Key Points
Using the largest set of patients with newly diagnosed myeloma, we identified 63 mutated driver genes.
We identified oncogenic dependencies, particularly relating to primary translocations, indicating a nonrandom accumulation of genetic hits.
Abstract
Understanding the profile of oncogene and tumor suppressor gene mutations with their interactions and impact on the prognosis of multiple myeloma (MM) can improve the definition of disease subsets and identify pathways important in disease pathobiology. Using integrated genomics of 1273 newly diagnosed patients with MM, we identified 63 driver genes, some of which are novel, including IDH1, IDH2, HUWE1, KLHL6, and PTPN11. Oncogene mutations are significantly more clonal than tumor suppressor mutations, indicating they may exert a bigger selective pressure. Patients with more driver gene abnormalities are associated with worse outcomes, as are identified mechanisms of genomic instability. Oncogenic dependencies were identified between mutations in driver genes, common regions of copy number change, and primary translocation and hyperdiploidy events. These dependencies included associations with t(4;14) and mutations in FGFR3, DIS3, and PRKD2; t(11;14) with mutations in CCND1 and IRF4; t(14;16) with mutations in MAF, BRAF, DIS3, and ATM; and hyperdiploidy with gain 11q, mutations in FAM46C, and MYC rearrangements. These associations indicate that the genomic landscape of myeloma is predetermined by the primary events upon which further dependencies are built, giving rise to a nonrandom accumulation of genetic hits. Understanding these dependencies may elucidate potential evolutionary patterns and lead to better treatment regimens.
Introduction
Multiple myeloma (MM) is characterized by the expansion of a population of clonally related plasma cells that compete for access to the bone marrow niche and that evolve into a complex spatiotemporal ecosystem.1 The clonal cells suppress normal plasma cell populations, leading to immunosuppression, impaired normal hematopoiesis, lytic bone lesions, and, via a number of different mechanisms, impaired renal function. The clinical outcome of MM is variable, and much of this variability is driven by acquired genetic factors, which immortalize and drive the subsequent progression of the disease. Current knowledge of drivers of disease comes from cytogenetic analyses, which have shown that the genome of MM is diverse and is characterized by structural rearrangements and copy number abnormalities.2-5 On the basis of these largely historical data, MM can be broadly split into cases with primary immunoglobulin translocations and those that are hyperdiploid with trisomies of the odd-number chromosomes. The 5 most frequent translocations are t(11;14), t(4;14), t(14;16), t(14;20), and t(6;14) at frequencies of 15%, 12%, 3%, 2%, and 1% of samples, respectively. Additional copy number gains and losses occur frequently, with the most frequent being del13q (59%), 1q+ (40%), del14q (39%), del6q (33%), del1p (30%), and del17p (8%).2 The genetic drivers of disease directly alter downstream biology and clinical behavior and as such can be used to classify disease subgroups and predict clinical outcome.
It is clear that there are many associations between genomic markers in myeloma, with the earliest example probably being where >90% of patients with t(4;14) also have deletion of 13q.6 As more samples are sequenced in MM4,5,7-10 and across different cancer types, there are increasing descriptions of cooccurrences, or oncogenic dependencies, between genomic markers.11-13 We have shown this previously with translocation groups and their partner genes, such as t(11;14) and mutation of CCND1.5 Equally, there are clear examples of mutual exclusivity of markers, mostly where abnormalities occur in genes of similar function or within the same pathway, such as KRAS and NRAS mutations in myeloma and other cancers.14,15 However, to date, the number of oncogenic dependencies known in myeloma has been limited by the data sets available. Identifying these dependencies is important to understand the biology of the tumors and their reliance on pathways and may help in identifying good drug targets.16,17
Here, in a genome-wide unbiased fashion, as part of the ongoing Myeloma Genome Project initiative to clinically exploit the genomic classification of MM, we used genomic data from the largest number of patients with genomically characterized newly diagnosed MM (NDMM) available to date to better define the mutational landscape, including chromosomal translocations, copy number abnormalities, and indel and single nucleotide variants. We identified the drivers of MM at a previously unattained resolution, determined the associations among them, and identified oncogenic dependencies between mutations, copy number changes, and translocation groups.
Methods
Patient characteristics
The Myeloma Genome Project is an ongoing initiative to assemble and analyze in a uniform and innovative fashion genetic data sets that have been generated on samples obtained from patients with MM who have been entered into clinical trials. The current work is based on an analysis of a set of NDMM cases with clinical and outcome data associated with whole-exome sequencing (N = 1273). The data were derived from the Myeloma XI trial,5 the Dana-Faber Cancer Institute/Intergroupe Francophone du Myelome,8 and the Multiple Myeloma Research Foundation CoMMpass study, which have been reported.18
Sequencing and identification of driver variants
Sequencing data were processed as described in the supplemental Methods (available on the Blood Web site) and have been deposited in the European Genome Archive under accession #EGAS00001001147 and #EGAS00001000036 or at dbGAP under accession #phs000748.v5.p4. Sequence analysis methods are described in detail in the supplemental methods and supplemental Figures 1 to 6.
Results
Identification of novel driver genes
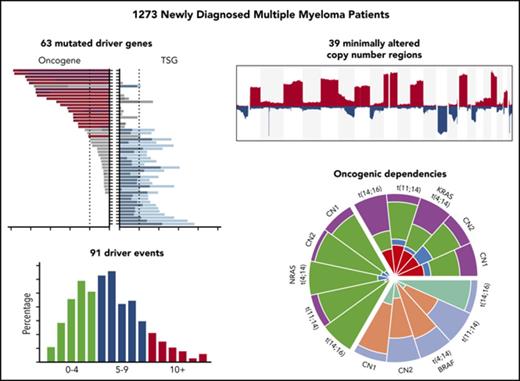
Four methods were used to identify driver genes in 1273 NDMM samples. The methods can be divided into those identifying drivers by a frequency-based approach (MutSigCV19 and dNdSCV20 ) and those using a functional based approach (20/20 rule21 and SomInaClust22 ). All methods were performed on both the whole data set and on individual cytogenetic subgroups (supplemental Tables 1-10). Using the frequency approach, MutSigCV identified 21 significantly mutated genes and dNdSCV identified 46 genes. For the functional approaches, the 20/20 rule identified 47 mutated genes and SomInaClust identified 21 genes. In total, 63 genes were identified by the 4 methods (Figure 1A-B; supplemental Tables 1-4 and 8-9; supplemental Figures 17-24).
The mutational driver landscape of newly diagnosed multiple myeloma. (A) Driver gene frequencies in the total data set (N = 1273) identified by either frequency-based (yellow) or functional-based (blue) methods or both (red). (B) Cancer clonal fraction (CCF) of driver genes colored by oncogene (ONC; red) or tumor suppressor gene (TSG; blue) score. Genes in gray did not score for either ONC or TSG. Genes are ordered by mean CCF (thick line). (C) ONC (red) and TSG (blue) scores determined by the 20/20 rule (dark) or SomInaclust (light). (D) Mean CCFs of ONCs and TSGs.
The mutational driver landscape of newly diagnosed multiple myeloma. (A) Driver gene frequencies in the total data set (N = 1273) identified by either frequency-based (yellow) or functional-based (blue) methods or both (red). (B) Cancer clonal fraction (CCF) of driver genes colored by oncogene (ONC; red) or tumor suppressor gene (TSG; blue) score. Genes in gray did not score for either ONC or TSG. Genes are ordered by mean CCF (thick line). (C) ONC (red) and TSG (blue) scores determined by the 20/20 rule (dark) or SomInaclust (light). (D) Mean CCFs of ONCs and TSGs.
Novel previously unidentified oncogenes included PTPN11 (activator of MEK/ERK signaling), PRKD2 (protein kinase D), SF3B1 (spliceosome factor), and IDH1 and IDH2 (DNA methylation). Other new tumor suppressor genes included UBR5, a ubiquitin ligase mutated in mantle cell lymphoma, and HUWE1, also a ubiquitin ligase.23 There were a high number of recurrent missense mutations in DIS3 and TP53 (69.4% and 45.7%, respectively). Of all 63 genes, only TP53, TRAF3, and TGDS had any impact on outcome in a univariate analysis.24
The cancer clonal fraction of the 63 driver genes was in general higher in oncogenes than tumor suppressor genes (P < .001; Figure 1C-D), consistent with oncogene activation either being an earlier event in progression to MM or asserting a greater selection pressure than tumor suppressor gene inactivation. This seems to be a novel finding, but it is likely to be reproduced in other cancers if this hypothesis is correct. TP53 was the notable exception, with a high cancer clonal fraction, and although it has a number of recurrently mutated codons, it is considered a tumor suppressor gene.
We show the importance of mutational activation of MEK/ERK signaling (KRAS, NRAS, BRAF, PTPN11, RASA2, NF1, PRKD2, and FGFR3; 50.0%). Other pathways identified included NF-κB signaling activation (TRAF2, TRAF3, CYLD, NFKB2, and NFKBIA; 14.0%), G1/S cell cycle transition (CCND1, RB1, CDKN2C, and CDKN1B; 5.0%), and epigenetic regulators (HIST1H1E, KMT2C, CREBBP, ARID1A, KMT2B, ATRX, EP300, SETD2, TET2, KDM5C, ARID2, DNMT3A, KDM6A, NCOR1, IDH1, and IDH2; 24.4%; supplemental Figure 8; supplemental Table 9). A univariate analysis by pathway did not identify any impact on survival (supplemental Figure 7).
Increased number of driver mutations is associated with poor outcome
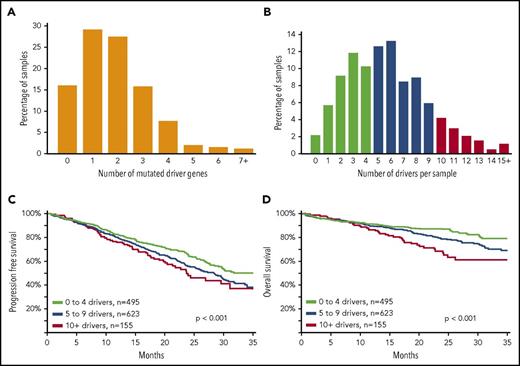
We determined the number of driver mutations per sample and saw that 15.9% of patients had no mutation in any of the 63 driver genes (Figure 2), 84.1% contained ≥1 mutation, 55.0% contained ≥2 mutations, 27.6% contained ≥3 mutations, and 11.9% contained ≥4 mutations. Because there were a sizable number of patients with no mutated driver genes, we integrated other drivers, including copy number and translocations (supplemental Table 7). Incorporating all potential drivers, the median number of drivers per sample was 5, with a range of 0 to 24 (Figure 2B). We grouped these patients according to the total number of drivers and saw that as the number of drivers increased, there was an association with worse progression-free and overall survival (P < .001; Figure 2C-D).
An increasing number of driver abnormalities is associated with poor prognosis. (A) Bar plot of number of mutated driver genes per sample; 203 samples (15.9%) did not contain any nonsynonymous single-nucleotide variants or indels in any of the 63 driver genes; 1070 samples (84.1%) contained ≥1 mutation, 700 samples (55.0%) contained ≥2 mutations, 351 samples (27.6%) contained ≥3 mutations, and 151 samples (11.9%) contained ≥4 mutations (N = 1273). (B) The distribution of all driver abnormalities identified per sample. The number of drivers per sample was calculated using the drivers listed in (supplemental Table 7). Each marker counts for a score of 1; score summed for each patient; maximum score = 91, because some drivers were mutually exclusive (eg, IG translocations), and some copy number features were summarized as a chromosomal arm alteration. The median number of drivers per sample was 5, with a range of 0 to 24. (C) Progression-free survival of patients was significantly negatively affected as the number of drivers increased (P < .001; N = 1273). (D) Overall survival of patients was significantly negatively affected as the number of drivers increased (P < .001; N = 1273).
An increasing number of driver abnormalities is associated with poor prognosis. (A) Bar plot of number of mutated driver genes per sample; 203 samples (15.9%) did not contain any nonsynonymous single-nucleotide variants or indels in any of the 63 driver genes; 1070 samples (84.1%) contained ≥1 mutation, 700 samples (55.0%) contained ≥2 mutations, 351 samples (27.6%) contained ≥3 mutations, and 151 samples (11.9%) contained ≥4 mutations (N = 1273). (B) The distribution of all driver abnormalities identified per sample. The number of drivers per sample was calculated using the drivers listed in (supplemental Table 7). Each marker counts for a score of 1; score summed for each patient; maximum score = 91, because some drivers were mutually exclusive (eg, IG translocations), and some copy number features were summarized as a chromosomal arm alteration. The median number of drivers per sample was 5, with a range of 0 to 24. (C) Progression-free survival of patients was significantly negatively affected as the number of drivers increased (P < .001; N = 1273). (D) Overall survival of patients was significantly negatively affected as the number of drivers increased (P < .001; N = 1273).
Measurements of DNA instability are associated with poor outcome subgroups
Given that the number of driver events may be related to genomic instability, we looked for mechanisms that may be responsible for it. We identified 2 surrogate markers of DNA instability associated with outcome, including an APOBEC mutational signature, which results in C>T/G>A transitions in a TCn context, present in 18.3% of cases and associated with the t(14;16) and t(14;20) subgroups (supplemental Figure 11) and the extent of loss of heterozygosity (LOH; >4.6%; supplemental Table 10), which is potentially a surrogate marker for homologous recombination deficiency. The extent of LOH was positively correlated with the APOBEC signature (P = .039), loss of TP53 (P < .001), and presence of mutation in at least 1 of 15 genes involved in homologous recombination deficiency (P < .001; supplemental Figures 12-13).
Copy number abnormalities are associated with mutational dependencies
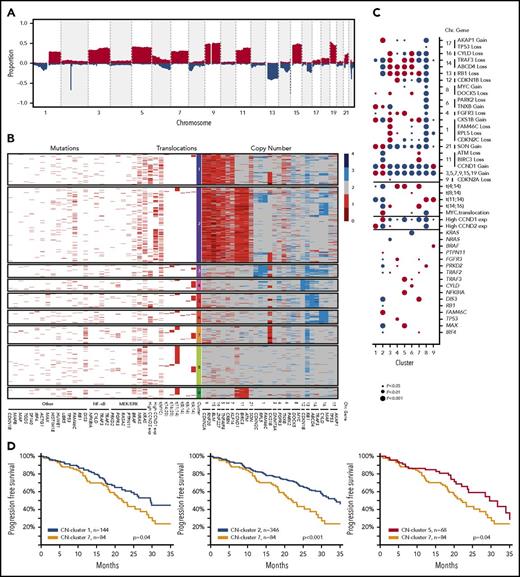
The comprehensive availability of mutational, structural, and copy number data prompted us to reevaluate the classification of myeloma. Copy number data were generated from the whole-exome data in an unbiased genome-wide fashion to identify minimally altered regions (n = 39), which theoretically contain either transcriptional units or single tumor suppressor or oncogenes. Genes of interest located at the peaks of change within these regions were identified (Figure 3A). Markers of the chromosomal gain of trisomic chromosomes were selected to identify these variables. The frequencies of copy number changes in these regions and derived biallelic events are shown in supplemental Tables 5 and 6.
The definition of copy number (CN) clusters using hierarchical clustering and their association with cytogenetic subgroups and significantly mutated genes. (A) CN data derived from 1074 whole-exome sequencing samples identify recurrent regions of gain and loss across the genome. (B) Hierarchical K-means clustering analysis of recurrent CN abnormalities identifies 9 CN clusters, of which 7 were predominantly translocation groups and 2 were hyperdiploid; additional details are listed in the supplemental methods. CN cluster 1 (13.7%) was hyperdiploid and associated with gains of 1q and 6p and expression of CCND2. CN cluster 2 (32.9%) was hyperdiploid with gain of 11q and expression of CCND1, but inversely associated with gain of 1q. CN cluster 3 (5.6%) was associated with t(14;16), deletions of 1p, 8p, 13q, 14q, and 16q, and gain of 1q. CN cluster 4 (5.2%) was associated with t(4;14), del13q, and del14q. CN cluster 5 (6.7%) was associated with t(4;14), del4p, del13q, and del14q as well mutations of NFKBIA, MAX, and TRAF3. CN cluster 6 (5.9%) was associated with deletions of 8p, 14q, and 16q, gain of 1q, and mutation of CYLD. CN cluster 7 (7.9%) was associated with t(4;14) and t(14;16), the APOBEC signature, deletions of 11q and 13q, gain of 1q, and mutation of DIS3. CN cluster 8 (17.3%) was associated with t(11;14) and mutations of CCND1 and PRKD2, but not with any deletions or gains. CN cluster 9 (4.8%) was associated with t(11;14) and gain of 11q as well as mutation of BRAF. The data plotted in this figure are listed in supplemental Table 11. (C) The associations of genetic markers with CN clusters illustrating significant associations and their directionality by the size of the circle; red, positive association; blue, negative association. (D) Progression-free survival Kaplan-Meier plots indicating differences in outcome between CN cluster 7 compared with clusters 1, 2, and 5.
The definition of copy number (CN) clusters using hierarchical clustering and their association with cytogenetic subgroups and significantly mutated genes. (A) CN data derived from 1074 whole-exome sequencing samples identify recurrent regions of gain and loss across the genome. (B) Hierarchical K-means clustering analysis of recurrent CN abnormalities identifies 9 CN clusters, of which 7 were predominantly translocation groups and 2 were hyperdiploid; additional details are listed in the supplemental methods. CN cluster 1 (13.7%) was hyperdiploid and associated with gains of 1q and 6p and expression of CCND2. CN cluster 2 (32.9%) was hyperdiploid with gain of 11q and expression of CCND1, but inversely associated with gain of 1q. CN cluster 3 (5.6%) was associated with t(14;16), deletions of 1p, 8p, 13q, 14q, and 16q, and gain of 1q. CN cluster 4 (5.2%) was associated with t(4;14), del13q, and del14q. CN cluster 5 (6.7%) was associated with t(4;14), del4p, del13q, and del14q as well mutations of NFKBIA, MAX, and TRAF3. CN cluster 6 (5.9%) was associated with deletions of 8p, 14q, and 16q, gain of 1q, and mutation of CYLD. CN cluster 7 (7.9%) was associated with t(4;14) and t(14;16), the APOBEC signature, deletions of 11q and 13q, gain of 1q, and mutation of DIS3. CN cluster 8 (17.3%) was associated with t(11;14) and mutations of CCND1 and PRKD2, but not with any deletions or gains. CN cluster 9 (4.8%) was associated with t(11;14) and gain of 11q as well as mutation of BRAF. The data plotted in this figure are listed in supplemental Table 11. (C) The associations of genetic markers with CN clusters illustrating significant associations and their directionality by the size of the circle; red, positive association; blue, negative association. (D) Progression-free survival Kaplan-Meier plots indicating differences in outcome between CN cluster 7 compared with clusters 1, 2, and 5.
The 39 regions of recurrent copy number alteration (CNA), including the markers for each of the trisomic chromosomes, were used in a K-means clustering approach to group the samples, which were then annotated with additional genetic information (Figure 3B). Clustering identified 9 copy number groups, of which 2 were hyperdiploid: cluster 1 with gains in chromosomes 3, 5, 9, 15, 19, and 21, and cluster 2 gains in the same chromosomes plus chromosome 11 and mutation of FAM46C. The remaining 7 groups were nonhyperdiploid, consisting of cluster 3, which is characterized by del1p; cluster 4 by del12p and 13q; cluster 5 by del13q and 14q and mutations in MAX, TRAF3, and NFKBIA; cluster 6 by del16q; cluster 7 by t(14;16), del11q, 1q gain, and mutation of DIS3; cluster 8 by t(11;14); and cluster 9 by t(11;14) with gain 11q (Figure 3C).
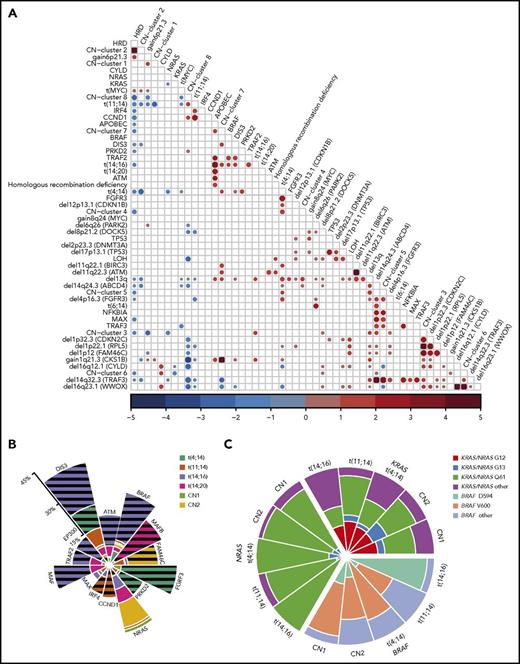
Oncogenic dependencies between mutations and CNAs were identified that distinguished each of the molecular subtypes of MM. These dependencies were exemplified by the presence of particular mutated genes in myeloma subgroups, as well as association with copy number changes and mutations (Figures 1, 3, and 4A). Mechanistically, we identified a critical relationship between acquired chromosomal CNAs and mutations on those chromosomes. In addition to the well-known association between del17p and mutation of TP53, resulting in biallelic inactivation of TP53, we describe an interaction of del13q with mutations in DIS3, del16q with mutations in CYLD and WWOX, and del14q with mutations in TRAF3 and MAX.
Oncogenic dependencies and their impact on survival. (A) The associations between acquired mutations, cytogenetic subgroups, CNAs, and copy number (CN) clusters. Positive (red) and negative (blue) associations and their odds ratios are shown, where the size of the circle represents the significance of the P value defined by the 2-sided Fisher’s exact test. Both gain- and loss-of-function mutations were associated with translocations. The t(4;14) group was associated with mutations of FGFR3, DIS3, and PRKD2, gain of 1q, and deletion of 13q, 14q, 4p16.3 (FGFR3), 1p22.1 (RPL5), 11q22.1 (BIRC3), and 12p13.1 (CDKN1B). The t(11;14) group was associated with mutations in IRF4 and CCND1, but not with gain of 1q or 6p or deletion of 13q, 14q, 8p, or 1p22.1 (RPL5). The t(14;16) group was associated with mutations in BRAF, DIS3, and TRAF2, del13q, gain of 1q, and the APOBEC signature. The t(14;20) group was associated with the APOBEC signature. Hyperdiploidy was associated with gain of 6p and translocations involving MYC, but not with mutations in MAX, DIS3, IRF4, or CCND1 or del13q or del14q. The most significant associations were between CN changes on the same chromosomes (del CDKN2C, RPL5, and FAM46C; del TRAF3 and ABCD4; del WWOX and CYLD), as well as CN cluster 2 with HRD, CN cluster 7 with gain of 1q, and CN cluster 6 with del16q. (B) Significant enrichment/depletion of mutated genes within translocation and hyperdiploid CN clusters. The percentage of samples with the gene mutated within the subgroup is shown. Significance is indicated by hatched lines. MAX mutations are significantly underrepresented in the CN-2 group. (C) The distribution of codon usage within KRAS, NRAS, and BRAF by molecular subgroup. Proportion of codon usage is indicated by the distance from the center. KRAS mutations are split between codons G12, G13, and Q61, whereas NRAS is predominantly mutated at codon Q61 (P < 2.2 × 10−16). BRAF mutations mostly affect codon V600, except in the t(14;16) group, where codon D594 is mutated (P = .003).
Oncogenic dependencies and their impact on survival. (A) The associations between acquired mutations, cytogenetic subgroups, CNAs, and copy number (CN) clusters. Positive (red) and negative (blue) associations and their odds ratios are shown, where the size of the circle represents the significance of the P value defined by the 2-sided Fisher’s exact test. Both gain- and loss-of-function mutations were associated with translocations. The t(4;14) group was associated with mutations of FGFR3, DIS3, and PRKD2, gain of 1q, and deletion of 13q, 14q, 4p16.3 (FGFR3), 1p22.1 (RPL5), 11q22.1 (BIRC3), and 12p13.1 (CDKN1B). The t(11;14) group was associated with mutations in IRF4 and CCND1, but not with gain of 1q or 6p or deletion of 13q, 14q, 8p, or 1p22.1 (RPL5). The t(14;16) group was associated with mutations in BRAF, DIS3, and TRAF2, del13q, gain of 1q, and the APOBEC signature. The t(14;20) group was associated with the APOBEC signature. Hyperdiploidy was associated with gain of 6p and translocations involving MYC, but not with mutations in MAX, DIS3, IRF4, or CCND1 or del13q or del14q. The most significant associations were between CN changes on the same chromosomes (del CDKN2C, RPL5, and FAM46C; del TRAF3 and ABCD4; del WWOX and CYLD), as well as CN cluster 2 with HRD, CN cluster 7 with gain of 1q, and CN cluster 6 with del16q. (B) Significant enrichment/depletion of mutated genes within translocation and hyperdiploid CN clusters. The percentage of samples with the gene mutated within the subgroup is shown. Significance is indicated by hatched lines. MAX mutations are significantly underrepresented in the CN-2 group. (C) The distribution of codon usage within KRAS, NRAS, and BRAF by molecular subgroup. Proportion of codon usage is indicated by the distance from the center. KRAS mutations are split between codons G12, G13, and Q61, whereas NRAS is predominantly mutated at codon Q61 (P < 2.2 × 10−16). BRAF mutations mostly affect codon V600, except in the t(14;16) group, where codon D594 is mutated (P = .003).
We identified that cluster 7 had an association with a worse outcome compared with clusters 1, 2, and 5 (Figure 3D). Cluster 7 was associated with gain or amplification of 1q, which is a key poor prognostic factor.24 Cluster 7 was also associated with the t(4;14) and t(14;16) cytogenetic groups but still performed worse than cluster 5, which was associated with t(4;14) and delFGFR3.
Translocation subgroups are associated with oncogenic dependencies
Significant associations between the primary translocation groups and mutations were seen, including FGFR3, PRKD2, and ACTG1 being only significantly mutated in t(4;14)s. CCND1, IRF4, LTB, and HUWE1 were only significantly mutated in t(11;14)s. MAF was only significantly mutated in t(14;16)s, and MAFB only in t(14;20)s. CDKN1B, FUBP1, NFKB2, PRDM1, PTPN11, RASA2, RFTN1, and SP140 were only significantly mutated in the hyperdiploid samples. The mutation of certain genes within particular myeloma subgroups indicates that oncogenic dependencies exist, where certain pathways are more important to particular subgroups than others.
Additionally, using Fisher’s exact test, we show that t(4;14) samples had more mutations in FGFR3, PRKD2, and DIS3; t(11;14) had more mutations in CCND1 and IRF4; t(14;16) had more mutations in ATM, BRAF, MAF, TRAF2, EP300, and DIS3; t(14;20) had more mutations in MAFB; and the hyperdiploid group with gain 11 (cluster 2) had more mutations in FAM46C (Figure 4A-B).
In addition to these dependencies, we also saw codon-specific mutations within genes that were reliant on the context of the myeloma subgroup, some of which we previously observed in a targeted panel data set.25 Key examples of these interactions include the variable distribution frequency and codon usage of KRAS, NRAS, and BRAF between subtypes (Figure 4C). There was a clear bias toward Q61 mutations in NRAS across all subgroups but a more equal distribution of mutations across codons 12, 13, and 61 in KRAS. There was a unique predominance of BRAFD594N variants in the t(14;16) subgroup compared with BRAFV600E in the other groups (Figure 3C), potentially indicating a different mechanism of action.
Expression of driver gene variants
To ensure that the proposed driver oncogenes are being expressed, we examined matched RNA sequencing (RNA-seq) variants from the Multiple Myeloma Research Foundation data set. We found a near linear correlation with exome-called variant allele frequency (VAF) and RNA-seq–called VAF, for both oncogenes and tumor suppressor genes (Figure 5A-D), indicating that DNA-level mutations in the coding sequence do not affect transcription, even for nonsense or frameshift mutations. There were some outliers in the analysis, where the RNA-seq VAF was increased over the exome VAF, indicating a disproportionate expression of the mutant allele. This was mostly due to translocation partner oncogenes (FGFR3, CCND1, MAF, and MAFB; Figure 5A) under the influence of the immunoglobulin superenhancer. It was possible to identify biallelic inactivation of TP53 using VAF for exome or RNA-seq, where either was >0.5 (Figure 5B). The oncogenes NRAS, KRAS, and BRAF also showed proportionate expression of alleles, but NRAS often had a high VAF because of frequent deletion of the locus, which is on 1p, indicating retention of the mutant allele. NF-κB genes were frequently inactivated because of nonsense or frameshift mutations, but this did not seem to alter expression at the RNA level.
Mutations in tumor suppressor genes and oncogenes are expressed. (A) Scatterplot with oncogene and tumor suppressor gene exome and RNA-seq VAFs (r2 = 0.585). Translocation partner oncogene mutations are colored and are generally outliers with increased expression. (B) TP53 mutations, including missense, nonsense, and frameshift, are detectable according to VAF. High VAF on either axis is due to deletion of TP53 and is indicative of biallelic inactivation. (C) KRAS, NRAS, and BRAF mutations are expressed according to VAF. NRAS has more high VAF-expressed mutations (>0.5), because it is on 1p, and the nonmutated allele is frequently deleted. (D) NF-κB genes CYLD, NFKBIA, TRAF2, and TRAF3.
Mutations in tumor suppressor genes and oncogenes are expressed. (A) Scatterplot with oncogene and tumor suppressor gene exome and RNA-seq VAFs (r2 = 0.585). Translocation partner oncogene mutations are colored and are generally outliers with increased expression. (B) TP53 mutations, including missense, nonsense, and frameshift, are detectable according to VAF. High VAF on either axis is due to deletion of TP53 and is indicative of biallelic inactivation. (C) KRAS, NRAS, and BRAF mutations are expressed according to VAF. NRAS has more high VAF-expressed mutations (>0.5), because it is on 1p, and the nonmutated allele is frequently deleted. (D) NF-κB genes CYLD, NFKBIA, TRAF2, and TRAF3.
Discussion
The advantage of integrating data sets is the power associated with this large study, which allowed us to identify a number of novel driver genes in MM. Here, we identified 63 mutational drivers in myeloma using 4 different methods in both the complete data set and translocation subgroups. New drivers in myeloma were identified, including the ubiquitin ligase UBR5, which plays a central role in the modulation of apoptosis,26 and HUWE1, which can affect the level of MYC expression via MIZ1.23 MYC activity was also potentially affected by mutations in MAX and via mutations in IRF4 and EGR1.27-29 A number of epigenetically active genes, which are potential therapeutic targets, were also identified as being significantly mutated, including IDH1 and IDH2,30 which had low-frequency gain-of-function mutations.31,32 The mutations seen in PRKD2 focused attention on protein kinase D inhibition as a potential novel therapeutic approach, especially in t(4;14) cases. Of the identified genes, 27% (17 of 63) were potentially actionable (supplemental Table 9). Few associations between mutations and survival were seen, with the most prominent being TP53. However, this may have been due to the short follow-up in this data set. Continued analysis of this and other data sets with longer follow-up may reveal additional associations between mutations and survival. Larger data sets will allow for deeper exploration of the genomic landscape of myeloma as a whole and be particularly important for probing cytogenetic groups that are less frequently seen. With so many known drivers, targeted sequencing approaches become possible that are faster and more cost effective than either whole-genome or whole-exome sequencing.
The predominant pathway affected by mutation was the MEK/ERK pathway, with 9 oncogenes affected (supplemental Figure 17). Two tumor suppressor genes were also involved in activating this pathway, RASA2 and NF1, which encode RasGAPs, the function of which is recycling activated Ras back to its inactive state.33 Another novel mechanism for activation of this pathway involved mutation of PTPN11, which encodes SHP2, known to be mutated in Rasopathy Noonan’s syndrome and in pediatric leukemia.34,35
In myeloma, the distribution of Ras mutations was 56% for KRAS and 44% for NRAS, with no HRAS mutations. This distribution is different to other cancers, where KRAS makes up 96% of Ras mutations in lung adenocarcinoma and 86% in colorectal adenocarcinoma, but only 27% in acute myeloid leukemia and 3% in melanoma. We see here that in myeloma, the codon usage for NRAS was biased toward Q61 mutations (81%), which is more similar to usage in melanoma (85%) than that seen in lymphoid tumors (21%). KRAS codon usage in myeloma was more evenly split between codons G12 (34.4%), G13 (12.8%), and Q61 (34.7%) compared with colorectal adenocarcinoma (83%; G12) and pancreatic ductal adenocarcinoma (97%; G12). In addition, myeloma had a high percentage of non-12/13/61 codons mutated in KRAS (18%), with the most common being Q22, Y64, K117, and A146. The different mutated codons between Ras genes may be indicative of subtle functional differences or preferences in mutational bias (eg, with mutational signatures).
Consistent with this hypothesis, a strikingly different mutational pattern of BRAF was seen in the t(14;16) group compared with the t(4;14) group, with 92% of mutations affecting codon D594 compared with 78% affecting codon V600, respectively (Figure 3C). This may have been related to the APOBEC mutational signature, which is dominant in t(14;16) and results in C>T/G>A transitions in a TCn context, because the BRAFD594N mutation conforms to this signature [T(C>T)A], whereas the BRAFV600E mutation does not [C(A>T)C]. BRAFD594 mutations have been reported in melanoma, and unlike the BRAFV600E mutants, which result in direct phosphorylation of MEK,36 they result in a kinase-dead BRAF, which indirectly activates the MEK pathway through binding CRAF in a Ras-independent manner.37 Functional work is required to determine if BRAF mutants behave the same in myeloma. Identifying which BRAF mutation a patient has is clinically important, because BRAF inhibitors are selective for BRAFV600E,38 and it has been suggested that BRAF inhibitors should not be used in patients with Ras activation (KRAS, NRAS, and BRAFD594 mutants).39
Mutations of DIS3, FAM46C, and SF3B1 were common, implicating RNA processing as a deregulated pathway.40-42 Interestingly, the mutational profiles of both DIS3 and SF3B1 suggest gain-of-function variants consistent with them being oncogenes. Another key pathway deregulated was NF-κB, where TRAF2, TRAF3, CYLD, NFKB2, and NFKBIA were mutated. The mutations in NFKBIA identify it as a novel negative regulator of the noncanonical NF-κB pathway.43,44 Despite the size of the study, we were unable to show any prognostic significance for pathway deregulation based on acquired mutations (supplemental Figure 7).
Genomic instability is key in myeloma, and we identified 3 markers of it through the APOBEC signature, homologous recombination deficiency, and increased LOH. There is a complex relationship between each of these markers and with alterations in TP53. Other markers of genomic instability, such as chromplexy and chromothripsis, may also be informative, but whole-genome sequencing studies to identify the rearrangements will be required.45,46 We also previously showed in a separate data set that homologous recombination deficiency–associated LOH is more frequent in those with high-risk disease and is associated with worse outcomes.47 In ovarian cancer, these mechanisms of genomic instability are used as key targets of PARP inhibitors, where sensitivity to PARP inhibition is induced either by chemically inhibiting the DNA damage response pathway or by inducing increased BRCAness with the proteasome inhibitor bortezomib.48,49
Significant associations between mutations, translocations, and CNA clusters were seen, consistent with the molecular subgroups of NDMM being biologically distinct (Figure 3C). The data are consistent with the etiologic events setting a genetic background against which subsequent events are superimposed dependent upon the processes they deregulate that collaborate to increase the survival of the cells of that background. There are several examples of abnormalities that are dependent on the primary translocation backgrounds, such as t(4;14) and mutations in FGFR3, DIS3, and PRKD2; t(11;14) and mutations in CCND1 and IRF4; hyperdiploidy and FAM46C mutations; and t(14;16) and the APOBEC mutational signature and mutations in MAF, BRAF, ATM, and DIS3. It is clear why some of these associations exist (eg, the translocation groups and the partner oncogenes [FGFR3, CCND1, and MAF]),50 but the other associations are less obvious and potentially more interesting. It remains unclear why mutations in FAM46C are associated with hyperdiploid myeloma, but deregulation of FAM46C in this group is important, because it is not only mutated but also inactivated through secondary MYC rearrangements.51,52 Determining the function of these abnormalities within their genetic backgrounds will be key to understanding the pathobiology of myeloma, along with integrating other data types such as RNA expression and DNA methylation.
Having examined copy numbers in >1000 patients, we were able to segment them according to common features. There were 2 clear hyperdiploid groups, which differed in the gain of chromosome 11. Apart from the obvious difference in CCND1 expression between the groups, because CCND1 is on chromosome 11, there were associations with other markers that were cluster specific. Those with gain of chromosome 11 were associated with FAM46C mutations and MYC rearrangements, but not with mutations in DIS3 or MAX, whereas the group without gain of chromosome 11 was associated with gain of 1q and high CCND2 expression. The latter was associated with poor prognosis, and using this division in a data set with longer follow-up will be of interest.
The oncogenic dependencies identified here indicate that the evolutionary processes occurring in a myeloma cell are more complex than previously thought. It seems that genetic progression and accumulation of events are dependent on the initiating event. The genomic landscape of the cell is predetermined by the primary event, upon which additional dependencies are built, giving rise to a nonrandom accumulation of key genetic hits. Understanding how these abnormalities interact within cellular pathways and processes will lead to the identification of new therapeutic options that can be used to exploit these evolutionary dependencies. Additional studies examining how these dependencies interact at the single-cell level, and in response to different treatment regimens, will be important in understanding the evolutionary processes in myeloma, particularly at relapse.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge continued support for the Myeloma Genome Project from colleagues at Celgene, especially Mark Alles, Michael Pehl, Rupert Vessey, Doug Bassett, Alec Reynolds, Andrew Dervan, and the Myeloma Disease Strategy Team.
Funding for data processing and storage was provided by Celgene Corporation.
Authorship
Contribution: The project was conceived and designed by A.T., N.M., and G.J.M.; funding was acquired by E.F. and A.T.; methodology development and oversight (computational pipeline/tools) were performed by K.M. and M.T.; project administration was performed by F.T. and E.F.; oversight and management of resources (data generation, collection, transfer, infrastructure, data processing) were performed by K.M., M.T., J.K., D.A., M.S., H.A.-L., N.M., S.L., K.C.A., G.H.J., E.F., F.T., M.F., N.B., and R.S.; data were curated by D.R. and J.O.; analyses and interpretation were performed by B.A.W., K.M., C.P.W., T.C.A., M.B., A.R., H.W., P.Q., A.H., F.E.D., M.S., F.T., M.O., E.F., Z. Yu, Z. Yang, M.T., D.A., J.K., P.M., B.D., A.K.S., H.G., M.S.R., H.E., P.S., J.S.M., G.H.J., K.C.A., H.A.-L., N.M., A.T., and G.J.M.; data visualization was performed by B.A.W., F.T., C.P.W., T.C.A., A.R., D.R., and A.H.; supervision and scientific direction were provided by A.T., N.M., M.T., and G.J.M.; and the manuscript was written by B.A.W., E.F., F.T., A.T., A.H., A.R., C.P.W., T.C.A., and G.J.M.
Conflict-of-interest disclosure: K.M., F.T., E.F., M.O., Z. Yu, Z. Yang, M.T., and A.T. are employed by or have equity ownership in Celgene Corporation. The remaining authors declare no competing financial interests.
Correspondence: Gareth J. Morgan, Myeloma Institute, University of Arkansas for Medical Sciences, 4301 W. Markham St, Little Rock, AR; e-mail: gjmorgan@uams.edu.
References
Author notes
B.A.W., K.M., and C.P.W. contributed equally to this work as joint lead authors.
N.M., A.T., and G.J.M. contributed equally to this work as joint senior authors.