Key Points
JNJ-75276617 shows preclinical activity in KMT2A- or NPM1-altered leukemia, synergizing with gilteritinib, venetoclax, and azacitidine.
Antiproliferative activity in cells harboring menin mutations M327I and T349M is due to a unique binding mode.
Visual Abstract
The interaction between menin and histone-lysine N-methyltransferase 2A (KMT2A) is a critical dependency for KMT2A- or nucleophosmin 1 (NPM1)–altered leukemias and an emerging opportunity for therapeutic development. JNJ-75276617 (bleximenib) is a novel, orally bioavailable, potent, and selective protein-protein interaction inhibitor of the binding between menin and KMT2A. In KMT2A-rearranged (KMT2A-r) and NPM1-mutant (NPM1c) acute myeloid leukemia (AML) cells, JNJ-75276617 inhibited the association of the menin-KMT2A complex with chromatin at target gene promoters, resulting in reduced expression of several menin-KMT2A target genes, including MEIS1 and FLT3. JNJ-75276617 displayed potent antiproliferative activity across several AML and acute lymphoblastic leukemia (ALL) cell lines and patient samples harboring KMT2A or NPM1 alterations in vitro. In xenograft models of AML and ALL, JNJ-75276617 reduced leukemic burden and provided a significant dose-dependent survival benefit accompanied by expression changes of menin-KMT2A target genes. JNJ-75276617 demonstrated synergistic effects with gilteritinib in vitro in AML cells harboring KMT2A-r. JNJ-75276617 further exhibited synergistic effects with venetoclax and azacitidine in AML cells bearing KMT2A-r in vitro, and significantly increased survival in mice. Interestingly, JNJ-75276617 showed potent antiproliferative activity in cell lines engineered with recently discovered mutations (MEN1M327I or MEN1T349M) that developed in patients refractory to the menin-KMT2A inhibitor revumenib. A cocrystal structure of menin in complex with JNJ-75276617 indicates a unique binding mode distinct from other menin-KMT2A inhibitors, including revumenib. JNJ-75276617 is being clinically investigated for acute leukemias harboring KMT2A or NPM1 alterations, as a monotherapy for relapsed/refractory acute leukemia (NCT04811560), or in combination with AML-directed therapies (NCT05453903).
Introduction
Acute myeloid leukemia (AML) is a genetically heterogeneous hematologic malignancy characterized by impaired differentiation and accumulation of clonal, abnormal myeloid progenitor cells known as myeloblasts.1,2 Likewise, acute lymphoblastic leukemia (ALL) is a disease characterized by accumulation of lymphoid progenitor cells in the bone marrow or extramedullary sites.3 Relapsed/refractory (R/R) AML and ALL are difficult-to-treat diseases because of their poor response to chemotherapy and the myriad of cooperating genetic and epigenetic events. Mutations, chromosomal translocations, and epigenetic changes affect gene expression programs that are involved in the regulation of stem cell properties, ultimately playing a role in leukemia pathogenesis.4-8
Histone-lysine N-methyltransferase 2A (KMT2A; alias mixed lineage leukemia) is a histone methyltransferase that plays an essential role in regulating gene expression during early development and hematopoiesis.9 Menin is a scaffolding protein that interacts with KMT2A and is essential for formation of a chromatin complex that maintains oncogenic gene expression in KMT2A-rearranged (KMT2A-r) leukemias.10 Menin binds to KMT2A and recruits additional proteins, including histone modifiers and transcription factors, to target genes, promoting gene transcription. KMT2A alterations, including rearrangements, amplifications, or partial tandem duplications (PTDs), are observed in ≈15% of AML cases and 15% of B-cell ALL (B-ALL) cases, including up to 80% of infant B-ALL cases.11-14KMT2A-altered acute leukemias are associated with a high level of resistance to chemotherapy.11,15,16 Translocations of KMT2A lead to oncogenic KMT2A-fusion proteins that enhance proliferation and block hematopoietic differentiation, ultimately driving the development of leukemia.17 Leukemias bearing KMT2A alterations are characterized by upregulated expression of menin-KMT2A target genes including the HOXA/B clusters, as well as the homeobox gene (HOX) transcription factor MEIS1.9,18-22
Genetic or pharmacologic disruption of the interaction between menin and KMT2A has been shown to block disease development and progression in human and mouse models of KMT2A-r AML and ALL in vivo.23-25 Studies have also revealed the importance of the menin-KMT2A interaction in AML with mutations in the nucleophosmin 1 (NPM1) gene.26NPM1 mutations are the second most common mutation type and found in ≈30% of AML cases. Like KMT2A-r, NPM1 mutations are associated with the upregulated expression of HOX gene clusters and MEIS1.27-29
Disruption of the menin-KMT2A interaction is an attractive therapeutic approach for KMT2A- or NPM1-altered acute leukemia with the potential to induce differentiation and leukemic cell death. Two menin-KMT2A inhibitors completing phase 1 clinical trials have provided proof of relevance for targeting the menin-KMT2A protein-protein interaction in KMT2A and NPM1 altered R/R acute leukemia. At their recommended phase 2 doses, revumenib achieved 30% complete remission/complete remission with partial recovery of peripheral blood counts in both KMT2A-r and NPM1-altered leukemia, whereas ziftomenib achieved 35% complete remission/complete remission with partial recovery of peripheral blood counts in NPM1-altered disease but did not reach proof of relevance in KMT2A-r leukemia.30
Recent studies have demonstrated that clinically relevant acquired resistance to revumenib was associated with specific somatic point mutations in the MEN1 gene encoding key amino acids in the KMT2A-binding pocket (MEN1M327I, MEN1T349M).31 Recombinant mutant MEN1 binding with revumenib and other menin-KMT2A inhibitors showed a significant reduction in binding affinity. KMT2A-r leukemia cells engineered to express the point mutations recapitulated revumenib inhibitor resistance, with significantly decreased antiproliferative effects. Similar effects were observed on treatment with other menin-KMT2A inhibitors.31
Although clinical proof of relevance has been achieved with 2 menin-KMT2A inhibitors, improvements in potency in the absence of off-target adverse effects may provide additional clinical benefit. JNJ-75276617 (bleximenib) is a potent, selective small molecule that binds to the menin protein and disrupts its interaction with KMT2A. In preclinical studies, JNJ-75276617 displayed robust preclinical efficacy against NPM1- and KMT2A-altered AML and B-ALL in vitro and in vivo, either as a monotherapy or in combination with gilteritinib, azacitidine, and/or venetoclax. Furthermore, JNJ-75276617 retained potency against endogenously edited AML cells harboring the heterozygous MEN1M327I or MEN1T349M mutations, which may be due to its unique mode of menin binding.
Collectively, these data support the ongoing clinical trials investigating JNJ-75276617 as a therapy for acute leukemias with either KMT2A or NPM1 alterations, as monotherapy for R/R acute leukemia (NCT04811560), or in combination with AML-directed therapies (NCT05453903).
Materials and methods
Compound synthesis
The molecule JNJ-75276617 (compound 27, preparation method B) and the enantiomer JNJ-75303722 (compound 28) were prepared and characterized according to procedures reported in patent application WO2021121327. For xenograft studies in mice, the oxalate salt of JNJ-75276617 was prepared following the procedure described in WO2021121327 (compound 70). The purities of the different batches of JNJ-75276617 were >98%, as determined by liquid chromatography–mass spectrometry analysis.
Homogeneous time-resolved fluorescence assay
Human, dog, and mouse menin (NP_000235, XP_038279381, and NP_001161961) were expressed with a C-terminal 6-His affinity tag in Escherichia coli BL21 DE3 and purified by nickel affinity chromatography. The ability of JNJ-75276617 to displace a fluorescein isothiocyanate (FITC)–labeled menin-binding motif (MBM) 1 peptide (derived from KMT2A) from binding to Tb-labeled human, dog, and mouse menin was quantified. The assay involved incubating JNJ-75276617 with menin and FITC-MBM1 peptide, followed by monitoring the time-resolved fluorescence energy transfer (TR-FRET) signal using an EnVision (PerkinElmer) or PHERAStar (BMG LABTECH) plate reader. Inhibitor potency was reported at the 300- or 420-minute time point when equilibrium was reached. The assay was performed 3 to 10 times for each condition. Percentage inhibition and potency were calculated using Prism software.
Quantigene multiplex assay
Quantigene multiplex assay was performed for expression profiling of menin-KMT2A target genes and differentiation markers in leukemia cells, according to the manufacturer’s protocol (ThermoFisher Scientific). A range of leukemia cell lines was treated with JNJ-75276617 or dimethyl sulfoxide (DMSO) for 48 to 72 hours. After lysing the cells, target-specific probes were mixed with cell lysates, and the signals from the probes were measured using a FLEXMAP 3D system (Luminex Corporation). Absolute 50% inhibitory concentration (IC50) values were calculated by dose-response modeling, and fold changes were calculated to determine the effect of JNJ-75276617 on gene expression.32
AML patient sample proliferation assay
AML blasts from peripheral blood or bone marrow from untreated patient samples were studied after informed consent, and the protocol was approved by the Medical Ethical Committee in accordance with the Declaration of Helsinki. JNJ-75276617 was tested for its antiproliferative activity in cocultures of primary AML cells and MS5 murine stromal cells.8,33,34 Cocultures were demidepopulated 7 days after the addition of JNJ-75276617 and analyzed by flow cytometry for absolute cell number quantification and differentiation using CD11b staining.
Animal studies
JNJ-75276617 was evaluated in human cell line–derived or patient-derived (PDX) AML or B-ALL xenografts from young adult or pediatric patients with KMT2A-r (MOLM14 AML, CBAM-68552, and CBAB-62871 B-ALL) or from adult patients with AML with NPM1 alterations (LEXFAM-2734, AM7577) in female 6- to 8-week-old immune-compromised mice. JNJ-75276617 at 10 mg/kg or venetoclax at 100 mg/kg was orally dosed daily for 4 weeks. Azacitidine was dosed at 2 mg/kg by intraperitoneal injection daily for the first week only. All experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals, the European Communities Council Directives 2010/63/EU, and the US Animal Welfare Act, and were approved by local ethics committees.
