHN0~yD7uZC2lRk0k~s6jn1zAeWuwZWAXiGqWgleyEYZTynqswj0nSqyo5hqNxc7FBgK1wDutbCpYnQGj1VGyE7K-0Vza1f7Rb69KiUWFaiIHhivmtd1vIoThajMDUiaGORjFmsnwAd4PGJXFS-2WvTiC8yncCGWI-ZNw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal