Key Points
A gene expression profile consistent with activated JAK2 signaling is seen in all MPN patients, including in patients with CALR mutations.
Transcriptional profiling discriminates subsets of MPNs based on JAK2V617F allele burden and on the presence of CALR and TET2 mutations.
Abstract
Genomic studies have identified somatic alterations in the majority of myeloproliferative neoplasms (MPN) patients, including JAK2 mutations in the majority of MPN patients and CALR mutations in JAK2-negative MPN patients. However, the role of JAK-STAT pathway activation in different MPNs, and in patients without JAK2 mutations, has not been definitively delineated. We used expression profiling, single nucleotide polymorphism arrays, and mutational profiling to investigate a well-characterized cohort of MPN patients. MPN patients with homozygous JAK2V617F mutations were characterized by a distinctive transcriptional profile. Notably, a transcriptional signature consistent with activated JAK2 signaling is seen in all MPN patients regardless of clinical phenotype or mutational status. In addition, the activated JAK2 signature was present in patients with somatic CALR mutations. Conversely, we identified a gene expression signature of CALR mutations; this signature was significantly enriched in JAK2-mutant MPN patients consistent with a shared mechanism of transformation by JAK2 and CALR mutations. We also identified a transcriptional signature of TET2 mutations in MPN patent samples. Our data indicate that MPN patients, regardless of diagnosis or JAK2 mutational status, are characterized by a distinct gene expression signature with upregulation of JAK-STAT target genes, demonstrating the central importance of the JAK-STAT pathway in MPN pathogenesis.
Introduction
The Philadelphia-negative myeloproliferative neoplasms (MPN) are hematopoietic disorders characterized by clonal expansion of mature myeloid elements. These include polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF). Genomic analysis of the MPNs has shown frequent mutational events in the JAK-STAT signaling pathway including JAK2V617F mutations in 90% to 95% of patients with PV, and in 50% to 60% of patients with ET and PMF.1-5 In addition, somatic mutations in the thrombopoietin receptor MPL in a subset of patients with JAK2V617F-negative ET and PMF, JAK2 exon 12 mutations6,7 in JAK2V617F-negative PV,8,9 and LNK mutations in JAK2V617F-negative MPN10-12 have been identified. These genetic data indicate that mutations that lead to constitutive JAK-STAT signaling are a common genetic event in the different MPNs.
The identification of mutations in the JAK-STAT pathway in the majority of MPN patients led to the development of JAK inhibitors, and approval by the US Food and Drug Administration of the JAK1/2 inhibitor ruxolitinib for the treatment of myelofibrosis (MF).13,14 Treatment with ruxolitinib and other JAK kinase inhibitors leads to substantive clinical benefit including marked reduction in splenomegaly and disease-associated symptoms. Notably, however, clinical responses are observed in JAK2V617F-positive and JAK2V617F-negative MF patients.13 This observation strongly implicates involvement of the JAK-STAT pathway in JAK2V617F-negative MF. However, the role and extent of JAK-STAT pathway activation has not been formally delineated in patients with varying JAK2V617F allele burdens and in MPN patients without known mutations in the JAK-STAT pathway.
More recently, 2 studies used whole exome sequencing to identify mutations in calreticulin (CALR) in the majority of JAK2V617F-negative MPN patients.15,16 These mutations were found to be exclusive of JAK2 and MPL mutations, suggesting that these mutations lead to activation of JAK-STAT signaling or of a critical pathway downstream of JAK-STAT signaling through a novel, alternate mechanism. Furthermore, whether CALR mutations lead to generalized JAK-STAT pathway activation similar to that seen with JAK2 kinase mutations or whether CALR mutations activate a discrete downstream signaling effector has not yet been delineated.
Although the mutational events involved in MPN pathogenesis have now been extensively delineated, the impact of different somatic alterations on transcriptional output has not been extensively evaluated. To answer these questions, we performed gene expression profiling in granulocytes from normal individuals and a cohort of patients with chronic MPNs and integrated these data with detailed molecular characterization to understand how transcriptional output in MPN cells relates to the clinical phenotype and molecular genotype of MPN patients. Our data indicate that transcriptional activation of the JAK-STAT pathway is a common molecular feature of the different MPNs regardless of JAK2 mutational status and that CALR-mutant MPN patients are characterized by expression of a JAK-STAT pathway signature. Finally, we present data showing that CALR and TET2 mutations in MPN patients have specific effects on transcriptional output that may potentially contribute to the phenotypic variability observed in MPN patients.
Methods
Patients
DNA and RNA were isolated from peripheral blood granulocytes from 97 MPN patients (as part of the 345 unique patient samples included in the Harvard Myeloproliferative Disorders Study5 ) as well as from 11 age-matched normal subjects. Approval was obtained from the institutional review boards at the Dana-Farber Cancer Institute and at Memorial Sloan-Kettering Cancer Center for these studies, and informed consent was provided according to the Declaration of Helsinki.
Human androgen-receptor gene clonality assay and X-inactivation ratio determination
Polymerase chain reaction (PCR) amplification of the polymorphic CAG repeat at the human androgen-receptor gene locus was used to determine the degree of skewing (DS) as previously described.17 Allelic skewing consistent with clonal granulopoiesis was defined as a 3:1 ratio between X-linked alleles, which is equivalent to a DS of at least 0.25.17
Mutational analysis and single nucleotide polymorphism (SNP) arrays
The JAK2V617F allele burden was determined in granulocyte DNA using a quantitative real-time PCR assay as described previously.17 Mutational analysis of ASXL1, CALR, JAK2, IDH1/2, and TET2 were performed by DNA resequencing of all coding exons of ASXL1 and TET2 as well as regions of known mutation in CALR, JAK2, IDH1, and IDH2 as previously described.16-18 In addition, genotyping of an additional 953 mutations representing 111 genes was performed on amplified DNA with the use of iPlex extension-chemistry methods (Sequenom) and mass spectrometry, as previously described for the complete set of OncoMap assays.19 All somatic mutations were validated by resequencing nonamplified DNA. In addition to mutational analyses, a total of 207 MPN tumor samples were analyzed using Affymetrix 250K StyI Arrays.20
RNAi-mediated knockdown of JAK2
The JAK2V617F-mutant acute myeloid leukemia cell line HEL was treated with 2 independent shRNA lentiviruses for JAK2 in a pLKO-puromycin selectable vector as described previously21 as well as 2 nontargeting shRNA controls (sh-Luciferase and green fluorescent protein vector control). Following selection of stable cell lines, RNA was purified from HEL cells using Trizol (Invitrogen). Processing of RNA and hybridization to oligonucleotide microarrays was then performed as described in the following text.
Oligonucleotide microarrays
RNA was purified from granulocytes using Trizol (Invitrogen). Linear amplification of 20 ng of total RNA was performed using the Ovation Biotin RNA Amplification and Labeling System (Nugen). Fragmented, labeled cDNA was hybridized to Affymetrix HG_U133AAofAv2 microarrays as described previously.22 Raw expression values were normalized using Robust Multiarray Averaging.23 All microarray data used in this manuscript are deposited in Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with this reference series number: GSE54646.
Marker gene selection
Raw gene expression values were preprocessed and normalized using Robust Multiarray Averaging.23 Genes with minimal variation across the dataset were excluded by discarding genes for which the maximum gene expression value, divided by the minimum value across all samples, was less than 2, or if the difference between maximum and minimum values was less than 100. Calculating a Student t test identified the marker genes. Statistical significance was determined by random permutation of the class labels.24 Significant markers were selected using a false-discovery rate (FDR) threshold of 0.05 (or 0.01 in some instances) computed using the Benjamini and Hochberg procedure.25 Analyses were performed using the GenePattern software package26 using the Comparative Marker Selection module.27
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed as described previously.28 The dataset was converted from probe sets to gene symbols, and the gene expression signatures were analyzed using the java GSEA package. The most differentially expressed genes for each comparison were used to generate a signature for GSEA analysis. The input motif gene sets were extracted from the Molecular Signature Database, version 4 (MSigDBv2).28 For STAT5A/B target gene analysis, STAT5A signature was obtained from previously published data,29 and STAT5B signature was obtained from MsigDB.
Results
Gene expression profiling distinguishes MPN patients from normal controls
Previous studies have characterized gene expression in CD34+ cells and granulocytes from MPN patients. However, these studies have not identified consistent distinct gene expression patterns that are characteristic of MPNs as a whole, or that are characteristic of specific MPN disease alleles.30-33 For example, previous studies have not shown success in determining whether genetically defined subtypes of MPN patients (such as JAK2V617F-mutant vs JAK2-wild-type [WT] MPN) have distinct gene expression signatures.34-37 CD34+ cells and neutrophils isolated from MPN patients are an admixture of clonal MPN cells and normal hematopoietic cells. We hypothesized that the limited insights from previous gene expression studies in MPN patient samples were a result of contaminating normal cells, which limits the ability to identify transcriptional features specific to the MPN clone. To define a set of MPN samples that included a predominance of clonal cells, we performed quantitative genotyping for the JAK2V617F mutation in all patients and clonality assays in informative female patients through analysis of X-inactivation. We defined clonal predominance as the presence of >50% imputed MPN cells, as defined by JAK2V617F allele burden >51%, X-inactivation DS >0.25, or the presence of another mutant disease allele by Sanger sequencing or with a detectable copy number alteration on Affymetrix SNP array analysis. We defined patients as having homozygous JAK2V617F mutations when the quantitative allele burden for JAK2V617F was greater than 60%, and we defined patients as having heterozygous mutations when the JAK2V617F allele burden was less than 60% with X-allele skewing greater than 0.25. With this approach, samples in which 50% or more of the sample was derived from admixed cells without a clonal marker were excluded from gene expression analysis.
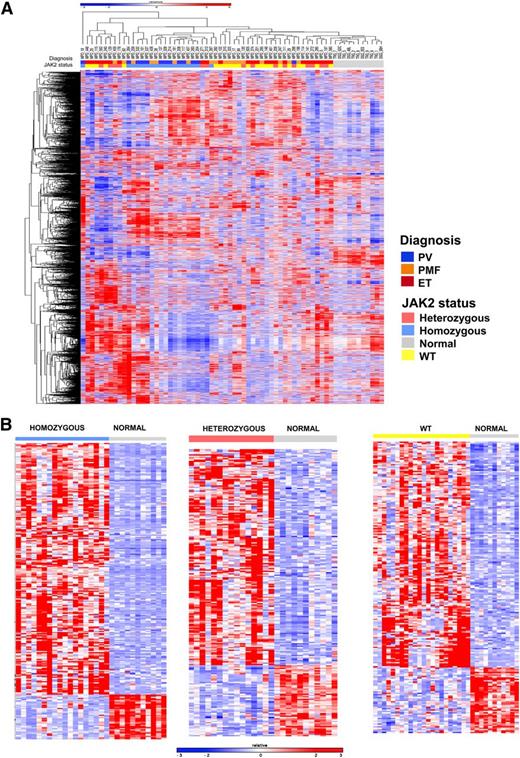
We then performed microarray gene expression analysis in 93 patients with MPNs (28 PV, 47 ET, 18 MF). Similar analysis was performed in parallel on granulocytes from 11 age-matched normal donors. In all, the levels of expression of 14 500 genes were assessed in each sample. As shown in Figure 1A, hierarchical clustering showed that MPN patients had a distinct gene expression profile compared with normal subjects. These data demonstrate that analysis of MPN patient samples without a large proportion of admixed normal cells can be used to identify MPN-specific gene expression signatures.
Gene expression profiling distinguishes patients with MPN from normal controls and homozygous JAK2V617F-mutant MPN patients from others. (A) Hierarchical clustering was performed on gene expression microarray data from the granulocytes of 55 MPN patients and 11 normal subjects. MPN patients were characterized by a distinct gene expression profile compared with normal subjects. Correlation of patient samples with clinical MPN subtype and JAK2 genotype showed that MPN patients with homozygous JAK2V617F mutations were characterized by a unique cluster of differentially expressed genes among MPN patients. (B) Heatmap representation of significant differentially expressed genes between normal subjects and MPN patients with homozygous JAK2V617F (265 genes FC >3 and FDR <0.01), heterozygous JAK2V617F (222 genes FC >2 and FDR <0.05), and JAK2 WT genotypes (209 genes FC >2 and FDR <0.05). A red-blue color scale depicts normalized gene expression levels (red: high; blue: low).
Gene expression profiling distinguishes patients with MPN from normal controls and homozygous JAK2V617F-mutant MPN patients from others. (A) Hierarchical clustering was performed on gene expression microarray data from the granulocytes of 55 MPN patients and 11 normal subjects. MPN patients were characterized by a distinct gene expression profile compared with normal subjects. Correlation of patient samples with clinical MPN subtype and JAK2 genotype showed that MPN patients with homozygous JAK2V617F mutations were characterized by a unique cluster of differentially expressed genes among MPN patients. (B) Heatmap representation of significant differentially expressed genes between normal subjects and MPN patients with homozygous JAK2V617F (265 genes FC >3 and FDR <0.01), heterozygous JAK2V617F (222 genes FC >2 and FDR <0.05), and JAK2 WT genotypes (209 genes FC >2 and FDR <0.05). A red-blue color scale depicts normalized gene expression levels (red: high; blue: low).
We also asked whether gene expression profiling could distinguish MPN patients based on clinical diagnosis or on JAK2V617F mutational status. We did not observe a PV, ET, or MF-specific gene expression signature in MPN neutrophils. However, we were able to detect significant shifts in previously defined PV- and ET-related gene expression signatures.38 Chen et al performed gene expression profiling in erythroid methylcellulose colonies from ET and PV patients, and thereby derived signatures of PV and ET.38 We also tested these PV and ET gene expression signatures and found significant enrichment of the PV erythroid signature in PV neutrophils from patients studied here (26 of 40 genes commonly upregulated and 20 of 24 genes commonly downregulated; FDR q-value 0.007 and 0.006, respectively) as well as significant enrichment of the ET erythroid signature in the ET neutrophils studied here (72 of 83 genes commonly upregulated and 20 of 21 genes commonly downregulated; FDR q-value 0.25 and 0.08, respectively) (supplemental Figure 1 and supplemental Table 1, available on the Blood Web site). In their previous study, Chen and colleagues hypothesized that this difference was a result of differential STAT1 activation based on the observation of differential expression of an interferon gene expression signature; by contrast we did not note enrichment of interferon or STAT1 signatures in PV or ET patients compared with each other or compared with normal donors. In addition, we did not observe a distinct gene expression signature based on the binary presence or absence of the JAK2V617F mutation.
Gene expression signature in MPN patients with homozygous JAK2V617F mutations
We noted that samples from patients with PV or MF with homozygous JAK2V617F mutations clustered separately from MPN patient heterozygous or WT for JAK2V617F, suggesting the presence of a specific gene expression signature in homozygous JAK2V617F neutrophils. Copy neutral loss of heterozygosity at chromosome 9p, which includes the JAK2 gene, has long been recognized in patients with MPNs39 and has led to the discovery of JAK2V617F mutations in MPN patients.2 Moreover, the close association between JAK2V617F homozygous mutations and acquired 9p loss of heterozygosity strongly suggests that acquired homozygosity for the JAK2 mutation provides a clonal advantage that is greater than heterozygous JAK2V617F-mutant cells. Despite these genetic observations, the effects of JAK2V617F gene dosage on transcriptional output have not been well delineated in primary patient samples. To address this question, we performed supervised analysis to identify specific differentially expressed genes in patients with homozygous JAK2V617F-mutant cells, heterozygous JAK2V617F mutation, or who were WT for JAK2V617F, compared with neutrophils from normal individuals. We found a robust difference in gene expression between homozygous JAK2V617F-mutant MPN patients and normal subjects, with the majority of differentially expressed genes showing increased expression in homozygous JAK2V617F-mutant MPN patients (Figure 1B). We identified 265 genes that were differentially expressed (FDR <0.01 and fold-change [FC] >3) in homozygous JAK2V617F-mutant MPN patients compared with normal subjects (Figure 1B; supplemental Table 2). We also compared gene expression in MPN patients with heterozygous JAK2V617F mutations or WT JAK2V617F and normal controls, respectively (Figure 1B). We identified 222 genes that were significantly differentially expressed (FC >2 and FDR <0.05) between MPN patients with heterozygous JAK2V617F mutations and normal controls (supplemental Table 3), and 209 genes with significant changes in gene expression between MPN patients with WT JAK2 and normal controls (supplemental Table 4).