Further details can be found in the supplemental Methods section (available on the Blood website).
Results
Development of JNJ-75276617, a highly potent, selective inhibitor targeting the interaction between menin and KMT2A characterized by strong binding affinity to the KMT2A binding pocket
To disrupt the interaction between menin and KMT2A, several scaffolds were explored, from which a central triazine scaffold was identified that unexpectedly provided a favorable combination of binding affinity to the menin protein and in vitro cardiovascular safety properties. Modifications, including the installation of a flexible tail region containing an additional basic center in the molecule, led to the discovery of JNJ-75276617. JNJ-75276617, an orally bioavailable, potent, and selective inhibitor of the binding between KMT2A and menin (Figure 1A), was found to have favorable nonclinical safety, drug disposition, and developability properties. In the menin-KMT2A homogeneous time-resolved fluorescence assay, the ability of test compounds to displace an FITC-MBM1 peptide from binding to Tb-labeled human menin was quantified. The average IC50 value of JNJ-75276617 against human menin across 10 independent experiments was 0.1 ± 0.05 nM (Table 1). Similarly, JNJ-75276617 exhibited potent inhibitory activity against mouse and dog menin, with IC50 values similar or slightly lower than those for human menin (Table 1). To identify any potential proteins interacting with JNJ-75276617 in cells, we evaluated JNJ-75276617 using the Evotec Cellular Target Profiling technology. Using compound affinity chromatography, proteins interacting with JNJ-75276617 were enriched from AML cell lines and identified using mass spectrometry analysis. Menin was identified as a high-affinity target protein with consistent affinities of ≈0.3 nM over replicates and different conditions in MOLM-14 and OCI-AML3 cells (supplemental Figure 1). Besides menin, only 1 of 5427 proteins (prolylcarboxypeptidase) was identified as having a potential weak interaction with JNJ-75276617 (Dissociation constant (Kd)> 10 μM). Furthermore, no significant activity was observed when JNJ-75276617 was assessed in a kinase panel (n = 109) at 1.0 μM at DiscoverX (KinomeScan). The binding profile of JNJ-75276617 was tested at 10 μM against 52 receptors, including nuclear receptors, voltage, and ligand-gated transporters (Eurofins CEREP). The percentage inhibition of the binding of a radioactively labeled ligand specific for each target was assessed. No significant interaction (ie, >50% inhibition of the control-specific binding) was observed for any of the receptors tested. The concentration-response relationship of the effect of JNJ-75276617 on the hERG (human ether-à-go-go-related gene) potassium channel current was evaluated in human embryonic kidney cells expressing the hERG gene. The IC50 for the inhibitory effect of JNJ-75276617 on hERG potassium current was >30 μM. In human induced pluripotent stem cell–derived cardiomyocytes, JNJ-75276617 did not cause any physiologically relevant effects on calcium transient duration 90%, beat rate, Ca2+ amplitude, incidence of cessation of beating, early afterdepolarization-like, or fibrillation-like events at a concentration of up to 5 μM.
Chemical and crystal structures of JNJ-75276617. (A) Chemical structure of JNJ-75276617. (B) Crystal structure of menin bound to JNJ-75276617 (Protein Data Bank Identifier [PDB ID]: in deposition). JNJ-75276617 is shown as ball and sticks, and selected side chains of menin are shown as sticks. Dotted lines represent key hydrogen-bonding interactions between the protein and the ligand, and ligand-binding pocket is depicted as gray surface. Menin residue numbering is according to transcript variant: NM_000244.
Chemical and crystal structures of JNJ-75276617. (A) Chemical structure of JNJ-75276617. (B) Crystal structure of menin bound to JNJ-75276617 (Protein Data Bank Identifier [PDB ID]: in deposition). JNJ-75276617 is shown as ball and sticks, and selected side chains of menin are shown as sticks. Dotted lines represent key hydrogen-bonding interactions between the protein and the ligand, and ligand-binding pocket is depicted as gray surface. Menin residue numbering is according to transcript variant: NM_000244.
JNJ-75276617–inhibited menin-KMT2A binding from various species in HTRF assays
| Species of menin . | IC50 ± SEM (nM) . | pIC50 ± SEM (nM) . |
|---|---|---|
| Human | 0.1 ± 0.016 (n = 10) | 10.084 ± 0.081 (n = 10) |
| Mouse | 0.045 ± 0.002 (n = 2) | 10.347 ± 0.019 (n = 2) |
| Dog | ≤0.066 ± 0.004 (n = 3) | ≥10.180 ± 0.025 (n = 3) |
| Species of menin . | IC50 ± SEM (nM) . | pIC50 ± SEM (nM) . |
|---|---|---|
| Human | 0.1 ± 0.016 (n = 10) | 10.084 ± 0.081 (n = 10) |
| Mouse | 0.045 ± 0.002 (n = 2) | 10.347 ± 0.019 (n = 2) |
| Dog | ≤0.066 ± 0.004 (n = 3) | ≥10.180 ± 0.025 (n = 3) |
HTRF, homogeneous time-resolved fluorescence; pIC50, negative log10 of the IC50; SEM, standard error of the mean.
A crystal structure was solved to elucidate the binding mode of JNJ-75276617 to menin (Figure 1B). The 3-Å resolution data showed good electron density for JNJ-75276617 and allowed definitive placement of JNJ-75276617 in the KMT2A-binding site of menin occupying the F9, P10, and P13 pockets (Figure 1B).35 In addition to a well-established hydrogen bond with Tyr281,36 a hydrogen bond (salt-bridge) with Asp290 side chain represents a unique feature of the binding mode of JNJ-75276617. Additionally, there is extensive shape complementarity between JNJ-75276617 and the peptide binding site, driving numerous hydrophobic interactions.
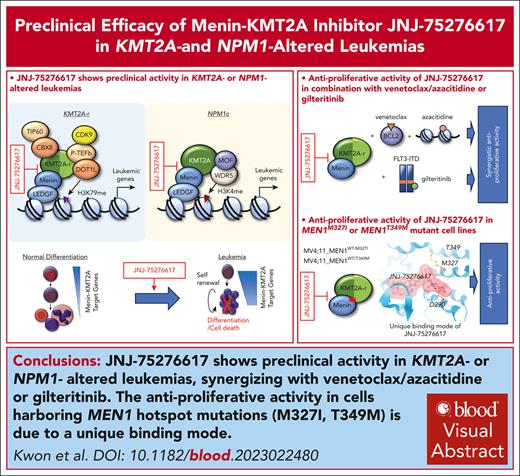
JNJ-75276617 decreased menin binding to target promoters and reduced menin-KMT2A target gene expression
The effect of JNJ-75276617 on menin occupancy of selected target gene promoters was evaluated by chromatin immunoprecipitation–quantitative polymerase chain reaction in cell lines bearing either KMT2A-r (MOLM-14) or NPM1c mutation (OCI-AML3). Signals for menin showed strong enrichment over background at all menin-KMT2A target gene promoters tested. Compared with DMSO control, menin binding was decreased in MOLM-14 and OCI-AML3 cells treated with JNJ-75276617 at concentrations from 0.1 to 1.0 μM. In all cases, the fold decreases fell outside the 95% confidence interval of the DMSO control, confirming inhibition of menin-KMT2A complex formation on chromatin (Figure 2A).
JNJ-75276617 inhibits menin-KMT2A complex association with chromatin, downregulates menin-KMT2A target genes, and induces myeloid cell differentiation in KMT2A-altered and NPM1-mutant AML cells. (A) A chromatin immunoprecipitation–quantitative polymerase chain reaction (qPCR) assay was performed to assess the binding of menin to target gene promoters (MEIS1, HOXA9, and HOXA10) in response to treatment with JNJ-75276617 for 48 hours. All qPCRs were performed in triplicate. Signals for menin binding in the KMT2A-r MOLM-14 or NPM1c OCI-AML3 samples were normalized to input values, and binding events per 1000 cells were calculated. Compared with dimethyl sulfoxide (DMSO) control, menin binding was decreased in MOLM-14 and OCI-AML3 cells treated with JNJ-75276617 at concentrations from 0.1 to 1.0 μM (MEIS1, 3.2- to 9.8-fold in MOLM-14, 1.4- to 2.6-fold in OCI-AML3; homeobox gene A9 [HOXA9], 2.4- to 3.4-fold in MOLM-14, 14.0- to 24.7-fold in OCI-AML3; homeobox gene A10 [HOXA10], 1.6- to 2.2-fold in MOLM-14, 5.2- to 10.3-fold in OCI-AML3); in all cases, the fold decreases fall outside the 95% confidence interval (CI) of the DMSO control. (B) JNJ-75276617 inhibited expression of menin-KMT2A target genes and increased expression of differentiation genes in KMT2A-altered and NPM1-mutant AML cells. Cells were incubated with the indicated concentrations of JNJ-75276617 for either 48 (MOLM-14) or 72 (OCI-AML3) hours. Relative expression of menin-KMT2A target genes and differentiation markers was calculated by dividing the normalized values of the treated samples by the normalized value of the DMSO control. The experiment was performed 3 times, and the error bars represent the mean ± standard deviation. (C,D) Differential expression of menin-KMT2A target genes and myeloid cell differentiation signature in response to JNJ-75276617 in various leukemia cells. Samples were prepared in duplicate. Microarray analysis was performed on leukemia cell lines in response to JNJ-75276617 treatment for 48 hours. Representative menin-KMT2A PD markers are shown in volcano plots (C). Gene sets involved in myeloid cell differentiation were enriched on treatment of JNJ-75276617 in KMT2A-r (MOLM-14, MV4-11) and NPM1c (OCI-AML3) cell lines as denoted by enrichment score (red), whereas no gene set enrichment was found in KMT2A/NPM1-WT (HL-60, KO-52, and K-562) cells (D). The experiment was performed once.
JNJ-75276617 inhibits menin-KMT2A complex association with chromatin, downregulates menin-KMT2A target genes, and induces myeloid cell differentiation in KMT2A-altered and NPM1-mutant AML cells. (A) A chromatin immunoprecipitation–quantitative polymerase chain reaction (qPCR) assay was performed to assess the binding of menin to target gene promoters (MEIS1, HOXA9, and HOXA10) in response to treatment with JNJ-75276617 for 48 hours. All qPCRs were performed in triplicate. Signals for menin binding in the KMT2A-r MOLM-14 or NPM1c OCI-AML3 samples were normalized to input values, and binding events per 1000 cells were calculated. Compared with dimethyl sulfoxide (DMSO) control, menin binding was decreased in MOLM-14 and OCI-AML3 cells treated with JNJ-75276617 at concentrations from 0.1 to 1.0 μM (MEIS1, 3.2- to 9.8-fold in MOLM-14, 1.4- to 2.6-fold in OCI-AML3; homeobox gene A9 [HOXA9], 2.4- to 3.4-fold in MOLM-14, 14.0- to 24.7-fold in OCI-AML3; homeobox gene A10 [HOXA10], 1.6- to 2.2-fold in MOLM-14, 5.2- to 10.3-fold in OCI-AML3); in all cases, the fold decreases fall outside the 95% confidence interval (CI) of the DMSO control. (B) JNJ-75276617 inhibited expression of menin-KMT2A target genes and increased expression of differentiation genes in KMT2A-altered and NPM1-mutant AML cells. Cells were incubated with the indicated concentrations of JNJ-75276617 for either 48 (MOLM-14) or 72 (OCI-AML3) hours. Relative expression of menin-KMT2A target genes and differentiation markers was calculated by dividing the normalized values of the treated samples by the normalized value of the DMSO control. The experiment was performed 3 times, and the error bars represent the mean ± standard deviation. (C,D) Differential expression of menin-KMT2A target genes and myeloid cell differentiation signature in response to JNJ-75276617 in various leukemia cells. Samples were prepared in duplicate. Microarray analysis was performed on leukemia cell lines in response to JNJ-75276617 treatment for 48 hours. Representative menin-KMT2A PD markers are shown in volcano plots (C). Gene sets involved in myeloid cell differentiation were enriched on treatment of JNJ-75276617 in KMT2A-r (MOLM-14, MV4-11) and NPM1c (OCI-AML3) cell lines as denoted by enrichment score (red), whereas no gene set enrichment was found in KMT2A/NPM1-WT (HL-60, KO-52, and K-562) cells (D). The experiment was performed once.
In addition, we used a Quantigene multiplex assay to measure the effects of JNJ-75276617 on the mRNA expression of multiple menin-KMT2A target and myeloid differentiation genes in 6 leukemia cell lines: 2 KMT2A-r (MOLM-14, MV4-11); 1 NPM1c (OCI-AML3); and 3 KMT2A/NPM1-wild type (WT) (KO-52, HL-60, and K562) (Figure 2B; supplemental Figure 2). JNJ-75276617 strongly suppressed expression of MEIS1 in all KMT2A-r and NPM1c AML cell lines; only minor changes were observed at higher doses in KMT2A/NPM1-WT cell lines. Moreover, JNJ-75276617 led to the reduction of other menin-KMT2A target genes (myocyte-specific enhancer factor 2C [MEF2C], Pre-B-cell leukemia homeobox 3 [PBX3], FLT3, jumonji domain-containing 1C [JMJD1C]), and increased expression of differentiation markers (myeloid cell nuclear differentiation antigen [MNDA] and integrin subunit α M [ITGAM/CD11b]) in KMT2A-r and NPM1c cell lines.
To validate the specificity and on-target activity of JNJ-75276617, we tested the less active enantiomer, JNJ-75303722. JNJ-75303722 demonstrated modest decreases in expression of menin-KMT2A target genes at μM concentrations (supplemental Figure 3), consistent with a 28-fold reduced affinity in the homogeneous time-resolved fluorescence assay for JNJ-75303722 compared with JNJ-75276617.
To investigate gene and pathway differential expression after treatment with JNJ-75276617, we analyzed broad gene expression changes across different leukemia cell lines using a whole-genome microarray platform. JNJ-75276617 reduced various menin-KMT2A target genes, including MEIS1 and PBX3 in KMT2A-r and NPM1c AML cells compared with KMT2A/NPM1-WT cell lines (Figure 2C; supplemental Figure 4). Gene set enrichment analysis was used for interpreting differential gene expression using the Gene Ontology pathways of the Molecular Signature Database (Figure 2D). Among the top differentially expressed genes, gene set enrichment analysis identified gene sets involved in myeloid cell differentiation in KMT2A-r and NPM1c AML cells (in contrast to KMT2A/NPM1-WT cell lines), consistent with the observed increase in myeloid differentiation markers (Figure 2B). On the basis of these findings, it was hypothesized that JNJ-75276617 may promote myeloid differentiation of leukemic cells.
JNJ-75276617 inhibited proliferation and induced apoptosis in AML and B-ALL cell lines
The antiproliferative activity of JNJ-75276617 was determined in a panel of 8 AML cell lines, including 4 from pediatric patients (KMT2A-r [MOLM-14, MOLM-13, MV4-11, and THP-1], KMT2A-PTD [EOL-1], NPM1c [OCI-AML3], and KMT2A/NPM1-WT [KO-52, HL-60]), 1 KMT2A-r B-ALL (RS4:11), and 1 KMT2A/NPM1-WT chronic myeloid leukemia cell line (K562). JNJ-75276617 led to antiproliferative activity in all KMT2A-r AML cell lines (IC50 <0.1 μM), except THP-1 (IC50 >15 μM) (Figure 3A,B). Interestingly, THP-1 cells showed notably low expression of menin-KMT2A target genes (data not shown) and harbor a neuroblastoma RAS (NRAS) G12D mutation leading to constitutive activation of the MEK/extracellular signal-regulated kinase pathway.37 Antiproliferative activity of JNJ-75276617 was observed in the NPM1c OCI-AML3 (IC50 = 0.045 μM), KMT2A-PTD EOL-1 (IC50 = 0.116 μM), and KMT2A-r B-ALL RS4:11 (IC50 = 0.040 μM) cell lines (Figure 3B). Normal nonproliferating cells, such as peripheral blood mononuclear cells, were not impacted by JNJ-75276617 up to 10 μM for 7 days (supplemental Table 2).
JNJ-75276617 inhibits proliferation and induces differentiation and apoptosis of AML and B-ALL cells with KMT2A alteration and NPM1 mutations and morphologic differentiation of KMT2A-AF9–transduced mouse bone marrow cells. (A) The antiproliferative activity of JNJ-75276617 was determined in a panel that included 8 AML cell lines (KMT2A-r [MOLM-14, MOLM-13, MV4-11, and THP-1], KMT2A–partial tandem duplication [PTD; EOL-1], NPM1c [OCI-AML3], KMT2A/NPM1-WT [KO-52 and HL-60]), 1 leukemia B-ALL cell line with KMT2A-r (RS4:11), and a KMT2A/NPM1-WT chronic myeloid leukemia (CML) cell line (K562). MOLM-14, MOLM-13, MV4-11, and THP-1 were originally derived from pediatric patients. Cells were treated with JNJ-75276617 for 8 days, and spheroid-like growth was measured in real time by live-cell imaging. Results are shown from a representative experiment in which all cell lines were evaluated in parallel (A). Absolute IC50 and the mean ± standard deviation (SD) values were calculated as percentage change in confluence to DMSO-treated cells. The experiment was performed at least 3 times for each cell line (B). (C,D) Flow cytometry analysis of differentiation and apoptosis in KMT2A-r and NPM1c AML cells following treatment with various concentrations of JNJ-75276617 for 7 days (C). Expression of differentiation markers CD14 and CD11b was evaluated in the viable cell population only, and apoptosis was evaluated in the total cell population (D). Duplicate samples were tested for each condition, and bars represent the mean ± SD. The experiment was performed 3 times. (E) Effect of JNJ-75276617 on morphologic differentiation of KMT2A-AF9–transformed mouse BM cells was examined by May-Grünwald Giemsa staining. KMT2A-AF9–transformed mouse BM cells were treated with DMSO or 200 nM JNJ-75276617 for 10 days. Condensed nuclei, a readout of neutrophil-like morphologic differentiation, were counted after 10-day JNJ-75276617 treatment from 3 independently captured images. Representative images are shown. The experiment was performed once.
JNJ-75276617 inhibits proliferation and induces differentiation and apoptosis of AML and B-ALL cells with KMT2A alteration and NPM1 mutations and morphologic differentiation of KMT2A-AF9–transduced mouse bone marrow cells. (A) The antiproliferative activity of JNJ-75276617 was determined in a panel that included 8 AML cell lines (KMT2A-r [MOLM-14, MOLM-13, MV4-11, and THP-1], KMT2A–partial tandem duplication [PTD; EOL-1], NPM1c [OCI-AML3], KMT2A/NPM1-WT [KO-52 and HL-60]), 1 leukemia B-ALL cell line with KMT2A-r (RS4:11), and a KMT2A/NPM1-WT chronic myeloid leukemia (CML) cell line (K562). MOLM-14, MOLM-13, MV4-11, and THP-1 were originally derived from pediatric patients. Cells were treated with JNJ-75276617 for 8 days, and spheroid-like growth was measured in real time by live-cell imaging. Results are shown from a representative experiment in which all cell lines were evaluated in parallel (A). Absolute IC50 and the mean ± standard deviation (SD) values were calculated as percentage change in confluence to DMSO-treated cells. The experiment was performed at least 3 times for each cell line (B). (C,D) Flow cytometry analysis of differentiation and apoptosis in KMT2A-r and NPM1c AML cells following treatment with various concentrations of JNJ-75276617 for 7 days (C). Expression of differentiation markers CD14 and CD11b was evaluated in the viable cell population only, and apoptosis was evaluated in the total cell population (D). Duplicate samples were tested for each condition, and bars represent the mean ± SD. The experiment was performed 3 times. (E) Effect of JNJ-75276617 on morphologic differentiation of KMT2A-AF9–transformed mouse BM cells was examined by May-Grünwald Giemsa staining. KMT2A-AF9–transformed mouse BM cells were treated with DMSO or 200 nM JNJ-75276617 for 10 days. Condensed nuclei, a readout of neutrophil-like morphologic differentiation, were counted after 10-day JNJ-75276617 treatment from 3 independently captured images. Representative images are shown. The experiment was performed once.
To validate the specificity and on-target activity of JNJ-75276617, we explored the antiproliferative activity of the enantiomer, JNJ-75303722. Notably, JNJ-75303722 exhibited significantly less antiproliferative effects, with IC50s > 1μM (supplemental Figure 3C).
To assess if JNJ-75276617 treatment resulted in AML and B-ALL cell death, flow cytometric analysis of apoptosis was conducted by annexin V and live/dead staining. JNJ-75276617 treatment induced apoptosis in MOLM-14 and OCI-AML3 AML cells, as well as RS4;11 B-ALL cells, in a dose-dependent manner after 7 days of treatment (Figure 3C).
JNJ-75276617 induced AML cell differentiation
To evaluate myeloid differentiation of AML cells on treatment with JNJ-75276617, we performed immunophenotypic analysis in KMT2A-r and NPM1c cells treated with increasing concentrations of JNJ-75276617 for up to 7 days. Both AML cell lines (MOLM-14, OCI-AML3) showed increased expression of the CD11b and CD14 differentiation markers after 3 days of treatment; the number of cells expressing CD11b or CD14 was further increased after 7 days of treatment (Figure 3D; supplemental Figure 5). The absence of apoptosis induction at day 3 suggests that JNJ-75276617 may induce differentiation before apoptosis (Figure 3D; supplemental Figure 5). Myeloid differentiation markers were not observed in B-ALL RS4;11 cells (Figure 3D; supplemental Figure 5).
To further investigate the effects of JNJ-75276617 on myeloid differentiation, experiments were conducted using May-Grünwald Giemsa staining to assess morphologic changes in KMT2A-AF9 fusion gene-transduced mouse bone marrow cells. After 10 days of treatment, cells treated with 200 nM JNJ-75276617 exhibited a significant increase in neutrophil-like morphology compared with the DMSO control group, as evidenced by condensed nuclei indicated by arrows (JNJ-75276617: 28.9% ± 2.6% of total analyzed cells; DMSO: 10.65% ± 1.35%) (Figure 3E). Collectively, the in vitro data across human AML cell lines and KMT2A-AF9–transduced mouse bone marrow cells indicate that JNJ-75276617 mediates antileukemic activity by induction of myeloid differentiation.
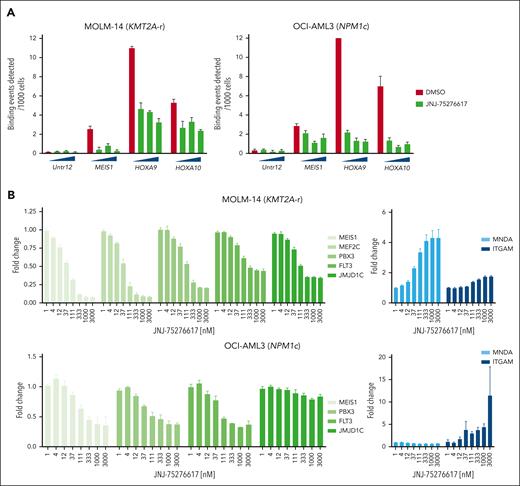
JNJ-75276617 inhibited proliferation and induced apoptosis in AML and B-ALL patient samples
The antiproliferative activity of JNJ-75276617 against primary AML patient samples was tested in cocultures of mononuclear cells enriched by CD34+ or CD117+ and MS5 murine stromal cells. Cocultures were demidepopulated 7 or 14 days after the addition of JNJ-75276617 and analyzed by flow cytometry. JNJ-75276617 demonstrated potent antiproliferative activity against 6 of 13 NPM1c patient samples at days 7 and 14. Furthermore, JNJ-75276617 induced the expression of CD11b in NPM1c samples (representative result shown in Figure 4A [S. M. Hogeling, M. Le Duy, N. La Rose, M. C. Kwon, A. T. J. Wierenga, F. A. J. v. d. Heuvel, V. v. d. Boom, A. Kuchnio, U. Philippar, G. Huls, and J. J. Schuringa, unpublished data, September 2023]).
JNJ-75276617 inhibits proliferation and induces differentiation and apoptosis of KMT2A-altered and NPM1-mutant primary AML or B-ALL patient samples. (A) The effect of JNJ-75276617 on viability of primary AML patient samples was tested in cocultures of mononuclear cells from primary AMLs, enriched by CD34+ or CD117+ selection, and MS5 murine stromal cells. Cell number was counted after 5 different human AML patient samples (with the noted mutations) were treated with JNJ-75276617 at the indicated concentrations for 7 days. CD11b staining was performed to assess differentiation in response to treatment. Test conditions were run in triplicate. Average values of DMSO control are indicated by horizontal dashed lines. Significant difference compared with DMSO control is indicated (∗P < .001, ∗∗P < .01, ∗∗∗P < .05). (B) The effect of JNJ-75276617 on viability of primary KMT2A-r B-ALL patient sample was tested in liquid culture. Cell number was counted after treatment of JNJ-75276617 at the indicated concentrations for 8 days. Test conditions were run in triplicate. Average values of DMSO control are indicated by horizontal dashed lines. Significant difference compared with DMSO control is indicated (∗P < .001). (C) Flow cytometry analysis of apoptosis in primary KMT2A-r B-ALL patient sample following treatment with various concentrations of JNJ-75276617 for 8 days. On day 8, cells were stained with CD45-PECy7 and annexin V APC, in a 96-well plate, and were incubated for 30 minutes at 4°C. Apoptosis was evaluated in the total cell population. Triplicate samples were tested for each condition, and bars represent the mean ± standard deviation. The experiment was performed once.
JNJ-75276617 inhibits proliferation and induces differentiation and apoptosis of KMT2A-altered and NPM1-mutant primary AML or B-ALL patient samples. (A) The effect of JNJ-75276617 on viability of primary AML patient samples was tested in cocultures of mononuclear cells from primary AMLs, enriched by CD34+ or CD117+ selection, and MS5 murine stromal cells. Cell number was counted after 5 different human AML patient samples (with the noted mutations) were treated with JNJ-75276617 at the indicated concentrations for 7 days. CD11b staining was performed to assess differentiation in response to treatment. Test conditions were run in triplicate. Average values of DMSO control are indicated by horizontal dashed lines. Significant difference compared with DMSO control is indicated (∗P < .001, ∗∗P < .01, ∗∗∗P < .05). (B) The effect of JNJ-75276617 on viability of primary KMT2A-r B-ALL patient sample was tested in liquid culture. Cell number was counted after treatment of JNJ-75276617 at the indicated concentrations for 8 days. Test conditions were run in triplicate. Average values of DMSO control are indicated by horizontal dashed lines. Significant difference compared with DMSO control is indicated (∗P < .001). (C) Flow cytometry analysis of apoptosis in primary KMT2A-r B-ALL patient sample following treatment with various concentrations of JNJ-75276617 for 8 days. On day 8, cells were stained with CD45-PECy7 and annexin V APC, in a 96-well plate, and were incubated for 30 minutes at 4°C. Apoptosis was evaluated in the total cell population. Triplicate samples were tested for each condition, and bars represent the mean ± standard deviation. The experiment was performed once.
Furthermore, JNJ-75276617 was evaluated in a primary KMT2A-r B-ALL patient sample, demonstrating significant antiproliferative effects and dose-dependent induction of apoptosis (Figure 4B,C).
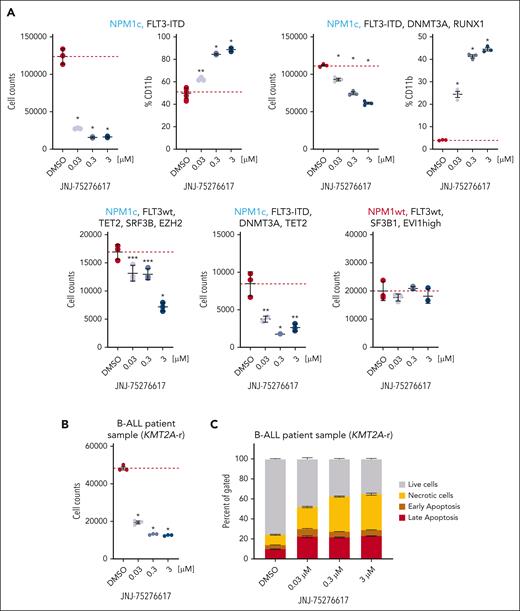
JNJ-75276617 treatment resulted in tumor regressions and modulated menin-KMT2A target gene expression in mice bearing human KMT2A-r AML
Following oral dosing in mice, JNJ-75276617 (as oxalate salt) showed adequate oral bioavailability (34%) and plasma half-life (≈9 hours) to explore dose-response relationships in vivo. Representative pharmacokinetic profiles are shown in Figure 5A. The antitumor activity of JNJ-75276617 was evaluated in subcutaneous MOLM-14 AML xenograft bearing mice. Daily oral JNJ-75276617 treatment resulted in dose-dependent tumor regressions of 70%, 97%, and 99% at 30, 50, and 100 mg/kg, respectively, after 5 weeks of treatment (P < .0001 each) (Figure 5B). At the 50- and 100-mg/kg JNJ-75276617 dose level, 10 of 10 and 9 of 9 tumors failed to regrow after 3 months off treatment, respectively. Tumor regression in the subcutaneous MOLM-14 model was associated with a reduction of mRNA expression of the menin-KMT2A target gene MEIS1 after 3 or 11 days of dosing (Figure 5C). Furthermore, JNJ-75276617 treatment decreased menin-KMT2A target genes FLT3, MEF2C, and PBX3 as well as increased expression of the differentiation genes MNDA and ITGAM/CD11b (Supplementary Figure 6A). The antitumor efficacy of JNJ-75276617 was further assessed using PDX models of KMT2A-r AML in mice. JNJ-75276617 treatment led to a statistically significant increased lifespan (ILS) by >89% across all dose levels (P < .05) in the KMT2A-AF6 AML PDX model (CBAM-68552) (Figure 5D). This effect correlated with a significant decrease in human CD45+ (hCD45+) leukemic cells and an increased expression of differentiation markers (eg, CD11b, CD13, and CD14) (Figure 5E; supplemental Figure 6B).
JNJ-75276617 alters menin-KMT2A target gene expression and prolongs survival of mice bearing NPM1c and KMT2A-r xenografts. (A) Plasma concentrations following a single oral administration of 5 mg/kg (gray squares) or 50 mg/kg (black squares) in mice. The dashed black line represents the IC50 from the in vitro MEIS1 expression assay (45 ± 6 nM, supplemental Figure 3B). (B) Mice bearing subcutaneous (SC) KMT2A-r MOLM-14 xenografts were treated with JNJ-75276617 at the dose levels indicated for 5 weeks. Line underneath the x axis indicates the dosing period. Data are displayed while at least two-thirds of animals remained in the group (n = 9-10/group). ∗ denotes significant tumor regression (P ≤ .05) from initial tumor volume after JNJ-75276617 treatment. (C) Mice bearing SC MOLM-14 xenografts were treated with JNJ-75276617 at the dose levels indicated for 3 or 11 days (n = 2-5/group). Tumors were harvested 16 hours after the last dose, and MEIS1 mRNA levels were assessed. Expression values are calculated relative to tumors treated with vehicle. Bar graphs represent the mean ± standard deviation (SD). ∗P < .0001. (D-G) Mice engrafted with patient-derived AML xenografts harboring KMT2A-r mutation: CBAM-68552 (D,E) or NPM1c: LEXFAM-2734 (F) or AM7577 (G) were treated with JNJ-75276617 at the dose levels indicated. (E) Bone marrows from CBAM-68552–engrafted mice following 4 weeks of drug treatment were analyzed for differentiation markers (human CD11b, CD13, and CD14) by flow cytometry (n = 3). (H,I) Mice engrafted with B-ALL xenografts with KMT2A-r (CBAB-62871; n = 7/group) were treated with JNJ-75276617 for 6 weeks. (I) Bone marrows from CBAB-62871-engrafted mice following 3 weeks of drug treatment were analyzed for differentiation markers (human CD11b, CD13, and CD14) by flow cytometry (n = 3). Individual values are shown as a dot plot, whereas bar graphs represent the mean ± SD. (D,F,G,H) Line underneath the x axis indicates the dosing period. ∗ denotes significant difference (P ≤ .05) in survival between treatment with JNJ-75276617 and vehicle control (n = 7-10/group). (E,I) ∗ denotes a significant difference vs vehicle control (P < .05).
JNJ-75276617 alters menin-KMT2A target gene expression and prolongs survival of mice bearing NPM1c and KMT2A-r xenografts. (A) Plasma concentrations following a single oral administration of 5 mg/kg (gray squares) or 50 mg/kg (black squares) in mice. The dashed black line represents the IC50 from the in vitro MEIS1 expression assay (45 ± 6 nM, supplemental Figure 3B). (B) Mice bearing subcutaneous (SC) KMT2A-r MOLM-14 xenografts were treated with JNJ-75276617 at the dose levels indicated for 5 weeks. Line underneath the x axis indicates the dosing period. Data are displayed while at least two-thirds of animals remained in the group (n = 9-10/group). ∗ denotes significant tumor regression (P ≤ .05) from initial tumor volume after JNJ-75276617 treatment. (C) Mice bearing SC MOLM-14 xenografts were treated with JNJ-75276617 at the dose levels indicated for 3 or 11 days (n = 2-5/group). Tumors were harvested 16 hours after the last dose, and MEIS1 mRNA levels were assessed. Expression values are calculated relative to tumors treated with vehicle. Bar graphs represent the mean ± standard deviation (SD). ∗P < .0001. (D-G) Mice engrafted with patient-derived AML xenografts harboring KMT2A-r mutation: CBAM-68552 (D,E) or NPM1c: LEXFAM-2734 (F) or AM7577 (G) were treated with JNJ-75276617 at the dose levels indicated. (E) Bone marrows from CBAM-68552–engrafted mice following 4 weeks of drug treatment were analyzed for differentiation markers (human CD11b, CD13, and CD14) by flow cytometry (n = 3). (H,I) Mice engrafted with B-ALL xenografts with KMT2A-r (CBAB-62871; n = 7/group) were treated with JNJ-75276617 for 6 weeks. (I) Bone marrows from CBAB-62871-engrafted mice following 3 weeks of drug treatment were analyzed for differentiation markers (human CD11b, CD13, and CD14) by flow cytometry (n = 3). Individual values are shown as a dot plot, whereas bar graphs represent the mean ± SD. (D,F,G,H) Line underneath the x axis indicates the dosing period. ∗ denotes significant difference (P ≤ .05) in survival between treatment with JNJ-75276617 and vehicle control (n = 7-10/group). (E,I) ∗ denotes a significant difference vs vehicle control (P < .05).
JNJ-75276617 significantly increased the survival of mice bearing NPM1c AML
The antitumor efficacy of JNJ-75276617 was further evaluated in PDX models of NPM1c AML in mice. LEXFAM-2734 NPM1c leukemia-bearing mice treated with 30, 50, or 100 mg/kg JNJ-75276617 did not reach median survival by the end of the study observation period (median survival >133 days) compared with 74 days for the vehicle-treated group, and 123 days for the 10-mg/kg JNJ-75276617 group (Figure 5F). JNJ-75276617 treatment resulted in statistically significant ILS of LEXFAM-2734 tumor-bearing mice by >66% for the 10-mg/kg group, and >79% for the 30-, 50-, and 100-mg/kg JNJ-75276617–treated groups compared with control mice (P < .0001). AM7577 NPM1c leukemia-bearing mice treated with JNJ-75276617 also did not reach median survival by the end of the study observation period (median survival >147 days), compared with 53.5 days for the vehicle-treated control group (Figure 5G). JNJ-75276617 treatment resulted in statistically significant ILS of AM7577 leukemia-bearing mice by >169% at all dose levels compared with control mice (P < .001). Moreover, JNJ-75276617 treatment led to statistically significant ILS of OCI-AML3 NPM1c AML-disseminated mouse model and DFAM-22359 NPM1c leukemia-bearing mice across all dose levels (supplemental Table 3).
JNJ-75276617 significantly increased the survival of mice bearing KMT2A-r B-ALL
The antitumor efficacy of JNJ-75276617 was further tested in a KMT2A-AF4 B-ALL PDX model (CBAB-62871). Mice treated with daily, oral 30 or 100 mg/kg JNJ-75276617 showed dose-dependent and statistically significant ILS by >48% for the 30-mg/kg, and >160% for the 100-mg/kg JNJ-75276617–treated groups compared with control mice (P = .002) (Figure 5H). Tumor burden was assessed after 21 days of treatment, and JNJ-75276617 potently reduced the leukemia cell burden in the bone marrow and increased expression of differentiation markers (eg, CD11b, CD13, and CD14) (Figure 5I; supplemental Figure 6C).
Efficacy of JNJ-75276617 in combination with FLT3 inhibition in KMT2A-r AML
Previous studies indicated that the combination of menin-KMT2A and Fms Related Receptor Tyrosine Kinase 3 (FLT3) inhibition may be a promising therapeutic strategy for patients with NPM1c or KMT2A-r leukemia and concurrent FLT3 mutation.38-40 Therefore, we evaluated the combination of the FLT3 inhibitor gilteritinib with JNJ-75276617 in KMT2A-r/FLT3 mutant MOLM-13 AML. We observed a significant synergistic effect (P < .001) on MOLM-13 cell proliferation at low doses of gilteritinib (5-10 nM) in combination with JNJ-75276617 (0.05-10 μM) (Figure 6A). These results support the combination of gilteritinib with JNJ-75276617 and provide additional evidence for the combined inhibition of menin-KMT2A and FLT3 as a potential therapeutic option for KMT2A-r FLT3 mutant leukemia.
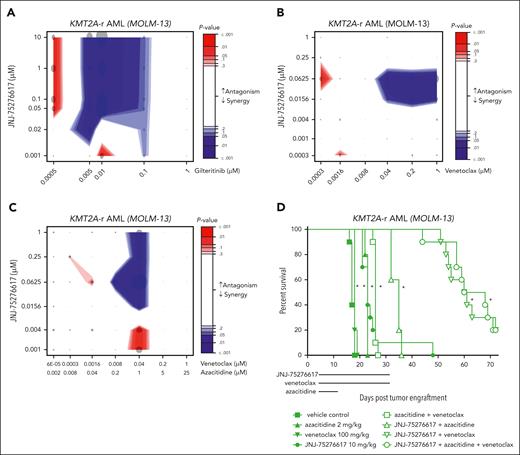
JNJ-75276617 exhibits synergistic effects with gilteritinib or venetoclax and azacitidine in AML. (A) KMT2A-r MOLM-13 cells were incubated with the indicated drug combination concentrations for 6 days (JNJ-75276617 and gilteritinib) in technical triplicates. The combination effect was calculated by extended Biochemically Intuitive Generalized Loewe (BIGL) package, with results shown under highest single agent (HSA) null model. Data from 3 independent experiments were pooled and analyzed. Combination effect is represented by the contour plots, with indicating antagonism (red area) and synergy (blue area) as represented by the intensity scale. Dot is proportional to the increase in effect compared with the null, and color intensity is statistical effect. (B) KMT2A-r MOLM-13 cells were treated with JNJ-75276617 for 8 days and venetoclax for 4 days in quadruplicate. The combination effect was calculated by extended BIGL package, with results shown under HSA null model. Data from 2 independent experiments were pooled and analyzed. Combination effect is represented by the contour plots, with indicating antagonism (red area) and synergy (blue area) as represented by the intensity scale. Dot is proportional to the increase in effect compared with the null and color intensity is statistical effect. (C) KMT2A-r MOLM-13 cells were incubated with the indicated drug combination concentrations for 8 days (JNJ-75276617 and azacitidine) and for 4 days (venetoclax) in quadruplicate. The combination effect was calculated by extended BIGL package, with results shown under HSA null model. Data from 2 independent experiments were pooled and analyzed. Combination effect is represented by the contour plots, with indicating antagonism (red area) and synergy (blue area) as represented by the intensity scale. Dot is proportional to the increase in effect compared with the null and color intensity is statistical effect. (D) Mice bearing disseminated MOLM-13 xenografts were treated with JNJ-75276617, venetoclax, azacitidine, or combinations at the dose levels indicated. Lines underneath the x axis indicate the dosing period for each drug. ∗ denotes significant difference (P ≤ .05) in survival compared with the vehicle control group.
JNJ-75276617 exhibits synergistic effects with gilteritinib or venetoclax and azacitidine in AML. (A) KMT2A-r MOLM-13 cells were incubated with the indicated drug combination concentrations for 6 days (JNJ-75276617 and gilteritinib) in technical triplicates. The combination effect was calculated by extended Biochemically Intuitive Generalized Loewe (BIGL) package, with results shown under highest single agent (HSA) null model. Data from 3 independent experiments were pooled and analyzed. Combination effect is represented by the contour plots, with indicating antagonism (red area) and synergy (blue area) as represented by the intensity scale. Dot is proportional to the increase in effect compared with the null, and color intensity is statistical effect. (B) KMT2A-r MOLM-13 cells were treated with JNJ-75276617 for 8 days and venetoclax for 4 days in quadruplicate. The combination effect was calculated by extended BIGL package, with results shown under HSA null model. Data from 2 independent experiments were pooled and analyzed. Combination effect is represented by the contour plots, with indicating antagonism (red area) and synergy (blue area) as represented by the intensity scale. Dot is proportional to the increase in effect compared with the null and color intensity is statistical effect. (C) KMT2A-r MOLM-13 cells were incubated with the indicated drug combination concentrations for 8 days (JNJ-75276617 and azacitidine) and for 4 days (venetoclax) in quadruplicate. The combination effect was calculated by extended BIGL package, with results shown under HSA null model. Data from 2 independent experiments were pooled and analyzed. Combination effect is represented by the contour plots, with indicating antagonism (red area) and synergy (blue area) as represented by the intensity scale. Dot is proportional to the increase in effect compared with the null and color intensity is statistical effect. (D) Mice bearing disseminated MOLM-13 xenografts were treated with JNJ-75276617, venetoclax, azacitidine, or combinations at the dose levels indicated. Lines underneath the x axis indicate the dosing period for each drug. ∗ denotes significant difference (P ≤ .05) in survival compared with the vehicle control group.
Efficacy of JNJ-75276617 in combination with venetoclax/azacitidine in KMT2A-r AML
Hypomethylating agents (eg, azacitidine) in combination with venetoclax have significantly improved clinical outcomes for patients with AML and have become a preferred frontline treatment for patients with AML ≥75 years of age, or for patients who have comorbidities that preclude intensive induction chemotherapy. Here, we evaluated preclinically whether JNJ-75276617 could enhance the mechanisms of action triggered by the venetoclax/azacitidine combination, which primarily drives apoptosis by displacing antiapoptotic proteins. Proliferation assays were performed in vitro to assess monotherapies and combinations using KMT2A-r MOLM-13 cells. The doublet combination of JNJ-75276617 plus venetoclax induced a significant increased and synergistic antiproliferative effect in vitro when compared with JNJ-75276617 or venetoclax monotherapy (Figure 6B). The triplet combination of JNJ-75276617 plus azacitidine and venetoclax also induced a synergistic antiproliferative effect that was significantly increased when compared with JNJ-75276617 monotherapy and the doublet combination of venetoclax plus azacitidine (Figure 6C).
In the disseminated KMT2A-r MOLM-13 model in mice, monotherapy treatment with JNJ-75276617 at 10 mg/kg and azacitidine at 2 mg/kg induced a significant ILS of 35% each compared with the vehicle control group (P < .0001 each), whereas 6% ILS was observed with 100 mg/kg venetoclax (P = .0076) (Figure 6D). The combination of azacitidine plus venetoclax resulted in a significant ILS of 53% compared with the vehicle control (P < .0001). Doublet combination treatment with JNJ-75276617 plus azacitidine or plus venetoclax resulted in a significant ILS of 106% or 256%, respectively, compared with the vehicle control (P < .0001 each). The JNJ-75276617 triplet combination with azacitidine plus venetoclax induced a significant ILS of 277%, compared with the vehicle control group (P < .0001). These results further showed that the doublet combinations of JNJ-75276617 plus venetoclax or azacitidine, or the triplet combination, provided a superior increase in survival compared with the clinical standard regimen of azacitidine plus venetoclax (P < .0001 each). In a disseminated NPM1c model (OCI-AML3), JNJ-75276617 doublet or triplet combinations with azacitidine and venetoclax also yielded superior survival vs the standard azacitidine plus venetoclax regimen; however, efficacy seemed most strongly driven by JNJ-75276617 (supplemental Figure 7). It is hypothesized that the triplet combination of JNJ-75276617 plus venetoclax and azacitidine could potentially provide a beneficial treatment option for KMT2A- or NPM1-altered AML.
Antiproliferative activity of JNJ-75276617 in MEN1M327I or MEN1T349M mutant cell lines
The menin-KMT2A inhibitor revumenib has shown promising clinical responses in KMT2A-r or NPM1-altered acute leukemias.30 However, some patients acquired resistance to revumenib because of somatic mutations in MEN1 impacting the inhibitor-menin interface.31KMT2A-r cell lines engineered to express mutant menin mutations (M327I or T349M) into the endogenous MEN1-coding sequence were found to be resistant to the antiproliferative effects of several menin-KMT2A inhibitors (eg, revumenib, MI3454, and DS-1594).31
To determine whether these menin mutations decreased binding of JNJ-75276617, a competitive KMT2A fluorescence polarization binding assay was performed. Interestingly, JNJ-75276617 binding affinity (IC50, inhibitor constant [Ki]) increased only minimally for the M327I mutant menin, whereas a more pronounced increase was observed for T349M mutant menin compared with WT menin. Overall, binding of JNJ-75276617 to WT, M327I, and T349M menin is potent with apparent inhibition constants (Ki) in the sub to double digit nM range (Figure 7A). Consistent with these results, the antiproliferative activity of JNJ-75276617 in MEN1 mutant M327I or T349M KMT2A-r MV4-11 AML cell lines demonstrated a robust decrease in viable cell number in MEN1M327I or MEN1T349M mutant cells, with IC50 values in the double-digit nM range (Figure 7B). Notably, JNJ-75276617 retained strong antiproliferative activity in cells expressing MEN1M327I or MEN1T349M mutations that conferred resistance to other menin-KMT2A inhibitors, such as revumenib. These data suggest limited impact of M327I or T349M MEN1 mutations on the binding affinity of JNJ-75276617 to menin (Figure 7A,B).31 Interestingly, within our initial in vitro CRISPR/Cas9 base-editing screens, M327 and T349 were not identified as sites of resistance to JNJ-75276617 (data not shown).
JNJ-75276617 exhibits potent antiproliferation activity in MEN1-mutant cell lines resistant to other menin-KMT2A inhibitors. (A) Fluorescence polarization assay demonstrating the dose-dependent displacement of a KMT2A peptide from WT, M327I-mutant, and T349M-mutant menin on treatment with JNJ-75276617. Data are represented as mean ± standard deviation (SD), n = 3 independent replicates. (B) Evaluation of JNJ-75276617 for antiproliferation activity in MEN1M327I and MEN1T349M mutant KMT2A-r MV4-11 cell lines. Drug response was calculated on the basis of the luminescence for each menin inhibitor concentration relative to the DMSO sample after 10 days of JNJ-75276617 treatment. Data are represented as mean ± SD, n = 3 independent experiments. (C) Superimposition of a docking model of JNJ-75276617 (green sticks; docked on M327I-mutant menin chain A; Protein Data Bank Identifier [PDB] ID: 8E90; second rotamer of W346 [chain B] is displayed in gray lines). JNJ-75276617 bound to wild-type menin (this study; PDB ID: in deposition) is shown for reference in orange and its binding pocket as gray surface. Side chain of M327/I327 is shown as sticks. The distance between selected atoms of T349 and JNJ-75276617 is shown as orange dashed line. Residue numbering is according to transcript variant: NM_000244.
JNJ-75276617 exhibits potent antiproliferation activity in MEN1-mutant cell lines resistant to other menin-KMT2A inhibitors. (A) Fluorescence polarization assay demonstrating the dose-dependent displacement of a KMT2A peptide from WT, M327I-mutant, and T349M-mutant menin on treatment with JNJ-75276617. Data are represented as mean ± standard deviation (SD), n = 3 independent replicates. (B) Evaluation of JNJ-75276617 for antiproliferation activity in MEN1M327I and MEN1T349M mutant KMT2A-r MV4-11 cell lines. Drug response was calculated on the basis of the luminescence for each menin inhibitor concentration relative to the DMSO sample after 10 days of JNJ-75276617 treatment. Data are represented as mean ± SD, n = 3 independent experiments. (C) Superimposition of a docking model of JNJ-75276617 (green sticks; docked on M327I-mutant menin chain A; Protein Data Bank Identifier [PDB] ID: 8E90; second rotamer of W346 [chain B] is displayed in gray lines). JNJ-75276617 bound to wild-type menin (this study; PDB ID: in deposition) is shown for reference in orange and its binding pocket as gray surface. Side chain of M327/I327 is shown as sticks. The distance between selected atoms of T349 and JNJ-75276617 is shown as orange dashed line. Residue numbering is according to transcript variant: NM_000244.