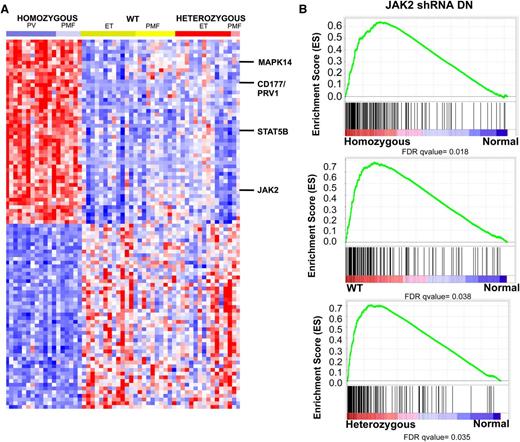
We next compared the specific gene expression signatures seen in MPN patients with homozygous JAK2V617F mutations, heterozygous JAK2V617F mutations, and WT JAK2 compared with normal controls. Analysis of the top 100 genes differentially expressed between homozygous JAK2V617F-mutant MPN patients vs normal controls demonstrated that this expression signature was specific for homozygous JAK2V617F-mutant MPN patients as compared with MPN patients with heterozygous JAK2V617F mutations or WT JAK2V617F. (Figure 2A; supplemental Table 5). The set of differentially expressed genes in MPN patients with homozygous JAK2V617F mutations included JAK2 itself, suggesting the possibility that JAK2 activation regulates its own expression as has been suggested in lymphoma cells with JAK2 amplification (supplemental Figure 2A).30,35 We also identified overexpression of CD177/PRV1 in homozygous JAK2V617F-mutant MPN patients; this is consistent with previous data demonstrating PRV1 overexpression in JAK2V617F-mutant MPN.40 We also observed increased expression of key signaling intermediates downstream of JAK2, including STAT5B and MAPK14, in homozygous JAK2V617F-mutant MPN patients (Figure 1A; supplemental Figure 2A). In addition, we noted increased expression of CSF3R and KRAS in JAK2V617F homozygous MPN patients vs control subjects. Mutation of these genes has been implicated in patients with JAK2V617F-negative MPNs41 and MPN/MDS overlap syndromes.42 We also noted increased expression of KRAS, JAK2, and JAK3 in JAK2V617F heterozygotes vs controls. Comparison of JAK2 WT MPN to normal controls again identified increased expression of KRAS and of TYK2. In addition, we observed significant increased expression of the Polycomb Repressive Complex 2 members EZH2 and SUZ12; notably increased expression of EZH2 has been shown to promote MPN in mice.43 Collectively, these data suggest that dysregulated expression of other oncogenic pathways aside from the JAK-STAT pathway may contribute to MPN pathogenesis, and that MPN patients with homozygous JAK2V617F mutations constitute a specific molecular subset of MPN with a characteristic gene expression signature.
MPN patients are characterized by a transcriptional signature of increased JAK2 activity regardless of JAK2 genotype. (A) Heatmap representation of differentially expressed genes (FC >3 and FDR <0.01) among MPN patients. A red-blue color scale depicts normalized gene expression levels (red: high; blue: low). Displayed are the top 100 differentially expressed genes derived from a supervised analysis comparing transcript expression in granulocytes from JAK2V617F homozygous mutant granulocytes vs normal subjects. The transcripts encoding JAK2, STAT5B, CD177 (PRV1), and MAPK214 are displayed. (B) GSEA showing enrichment of JAK2 shRNA signature in MPN patients relative to normal subjects regardless of JAK2 mutational status.
MPN patients are characterized by a transcriptional signature of increased JAK2 activity regardless of JAK2 genotype. (A) Heatmap representation of differentially expressed genes (FC >3 and FDR <0.01) among MPN patients. A red-blue color scale depicts normalized gene expression levels (red: high; blue: low). Displayed are the top 100 differentially expressed genes derived from a supervised analysis comparing transcript expression in granulocytes from JAK2V617F homozygous mutant granulocytes vs normal subjects. The transcripts encoding JAK2, STAT5B, CD177 (PRV1), and MAPK214 are displayed. (B) GSEA showing enrichment of JAK2 shRNA signature in MPN patients relative to normal subjects regardless of JAK2 mutational status.
MPN patients are characterized by a transcriptional signature of increased JAK2 activity regardless of JAK2 mutational status
Given that we could distinguish between MPN patients and normal controls based on gene expression profiling, we sought to determine if there were functional pathways that contribute to pathologic transcriptional output in MPN cells. Given the high prevalence of JAK2 pathway mutations in MPN patients, we hypothesized that a specific pattern of gene expression associated with constitutive JAK2 activity would distinguish MPN patients from normal controls. We performed gene expression profiling in the JAK2V617F homozygous mutant HEL cell line following treatment with 2 independent shRNAs targeting JAK2 or 2 different control shRNAs (supplemental Figure 3). Supervised analysis of gene expression in HEL cells with JAK2 knockdown relative to control cells showed 175 significantly differentially expressed probe sets (FDR <0.05) (supplemental Figure 3; supplemental Table 6). We then applied this JAK2 shRNA knockdown signature to MPN samples and controls. This analysis showed significant enrichment of the JAK2 shRNA signature in MPN patients relative to controls (Figure 2B; supplemental Figure 2B). Enrichment of the JAK2 shRNA signature in MPN samples was independent of JAK2V617F mutational status, consistent with a role for activated JAK2 signaling governing aberrant gene expression in MPN patients regardless of genotype or clinical phenotype. (Figure 2B). Consistent with this finding, GSEA did not show enrichment of the JAK2 shRNA signature in JAK2V617F homozygous MPN patients compared with MPN patients heterozygous or WT for JAK2V617F (supplemental Figure 3C). By contrast, GSEA showed that the JAK2 shRNA signature was significantly enriched in MPN samples relative to normal (supplemental Figure 3D). These data indicate that a gene expression signature derived from JAK2 activation is characteristic of MPN patients regardless of JAK2V617F mutational status.
CALR-mutant MPN patients are characterized by a gene signature associated with activated JAK2 signaling
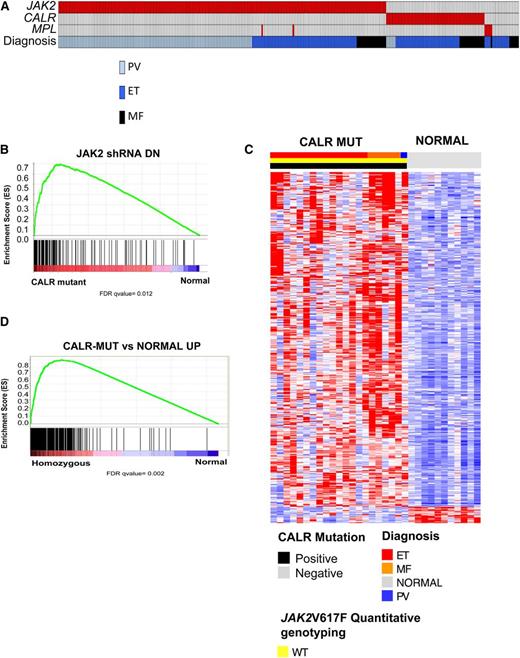
Two seminal articles, by Klampfl et al16 and by Nangalia et al,15 used exome sequencing to identify somatic CALR mutations in 70% to 80% of JAK2-WT ET and PMF patients.15,16 CALR mutations are mutually exclusive of JAK2 and MPL mutations, suggesting a convergent mechanism of transformation between JAK2/MPL mutations and CALR mutations that results in activation of signal transduction pathways. Given the identification of transcriptional markers of JAK-STAT signaling activity in MPN patients regardless of JAK2 mutations, we sought to explore the transcriptional profile of CALR-mutant MPN patients and to compare their gene expression signature to JAK2V617F-mutant MPN patients. Targeted sequencing of the regions of known mutations in CALR in our MPN patient cohort identified CALR mutations in 84% (63/75) of JAK2/MPL WT patient samples (Figure 3A). Consistent with previously reported observations, CALR mutations were exclusively observed in JAK2/MPL-WT ET/MF patients.15,16
CALR-mutant MPN patients are characterized by a gene signature associated with activated JAK2 signaling. (A) Mutational status of JAK2, CALR, and MPL mutational status as well as clinical MPN diagnosis in 290 MPN patients. An individual column represents each patient. (B) GSEA showing enrichment of JAK2 shRNA signature in MPN patients with CALR mutations relative to normal subjects. (C) Heatmap representation of the 433 significantly differentially expressed genes (413 genes upregulated and 20 downregulated; FDR <0.01 and FC >2) in granulocytes from CALR-mutant MPN patients relative to normal subjects (21 MPN patients and 11 normal subjects). A red-blue color scale depicts normalized gene expression levels (red: high; blue: low). (D) GSEA showing significant enrichment of CALR-mutant MPN signature in MPN patients with homozygous JAK2V617F mutations relative to normal subjects.
CALR-mutant MPN patients are characterized by a gene signature associated with activated JAK2 signaling. (A) Mutational status of JAK2, CALR, and MPL mutational status as well as clinical MPN diagnosis in 290 MPN patients. An individual column represents each patient. (B) GSEA showing enrichment of JAK2 shRNA signature in MPN patients with CALR mutations relative to normal subjects. (C) Heatmap representation of the 433 significantly differentially expressed genes (413 genes upregulated and 20 downregulated; FDR <0.01 and FC >2) in granulocytes from CALR-mutant MPN patients relative to normal subjects (21 MPN patients and 11 normal subjects). A red-blue color scale depicts normalized gene expression levels (red: high; blue: low). (D) GSEA showing significant enrichment of CALR-mutant MPN signature in MPN patients with homozygous JAK2V617F mutations relative to normal subjects.