A molecular model of JNJ-75276617 was built by computationally docking it on the MEN1M327I-mutant menin structure. In this modeled binding mode, JNJ-75276617 exhibited only a minor shift within the binding pocket (Figure 7C; supplemental Figure 8). Because the T349M residue is not in direct contact with JNJ-75276617, this mutation may result in repacking of the mutant menin protein to accommodate the bulkier side chain methionine residue and a minor shift of JNJ-75276617 within its binding site. It is possible that this minor shift would still maintain key van der Waals interactions as well as maintain the hydrogen bond (and salt bridge) with Asp290.
Discussion
The interaction between menin and KMT2A has been identified as a critical dependency in KMT2A- and NPM1-altered leukemias, and disrupting this interaction has emerged as a potential therapeutic strategy. Here, we demonstrate that JNJ-75276617 is a novel, potent, and selective inhibitor of the menin-KMT2A interaction, which displays significant preclinical activity against AML/B-ALL cell lines and primary patient samples bearing KMT2A- or NPM1 alterations.
JNJ-75276617 inhibits the interaction between menin-KMT2A and the association of the menin-KMT2A complex with the chromatin of target gene promoters, resulting in reduced expression of menin-KMT2A target genes, such as MEIS1 and FLT3. JNJ-75276617 treatment leads to increased expression of differentiation markers, such as integrin alpha M (ITGAM)/CD11b and myeloid cell nuclear differentiation antigen (MNDA), followed by apoptotic cell death. Additionally, JNJ-75276617 induced neutrophil-like morphologic differentiation in KMT2A-AF9 fusion gene-transduced mouse bone marrow cells. These findings suggest that JNJ-75276617 can overcome the blockade in differentiation that is characteristic of AML cells and induce apoptosis. As a result, JNJ-75276617 displayed potent antiproliferative activity across a range of AML/B-ALL cell lines and patient samples harboring KMT2A alterations or NPM1 mutations in vitro. Importantly, JNJ-75276617 had no/minimal impact on healthy counterpart cells. Moreover, JNJ-75276617 did not indicate any risks in hERG and induced pluripotent stem cell–derived cardiomyocyte assays, highlighting its selectivity.
In KMT2A- and NPM1-altered AML and B-ALL leukemia models in mice, JNJ-75276617 reduced leukemic burden and provided a significant dose-dependent survival benefit with accompanying decreased menin-KMT2A target gene and increased differentiation marker expression. Moreover, JNJ-75276617 demonstrated synergistic effects with gilteritinib in vitro in AML cells with KMT2A-r. Furthermore, the addition of JNJ-75276617 resulted in superior survival benefit vs the standard azacitidine plus venetoclax regimen. It is hypothesized that the triplet combination of JNJ-75276617, venetoclax, and azacitidine could potentially provide a beneficial treatment option for AML with KMT2A or NPM1 alterations.
The development of resistance to menin-KMT2A inhibitors by MEN1 mutations has emerged as a potential challenge in the treatment of acute leukemias with KMT2A or NPM1 alterations. Our studies revealed that MEN1 mutants MEN1M327I or MEN1T349M conferring resistance to other menin-KMT2A inhibitors were susceptible to JNJ-75276617. JNJ-75276617 has potent antiproliferative activity against MEN1M327I or MEN1T349M mutant cellular models in the double-digit nM range. Using a molecular model that computationally docked JNJ-75276617 on the MEN1M327I-mutant menin structure, JNJ-75276617 appeared to exhibit only a minor shift within the binding pocket. Similarly, only a moderate shift of JNJ-75276617 is expected within the MEN1T349M binding site, specifically as the T349 residue is not in direct contact with JNJ-75276617.
JNJ-75276617 appears to have a unique binding mode, especially in the flexible tail region of the compound, compared with other menin-KMT2A inhibitors (eg, revumenib).31 We hypothesize that such relative differences in binding modes can explain the differential relative impact of MEN1M327I or MEN1T349M mutations on the binding affinity of these menin-KMT2A inhibitors and resulting biological activity. Specifically, JNJ-75276617 does not depend on a key hydrogen bond with Trp346 (unlike revumenib and other menin-KMT2A inhibitors),31 and this key interaction is lost in the M327I mutant protein for revumenib and potentially other menin-KMT2A inhibitors engaging with a similar pharmacophore. According to our binding model and supporting experimental data, JNJ-75276617 would potentially be only slightly shifted in the binding site, without losing the key unique interactions in the binding pocket (eg, with Asp290), and therefore retaining antiproliferative activity.
In conclusion, JNJ-75276617 is a highly selective and potent menin-KMT2A inhibitor that disrupts the crucial menin-KMT2A protein complex and mediates significant preclinical activity against AML/B-ALL cell lines, primary patient samples, and cell line–derived xenograft and PDX models bearing KMT2A or NPM1 alterations. These data support the ongoing first-in-human trial investigating JNJ-75276617 as monotherapy for R/R acute leukemia harboring either KMT2A or NPM1 alterations (NCT04811560). Furthermore, the combination data of JNJ-75276617 plus azacitidine and venetoclax support the ongoing clinical trial investigating JNJ-75276617 in combination with AML-directed therapies (NCT05453903).
Acknowledgments
The authors thank Sebastian Karl Wandinger, Stephanie Blencke, and Tanja Wagner from Evotec GmbH for performing cellular target profiling assay with JNJ-75276617.
D.V.W. was supported by the German Research Foundation (DFG) grant WE 7304/1-1). F.P. was supported by DFG grant PE 3217/1-1), a Momentum Fellowship award by the Mark Foundation for Cancer Research, and a research grant from the Else Kröner-Fresenius-Stiftung (2021-EKEA.111). S.M.H. and J.J.S. were supported by a grant from the Dutch Cancer Foundation to J.J.S. (11013) and a strategic Public Private Partnership grant from Health Holland/TKI (LSHM200204). J.A.C. was supported by a K99/R00: Pathway to Independence Award (1K99CA279888-01) from the National Institutes of Health (NIH)/National Cancer Institute (NCI). S.A.A. was supported by NIH/NCI grants CA176745, CA259273, and CA066996.
Authorship
Contribution: M.C.K., J.W.T., O.Q., X.D., T.V., V.P., P.L.S., F.J., P.V., W.C., B.B., J.A.P., N.D., D.K., G.U., B.V., J.P.E., G.S.C., R.K., R.S., L.F., C.G., N.D., E.C.P., D.M.W., R.A., Y.E., J.J.S., S.A.A., P.J., L.B., K.P., and U.P designed the experiments; M.C.K., T.V., V.P., A.M., D.G., D.V.W., H.Y., J.A.C., C.J., F.P., S.M.H., W.C., V.K., F.E., B.B., P.J, L.B., K.V., J.A.P., N.D., G.U., B.V., and R.K. performed the experiments; M.C.K., J.W.T., O.Q., X.D., T.V., V.P., A.M., D.G., D.V.W., H.Y., J.A.C., F.P., S.M.H., P.L.S., F.J., P.V., W.C., V.K., F.E., B.B., P.J., L.B., K.V., S.E.A., J.A.P., A.K., N.D., D.K., G.U., B.V., J.P.E., G.S.C., R.K., R.S., L.F., C.G., N.D., E.C.P., D.M.W., R.A., Y.E., E.S.F., J.J.S., S.A.A., K.P., and U.P. analyzed and interpreted the data; and M.C.K., J.W.T., V.P., D.V.W, H.Y., D.G., P.L.S., F.J., W.C.,V.K., F.E., B.B., A.K., B.V., L.F., C.G., N.D., E.C.P., E.S.F., J.J.S., S.A.A., P.J., L.B., K.P., and U.P. wrote and edited the manuscript.
Conflict-of-interest disclosure: M.C.K., J.W.T., T.V., V.P., A.M., D.G., P.L.S., F.J., V.K., F.E., B.B., K.V., S.E.A., P.J., L.B., A.K., N.D., D.K., G.U., B.V., G.S.C., R.K., R.S., L.F., C.G., N.D., E.C.P., R.A., Y.E., K.P., and U.P. are currently employees of Janssen Research & Development and may own stock/stock options in Johnson & Johnson. O.Q., X.D., P.V., W.C., J.P.E., and D.M.W. are former employees of Janssen Research & Development and may own stock/stock options in Johnson & Johnson. J.W.T., O.Q., X.D., V.P., W.C., N.D., and G.U. are named as inventors on patent applications related to MENIN inhibition WO/2021/121327. M.C.K., J.W.T., O.Q., X.D., T.V., V.P., W.C., B.B., N.D., L.F., C.G., N.D., E.C.P., K.P., and U.P. are named as inventors on patent applications related to MENIN inhibition WO/2022/237719 and WO/2022/237720. S.A.A. has been a consultant and/or shareholder for Neomorph Inc, Imago Biosciences, Cyteir Therapeutics, C4 Therapeutics, Nimbus Therapeutics, and Accent Therapeutics. S.A.A. has received research support from Janssen and Syndax. S.A.A. is named as an inventor on a patent application related to MENIN inhibition WO/2017/132398A1. E.S.F. is a founder, scientific advisory board member, and equity holder of Civetta Therapeutics, Proximity Therapeutics, and Neomorph, Inc (also board of directors). He is an equity holder and scientific advisory board member for Avilar Therapeutics, Photys Therapeutics, and Ajax Therapeutics and an equity holder in Lighthorse Therapeutics. E.S.F. is a consultant to Novartis, EcoR1 capital, Odyssey, and Deerfield. The Fischer laboratory receives or has received research funding from Deerfield, Novartis, Ajax, Interline, Bayer, and Astellas. The remaining authors declare no competing financial interests.
Correspondence: Ulrike Philippar, Discovery Oncology, Janssen R&D, Turnhoutseweg 30, 2340 Beerse, Belgium; email: uphilipp@its.jnj.com.