We used GSEA analysis of the JAK2V617F homozygous, JAK2V617F heterozygous, JAK2 WT, and CALR-mutant signatures against the gene set collections in MSigDB (this includes the C2: curated gene sets, C5: Gene Ontology gene sets, C6: Oncogenic signatures gene sets) to assess for global differences in the signatures of these subgroups. This analysis showed that JAK2V617F-mutant homozygous MPN samples have significant enrichment in gene sets characterized by activation of JAK/STAT signaling, KRAS signaling, and MYC transcription, among others, compared with other subgroups (supplemental Tables 7 and 8). In addition, a supervised analysis of CALR-mutant MPN vs normal controls demonstrated upregulation in Polycomb Repressive Complex 2 core members EZH2 and SUZ12, as well as KRAS. Importantly, CALR-mutant MPN patients also demonstrated upregulation in JAK2 and STAT1 (supplemental Table 9).
We next used GSEA to interrogate whether the JAK2 shRNA signature was enriched in CALR-mutant MPN samples. We found significant enrichment of the JAK2 shRNA signature in CALR-mutant MPN patients relative to controls (Figure 3B). We then performed supervised gene expression analysis in CALR-mutant MPN patients relative to controls and identified 433 differentially expressed genes (413 genes upregulated and 20 downregulated; FDR <0.01 and FC >2) (Figure 3C; supplemental Table 9) in CALR-mutant MPN patients. We observed significant enrichment of the CALR-mutant MPN gene signature in JAK2V617F-mutant MPN patients relative to normal individuals (Figure 3D).
Studies have demonstrated that Stat5a/b are important for the development of an MPN phenotype in Jak2V617F-driven murine MPN models.44,45 We thus investigated whether STAT5A targets were differentially expressed in JAK2V617F-mutant (homozygous and heterozygous) MPN patients, as well as in CALR-mutant MPN patients. The STAT5A signature was derived from a previously published study,29 and the STAT5B signature was extracted from MsigDB. We found increased expression of STAT5A targets in all MPN genotype-defined subtypes compared with normal controls. We also found that STAT5B targets increased in expression in MPN patients compared with controls, including in CALR-mutant MPN patients compared with controls (supplemental Figure 4; supplemental Tables 7 and 8). Collectively, these data suggest a shared pattern of transcriptionally altered genes in CALR- and JAK2V617F-mutant MPN patients relative to control subjects, including increased expression of STAT5A/B direct targets.
Integrated genomic analysis of MPN patients
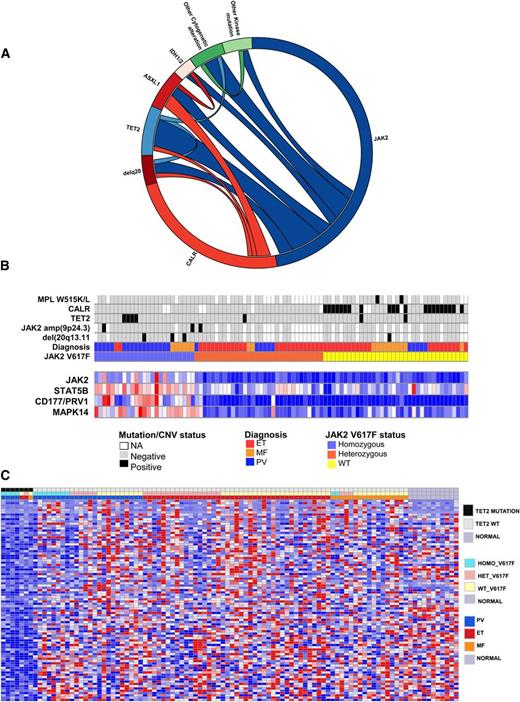
In addition to mutations in JAK2, MPL, and CALR, studies have identified recurrent somatic mutations in epigenetic regulatory proteins in MPN patients, including in TET2, ASXL1, and IDH1/2. To understand the impact of additional genetic alterations on transcriptional profile of CALR and JAK2V617F-mutant MPN patients, we performed targeted sequencing for TET2, ASXL1, IDH1/2, Affymetrix 250K SNP arrays, gene expression, and mass spectrometry-based genotyping for known mutations in 111 additional genes19 in 93 MPN (Figure 4A). The most common additional genomic alterations observed in MPN patients were TET2 mutations (7.2%), ASXL1 mutations (7.1%), amplifications/uniparental disomy of JAK2 (15.9%), and 20q deletions (8%). ASXL1 mutations occurred in 5.3% of JAK2-mutant MPNs and 11.6% of CALR-mutant MPNs (not significant), whereas TET2 mutations occurred in 10.7% of JAK2-mutant MPNs, and 4.7% of CALR-mutant MPNs (not significant). Correlating these data with JAK2V617F and CALR mutations, we observed that amplifications or uniparental disomy of the locus of JAK2 occurred exclusively in patients with heterozygous JAK2V617F mutations and were never observed in concert with CALR mutations. We validated each case of JAK2 amplification by real-time PCR and observed a JAK2V617F allele burden of 65% to 75% consistent with selective amplification of the JAK2V617F allele (Figure 4B). Given that TET2 mutations were the most common somatic mutations observed, aside from JAK2 or CALR, and the clear relevance of TET2 mutations to myeloid transformation,46,47 we investigated whether TET2 mutations had a characteristic gene expression signature in MPN patients. Supervised gene expression analysis of TET2-mutant MPN vs TET2 WT identified 78 significantly differentially expressed genes in TET2-mutant vs WT MPN patients (FDR <0.05; Figure 4C; supplemental Table 10). Notably, nearly all genes in this gene signature were transcriptionally silenced in TET2-mutant MPN patients, consistent with the known role of TET2 in negatively regulating DNA methylation through chemical modifications of DNA. In addition, GSEA analysis of hematopoietic differentiation signatures in TET2-mutant vs WT neutrophils from MPN patients showed enrichment of myeloid differentiation gene expression signature in neutrophils from TET2-mutant samples. (supplemental Table 7). These data are consistent with studies in primary human cells48,49 and in murine Tet2 knockout models50-53 showing that loss/mutation in TET2 leads to myeloid bias and increased myeloid colony output. These data demonstrate that mutations outside the JAK-STAT pathway can have distinct effects on gene expression that contribute to aberrant transcriptional output.
Integrative genomic analysis shows impact of mutations coexisting with CALR and JAK2 mutations on transcriptome of MPN patients. (A) Circos plots showing mutational frequencies and cooccurrences in 97 MPN patient samples. Genetic data regarding JAK2V617F allele burden, cytogenetic alterations, and mutations in CALR, TET2, ASXL1, and IDH1/2 as well as mass spectrometric-based genotyping data are displayed. (B) Integration of somatic genetic alterations, recurrent copy number alterations from SNP array data, and gene expression of key altered transcripts in MPN patients. Each patient is represented in an individual column in the top heatmap and relative level of gene expression of STAT5B, JAK2, MAPK14, and MET is shown in the bottom heatmap. (C) Significantly differentially expressed genes (FDR <0.05) based on supervised analysis of gene expression of TET2-mutant vs WT MPN patients (93 samples). Sixty-one genes were significantly differentially expressed.
Integrative genomic analysis shows impact of mutations coexisting with CALR and JAK2 mutations on transcriptome of MPN patients. (A) Circos plots showing mutational frequencies and cooccurrences in 97 MPN patient samples. Genetic data regarding JAK2V617F allele burden, cytogenetic alterations, and mutations in CALR, TET2, ASXL1, and IDH1/2 as well as mass spectrometric-based genotyping data are displayed. (B) Integration of somatic genetic alterations, recurrent copy number alterations from SNP array data, and gene expression of key altered transcripts in MPN patients. Each patient is represented in an individual column in the top heatmap and relative level of gene expression of STAT5B, JAK2, MAPK14, and MET is shown in the bottom heatmap. (C) Significantly differentially expressed genes (FDR <0.05) based on supervised analysis of gene expression of TET2-mutant vs WT MPN patients (93 samples). Sixty-one genes were significantly differentially expressed.
Discussion
Although JAK2V617F mutations are identified in the majority of MPN patients, the role of JAK2 mutations in disease initiation and maintenance remains incompletely delineated. We used gene expression profiling data to show that MPN patients, regardless of clinical phenotype, have a gene expression signature distinct from normal controls. This signature is based on expression of a JAK-STAT pathway activation signature in all MPN patients regardless of clinical phenotype or somatic genotype. These data suggest that despite the fact that the MPN disease-initiating cell is likely a hematopoietic stem/progenitor cell in all 3 diseases,54 the JAK-STAT pathway activation is transmitted to differentiated progeny consistent with a central role for JAK-STAT activation in the hematopoietic hierarchy of MPN patients. These data thus provide important insights into the biology of MPNs, by showing the central role of the JAK-STAT pathway across MPN genotypes and clinical phenotypes.
The JAK-STAT pathway activation signature is most evident in patients with a high JAK2V617F-mutant allele burden or with amplification of the mutant JAK2V617F allele, suggesting there are quantitative differences in signaling output based on the relative presence of heterozygous or homozygous JAK2V617F-mutant cells. These data are consistent with findings from mouse models,55 in which JAK2V617F gene dosage influences disease severity. These data are also consistent with clinical observations linking quantitative JAK2V617F allele burden in PV to clinical parameters, including an increased degree of polycythemia and an increased rate of fibrotic transformation.56 It will be important to delineate which downstream effectors of JAK2 contribute to increased disease severity in the setting of homozygous JAK2V617F mutations, and whether therapies targeting downstream signaling effectors show increased efficacy in MPN patients with a high JAK2V617F allele burden. This also provides a further rationale to pursue therapeutic strategies that can more effectively target JAK2, either through the design of more potent JAK2 kinase inhibitors or through combination approaches that allow for more potent inhibition of the JAK-STAT pathway.
The seminal studies that identified CALR mutations in JAK2V617F-negative ET and PMF patients provide strong genetic data indicating that CALR mutations represent an alternate mechanism to activate oncogenic signaling effectors in JAK2 WT MPN patients. This is supported by in vitro data demonstrating that overexpression of mutant CALR in Ba/F3 cells results in increased phosphorylation of STAT5. However, whether CALR mutations directly activate JAK2 signaling, and the mechanism(s) linking CALR to tyrosine kinase signaling activation have not been delineated. We show here that CALR-mutant MPN patients are characterized by the same transcriptional signature of activated JAK-STAT signaling observed in JAK2V617F-mutant MPN patients compared with normal controls, suggesting a common mode of transformation in both JAK2- and CALR-mutant MPN patients. Further supporting this hypothesis, we derived a CALR-mutant gene expression signature and showed that the CALR-mutant signature was significantly enriched in JAK2V617F-mutant MPN patients. It will be important to delineate whether there are specific transcriptional effectors downstream of CALR mutations that contribute to MPN pathogenesis, and to investigate whether the CALR-mutant gene expression signature is seen in other hematopoietic malignancies or disease states indicative of a broader role in oncogenic transformation.
Although our gene expression data here demonstrate an important role for JAK2 activation in altering transcriptional output in MPN cells, the presence of additional somatic mutations in MPN patients, including mutations in ASXL1,57 and TET2,20,58,59 suggest that there are other genomic events that contribute to altered gene expression in MPN cells. Our data suggest ASXL1 mutations are most common in CALR-mutant MPN patients, whereas TET2 mutations occur most often in JAK2V617F-mutant MPN patients. Although not statistically significant, further studies to delineate the cooccurrence of other mutations with JAK2 and CALR mutations using larger sample sizes may show distinct routes of pathogenesis and oncogenic cooperativity in JAK2-mutant and CALR-mutant MPNs.
TET2 mutations are the most common somatic mutations outside the JAK-STAT pathway observed in MPN patients.60-63 Here we show that TET2-mutant MPN patients exhibit a distinct gene expression signature compared with TET2-WT patients, indicating that this mutation has distinct effects on transcriptional output. The mechanisms by which TET2 mutations alter DNA hydroxymethylation/methylation at specific sites in hematopoietic cells, and the specific TET2 targets that are critical for its role in oncogenic transformation, remain to be elucidated. In addition, whether the TET2-mutant gene expression signature identified here extends to other TET2-mutant hematopoietic malignancies will need to be investigated in subsequent studies.
Our data also suggest that there are additional signaling and epigenetic pathways that contribute to MPN pathogenesis. We observed dysregulated expression of additional cytokine signaling pathways, including overexpression of KRAS and CSF3R, with a known role in the pathogenesis of myeloid malignancies. These data reinforce recent studies suggesting that targeting downstream or collateral signaling pathways in conjunction with JAK-STAT inhibition may improve therapeutic efficacy. For example, preclinical models using combined inhibition of PI3K/AKT/mTOR pathways and the JAK-STAT pathway have yielded encouraging results,64,65 and clinical trials using such approaches are being planned.66 The observation that KRAS and its signaling intermediates are highly expressed in all genetic subtypes of MPN suggest targeting the MAPK pathway in concert with JAK-STAT inhibition warrants further preclinical and clinical evaluation.
In addition to supporting our biologic understandings of these disorders, the results presented here are consistent with the notion that JAK-targeted therapies demonstrate clinical efficacy in MPN patients regardless of the observed genotype. Indeed, clinical data from studies with ruxolitinib and other JAK inhibitors demonstrated that MPN patients with and without JAK2V617F mutations experienced similar decreases in splenomegaly and constitutional symptoms.13 Our work demonstrates that the JAK-STAT pathway is fundamentally important to the pathogenesis of MPNs, regardless of MPN genotype. Further genomic alterations, such as changes in the JAK2V617F allele burden as well as the presence of other mutational events such as TET2 mutations, can alter the gene expression signature, and potentially phenotype, of MPNs. As such, the wide array of genomic alterations that cooccur with JAK2/MPL/CALR mutations likely play a role in the phenotypic heterogeneity observed in MPN patients, and in the heterogeneous response to MPN therapies.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank members of the Levine and Ebert laboratories for helpful comments and discussion.
This work was supported by grants from the Myeloproliferative Neoplasm Foundation (R.L.L., B.L.E.), the Starr Cancer Consortium (D.G.G., T.R.G., B.L.E., R.L.L.), and the National Institutes of Health, National Cancer Institute (grant 1R01CA151949-01) (R.L.L.). R.L.L. and B.L.E. are Leukemia and Lymphoma Society Scholars, and O.A.-W. and R.R. are American Society of Hematology Scholars.
Authorship
Contribution: B.L.E., R.L.L., T.R.G., and D.G.G. conceived the project; R.R., F.A.-S., O.A.-W., B.L.E., and R.L.L. wrote the manuscript with input from all other authors; R.R., F.A.-S., O.A.-W., J. P. Patel, J.-P.B., O.K., M.W., L.B., B.L.E., and R.L.L. analyzed data; and O.A.-W., J. P. Patel, C.H.M., A.J.B., J. Pretz, T.H., J.A., B.L.E., and R.L.L. performed experiments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Benjamin L. Ebert, Brigham and Women’s Hospital, 1 Blackfan Circle, Boston, MA 02115; e-mail: bebert@partners.org; and Ross L. Levine, Memorial Sloan Kettering Cancer Center, 1275 York Ave, New York, NY 10065; e-mail: leviner@mskcc.org.
References
Author notes
R.R., F.A.-S., and O.A.-W. contributed equally to this study.