References
Author notes
For original data, please contact uphilipp@its.jnj.com.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![Chemical and crystal structures of JNJ-75276617. (A) Chemical structure of JNJ-75276617. (B) Crystal structure of menin bound to JNJ-75276617 (Protein Data Bank Identifier [PDB ID]: in deposition). JNJ-75276617 is shown as ball and sticks, and selected side chains of menin are shown as sticks. Dotted lines represent key hydrogen-bonding interactions between the protein and the ligand, and ligand-binding pocket is depicted as gray surface. Menin residue numbering is according to transcript variant: NM_000244.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/144/11/10.1182_blood.2023022480/4/m_blood_bld-2023-022480-gr1.jpeg?Expires=1768346814&Signature=dw4lJkCkaazqn~SWiMbGI9QL8AIHwEF2ehfcs5t9ncKiuQNjC~mqrQRojuFkQQQf2vTtqo6QMdYpz2cenu40EukGiZW2npdhU1O-b2JzZalDo7FHOBNMGCh3ZADVSWOmhr2mw7-8Si1pFwbIzDLrYBT156Qt8fa522TX~xwM01OKq~G-KEGbXLy8OhdKVXZ98EIV5ahslHAJAYEoiOIi5qd4WjWrietA98ztKcp-6KjA6RfdDFW1QuWXGCU6X43BgKpzaApolu0-moUgySOQfqltC~EInRCdH76lUgwTG4AIR~9eCgvqGm8IOvOqhsfJ6pALzsrMOJleIrTKveHWlA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![JNJ-75276617 inhibits menin-KMT2A complex association with chromatin, downregulates menin-KMT2A target genes, and induces myeloid cell differentiation in KMT2A-altered and NPM1-mutant AML cells. (A) A chromatin immunoprecipitation–quantitative polymerase chain reaction (qPCR) assay was performed to assess the binding of menin to target gene promoters (MEIS1, HOXA9, and HOXA10) in response to treatment with JNJ-75276617 for 48 hours. All qPCRs were performed in triplicate. Signals for menin binding in the KMT2A-r MOLM-14 or NPM1c OCI-AML3 samples were normalized to input values, and binding events per 1000 cells were calculated. Compared with dimethyl sulfoxide (DMSO) control, menin binding was decreased in MOLM-14 and OCI-AML3 cells treated with JNJ-75276617 at concentrations from 0.1 to 1.0 μM (MEIS1, 3.2- to 9.8-fold in MOLM-14, 1.4- to 2.6-fold in OCI-AML3; homeobox gene A9 [HOXA9], 2.4- to 3.4-fold in MOLM-14, 14.0- to 24.7-fold in OCI-AML3; homeobox gene A10 [HOXA10], 1.6- to 2.2-fold in MOLM-14, 5.2- to 10.3-fold in OCI-AML3); in all cases, the fold decreases fall outside the 95% confidence interval (CI) of the DMSO control. (B) JNJ-75276617 inhibited expression of menin-KMT2A target genes and increased expression of differentiation genes in KMT2A-altered and NPM1-mutant AML cells. Cells were incubated with the indicated concentrations of JNJ-75276617 for either 48 (MOLM-14) or 72 (OCI-AML3) hours. Relative expression of menin-KMT2A target genes and differentiation markers was calculated by dividing the normalized values of the treated samples by the normalized value of the DMSO control. The experiment was performed 3 times, and the error bars represent the mean ± standard deviation. (C,D) Differential expression of menin-KMT2A target genes and myeloid cell differentiation signature in response to JNJ-75276617 in various leukemia cells. Samples were prepared in duplicate. Microarray analysis was performed on leukemia cell lines in response to JNJ-75276617 treatment for 48 hours. Representative menin-KMT2A PD markers are shown in volcano plots (C). Gene sets involved in myeloid cell differentiation were enriched on treatment of JNJ-75276617 in KMT2A-r (MOLM-14, MV4-11) and NPM1c (OCI-AML3) cell lines as denoted by enrichment score (red), whereas no gene set enrichment was found in KMT2A/NPM1-WT (HL-60, KO-52, and K-562) cells (D). The experiment was performed once.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/144/11/10.1182_blood.2023022480/4/m_blood_bld-2023-022480-gr2b.jpeg?Expires=1768346815&Signature=YU8421g5TdYXjF1dWRI3uE51N1NEmS5fh5JXRiDeUiMG7wjd7Qvk618TgVmckUWcK2ofDlq0RuQx2FhpUckaJd-H-tqvWHuayYBkr41hmwOmZlMk0Mml5lYHNSr3fpKd6oyv4QbJRd~CE4BCdvkOHI7agl7vQPHUoPECl1sT4p9854lqQRzTyx-u-oeU9ZSexYH94ya0qkpmtqMisbaNcIhDJfFg~Evzg9QuBmP0Ev5hv9RwoYCjFAguom~FI-La56UyH0~Fe24tD6KknuJk7WSKTUQHU47nCxw2ysG0zMKBv1Fh2ae8-j8bw2mPiySFtwhYjQZh-wX-3GhfoIXXbg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![JNJ-75276617 inhibits proliferation and induces differentiation and apoptosis of AML and B-ALL cells with KMT2A alteration and NPM1 mutations and morphologic differentiation of KMT2A-AF9–transduced mouse bone marrow cells. (A) The antiproliferative activity of JNJ-75276617 was determined in a panel that included 8 AML cell lines (KMT2A-r [MOLM-14, MOLM-13, MV4-11, and THP-1], KMT2A–partial tandem duplication [PTD; EOL-1], NPM1c [OCI-AML3], KMT2A/NPM1-WT [KO-52 and HL-60]), 1 leukemia B-ALL cell line with KMT2A-r (RS4:11), and a KMT2A/NPM1-WT chronic myeloid leukemia (CML) cell line (K562). MOLM-14, MOLM-13, MV4-11, and THP-1 were originally derived from pediatric patients. Cells were treated with JNJ-75276617 for 8 days, and spheroid-like growth was measured in real time by live-cell imaging. Results are shown from a representative experiment in which all cell lines were evaluated in parallel (A). Absolute IC50 and the mean ± standard deviation (SD) values were calculated as percentage change in confluence to DMSO-treated cells. The experiment was performed at least 3 times for each cell line (B). (C,D) Flow cytometry analysis of differentiation and apoptosis in KMT2A-r and NPM1c AML cells following treatment with various concentrations of JNJ-75276617 for 7 days (C). Expression of differentiation markers CD14 and CD11b was evaluated in the viable cell population only, and apoptosis was evaluated in the total cell population (D). Duplicate samples were tested for each condition, and bars represent the mean ± SD. The experiment was performed 3 times. (E) Effect of JNJ-75276617 on morphologic differentiation of KMT2A-AF9–transformed mouse BM cells was examined by May-Grünwald Giemsa staining. KMT2A-AF9–transformed mouse BM cells were treated with DMSO or 200 nM JNJ-75276617 for 10 days. Condensed nuclei, a readout of neutrophil-like morphologic differentiation, were counted after 10-day JNJ-75276617 treatment from 3 independently captured images. Representative images are shown. The experiment was performed once.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/144/11/10.1182_blood.2023022480/4/m_blood_bld-2023-022480-gr3.jpeg?Expires=1768346815&Signature=AZ5zp6KU8z~5PlbgwBu68CRoYDdj25ETmuNZqoj-pLz2J1BURPtkxp1ugPUS4kKq9dyxUvIiLjgUHO8Qznc8GWiS4jvS61Ez9EqkEeHy4kUolDtHnp83Kz37u3Gg3i-c1NPKcJ2OaGuemdTHp6hekFir2ujOmUAxzqYbUxoANVQGeW1GBrCnU9nG4XbY7UIRyQrC31u5FK4T4o4pNNu034hHFUmKyllEULn9TAqss6aCxL3WPuKOwXaAnwBR3Wc7bXkh6ijNmpLVjEq-bEMbo881hGRA0ZHr5e6KLYsD37I2DAQPLxA99Z8Jq9TCD7kte57yrpi8ujQ04eofCdvSIg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![JNJ-75276617 exhibits potent antiproliferation activity in MEN1-mutant cell lines resistant to other menin-KMT2A inhibitors. (A) Fluorescence polarization assay demonstrating the dose-dependent displacement of a KMT2A peptide from WT, M327I-mutant, and T349M-mutant menin on treatment with JNJ-75276617. Data are represented as mean ± standard deviation (SD), n = 3 independent replicates. (B) Evaluation of JNJ-75276617 for antiproliferation activity in MEN1M327I and MEN1T349M mutant KMT2A-r MV4-11 cell lines. Drug response was calculated on the basis of the luminescence for each menin inhibitor concentration relative to the DMSO sample after 10 days of JNJ-75276617 treatment. Data are represented as mean ± SD, n = 3 independent experiments. (C) Superimposition of a docking model of JNJ-75276617 (green sticks; docked on M327I-mutant menin chain A; Protein Data Bank Identifier [PDB] ID: 8E90; second rotamer of W346 [chain B] is displayed in gray lines). JNJ-75276617 bound to wild-type menin (this study; PDB ID: in deposition) is shown for reference in orange and its binding pocket as gray surface. Side chain of M327/I327 is shown as sticks. The distance between selected atoms of T349 and JNJ-75276617 is shown as orange dashed line. Residue numbering is according to transcript variant: NM_000244.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/144/11/10.1182_blood.2023022480/4/m_blood_bld-2023-022480-gr7.jpeg?Expires=1768346815&Signature=k07T6VMWoVV5v4gVa63yRi2hKto3ojJiAJH7~2PXELUxLQdyLB4jXGIQXIKJuKux1eq9HHLphH1GEz0iyX3rpMd~8Y1K2awvx9kibuf4iAfbWM4~KQc56TelrKYSSwXK9Zmsg6BtJc5cufu~jZQf9GY7wtDV4jm~j~XLEILg~SYiZYdqbCYmtkxvqNHjHqJSB42YaI79Y-qPGbA0SuZqSq8AoHEdWSYmD~HtcbdrhI4Cmf1JbMbNuRBD5-aXsM7EJEaBzfCoiYOZqYVzWZK6NWoYNd1LgvtyaDFGno30hjCb2-gIudfBdF88-pzMC0aH5x5C0pOm65zlqQefJrkORw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal