Key Points
RNA editing enzyme ADAR2, but not ADAR1 and ADAR3, was specifically downregulated in CBF AMLs.
The RNA editing capability of ADAR2 is essential for its suppression of leukemogenesis in an additional exon 9a–driven AML mouse model.
Abstract
Adenosine-to-inosine RNA editing, which is catalyzed by adenosine deaminases acting on RNA (ADAR) family of enzymes, ADAR1 and ADAR2, has been shown to contribute to multiple cancers. However, other than the chronic myeloid leukemia blast crisis, relatively little is known about its role in other types of hematological malignancies. Here, we found that ADAR2, but not ADAR1 and ADAR3, was specifically downregulated in the core-binding factor (CBF) acute myeloid leukemia (AML) with t(8;21) or inv(16) translocations. In t(8;21) AML, RUNX1-driven transcription of ADAR2 was repressed by the RUNX1-ETO additional exon 9a fusion protein in a dominant-negative manner. Further functional studies confirmed that ADAR2 could suppress leukemogenesis specifically in t(8;21) and inv16 AML cells dependent on its RNA editing capability. Expression of 2 exemplary ADAR2-regulated RNA editing targets coatomer subunit α and component of oligomeric Golgi complex 3 inhibits the clonogenic growth of human t(8;21) AML cells. Our findings support a hitherto, unappreciated mechanism leading to ADAR2 dysregulation in CBF AML and highlight the functional relevance of loss of ADAR2-mediated RNA editing to CBF AML.
Introduction
RNA processing, including alternative splicing and polyadenylation as well as RNA editing and modification, significantly contributes to the composition and complexity of the transcriptome. Specifically, adenosine-to-inosine (A-to-I) editing is the most prevalent type of RNA editing in mammals, contributing to multilevel regulation of gene expression and activity. This process is catalyzed by the adenosine deaminase acting on RNA (ADAR) family of enzymes that recognize double-stranded RNAs.1 In vertebrates, the ADAR family consists of 3 members: ADAR1, ADAR2, and ADAR3.2 ADAR1 and ADAR2 mediate the adenosine deamination editing reaction, whereas ADAR3 has no documented deaminase activity.3 A-to-I RNA editing does not simply alter the RNA sequence per se, but such sequence changes can further affect the cellular fate of RNA molecules. In principle, A-to-I editing sites can be found in both coding and noncoding regions. However, the vast majority of A-to-I editing sites are in introns and untranslated regions harboring double-stranded RNA structures formed by inverted Alu repetitive elements.4 Over the decades, accumulating evidence suggests dysregulated A-to-I editing as one of the key drivers for various cancer types, especially solid tumor cancers.5-11 In coding regions, RNA editing can lead to amino acid codon changes. Differential editing of these protein-recoding sites has been found to affect human diseases, such as neurological diseases and cancers.6,8,10,12-14 In cancer, only a few aberrant protein-recoding targets have been reported thus far, and they contribute to tumorigenesis largely by enhancing cancer-promoting activity or repressing tumor-suppressive activity.7,8,10,15
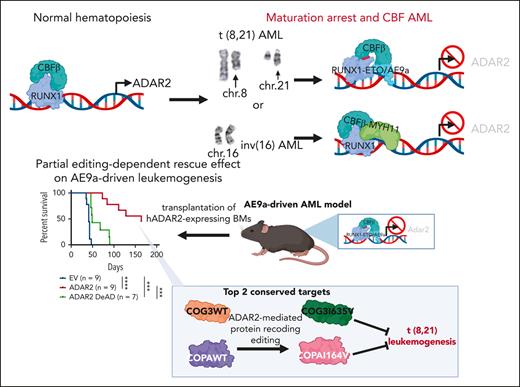
Although RNA editing has been demonstrated to influence solid tumors, its influence in hematological malignancies is less clear, especially in acute myeloid leukemia (AML), one of the most common hematological malignancies characterized by the inhibition of differentiation of myeloid progenitor cells, with high incidence and recurrence rates.5,9,16-21 Furthermore, although there are a number of studies on ADAR1 and ADAR1-mediated RNA editing events in multiple myeloma22,23 and chronic myeloid leukemia,24-26 the role of ADAR2 in AML and other hematological malignancies remains unknown. Core-binding factor (CBF) is a heterodimeric transcription factor complex necessary for normal hematopoiesis. The heterodimer is composed of a non–DNA-binding unit CBFβ and a DNA-binding unit CBFα, which can be either RUNX1, RUNX2, or RUNX3. CBF AMLs are characterized by recurrent chromosome translocations, including t(8;21)(q22;q22) and inv(16)(p13q22)/t(16;16), which result in the formation of RUNX1-ETO or CBFβ-MYH11 fusion genes, respectively. Chromosomal translocations can alter DNA-binding capability and create alternate binding sites of the heterodimer, leading to disruption of normal transcription programs and consequent maturation arrest. Unlike the wild-type (WT) RUNX1, RUNX1-ETO fusion proteins (also named AML1-ETO) drive leukemogenesis through assembling transcriptional regulatory complexes, either as repressors or activators.27-39 Although the presence of RUNX1-ETO defines a precursor stage of leukemia, additional molecular events are required for transformation.40,41 In this study, we analyzed the expression profiles of 3 ADAR enzymes in patients with AML of distinct molecular subtypes from publicly available complementary DNA microarray42 and the The Cancer Genome Atlas (TCGA) RNA sequencing (RNA-seq) data sets.43 We found that ADAR2, but not ADAR1 and ADAR3, was downregulated specifically in patients with CBF AML. In t(8;21) subtype, the downregulation of ADAR2 was mediated by the RUNX1-ETO fusion protein through its competition with WT RUNX1 for DNA binding. Importantly, ADAR2 can suppress leukemogenesis of t(8;21) AML through its RNA editing capabilities as evidenced in both human cell line and murine models. These results linked ADAR2 and its RNA editing function to the pathogenesis of CBF AMLs.
Materials and methods
Mice
C57BL/6J mice were obtained from The Jackson Laboratory. All mice were housed in a sterile barrier facility within the comparative medicine facility at the National University of Singapore under housing conditions of 22°C temperature, 50% humidity, and a 12:12 light:dark cycle. All mice experiments performed in this study were approved by institutional animal care and use committee of National University of Singapore. In this study, 8- to 16-week-old male mice were used for bone marrow (BM) harvesting or transplantation, and 8- to 16-week-old female mice were used for timed mating. See supplemental Materials and Methods, which is available on the Blood website, for additional details.
Analysis of RNA editing by Sanger sequencing
To amplify regions containing component of oligomeric Golgi complex 3 (COG3) I635V or coatomer subunit α (COPA) I164V sites, complementary DNA from different cells were used for Platinum Green Hot Start PCR (2X) Master mix (Thermo Fisher Scientific catalog #13001014). Purified polymerase chain reaction products were sent for direct sequencing, and the result was visualized with SnapGene software (SnapGene, San Diego, CA). The frequencies of COPA and COG3 editing were calculated based on the peak areas of adenosine and guanosine determined using SnapGene. The sequences of primers are listed in supplemental Table 1.
Patient samples
BM mononuclear cells that were collected from patients with AML demonstrating t(8;21) (n = 11) or normal karyotype (n = 13) were used for the analysis of expression of ADAR2 and A-to-I editing level of COG3. Each patient gave informed consent to this study based on the tenets of the revised Helsinki protocol produced by the institutional committees for the protection of human subjects and analysis of the human genome. This study was approved by the institutional review board of Kumamoto University Hospital. CD34+ cells were obtained from BM samples of patients undergoing total knee replacements (healthy, n = 16). Each patient gave informed consent. This study complies with all relevant ethical regulations. The human studies were approved by the following institutional review boards: the Domain Specific Review Board under the National Healthcare Group in Singapore, the Central Institution Review Board of SingHealth, of which the National Cancer Center Singapore, Singapore General Hospital, and National University Hospital were all constituent members (Central Institution Review Board Reference, 2016/2626 and 2018/2112).
Statistical analysis
Statistical significance was assessed using two-tailed Student t test, unless otherwise specified. Additional details are described in supplemental Materials and Methods.
Results
ADAR2 is selectively downregulated in CBF AMLs
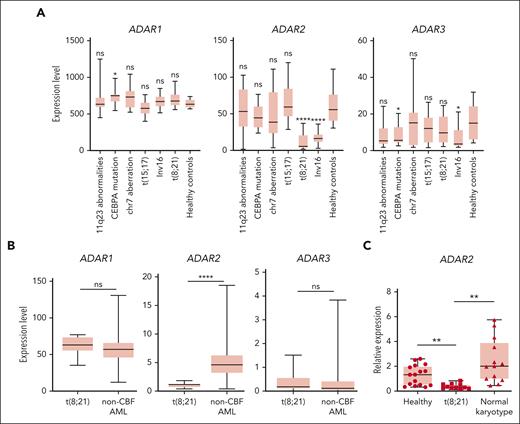
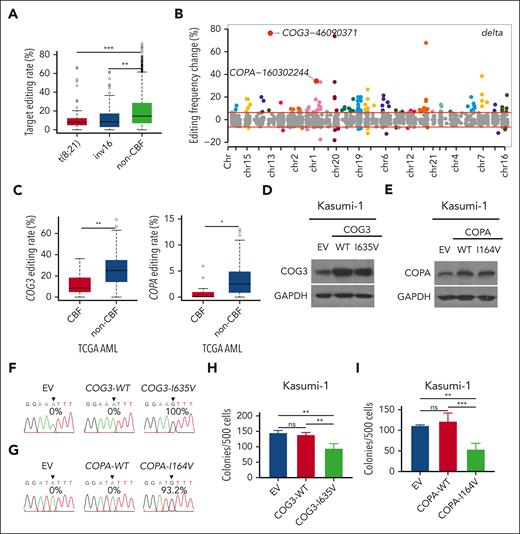
To understand the role of ADARs in AML, we first analyzed gene expression microarray data from 285 patients with AML.42 We found that ADAR2 (but not ADAR1 or ADAR3) was significantly downregulated specifically in patients with CBF AML (Figure 1A). Intriguingly, we did not observe the differential expression of ADAR1 in any AML subtypes from this cohort, although previous studies reported the role of ADAR1 in hematological malignancies.24-26 To further support this finding, we also analyzed the RNA-seq data of patients with t(8,21) CBF AML from TCGA. Consistently, the expression level of ADAR2, but not ADAR1 or ADAR3, was significantly lower in t(8;21) and inv16 AML (Figure 1B; supplemental Figure 1) than that in other patients with AML. In addition, we examined the level of ADAR2 messenger RNA (mRNA) in mononuclear cells isolated from our in-house samples from patients with AML [patients with either t(8,21) (n = 11) or normal karyotype (n = 13)] and CD34+ BM cells from healthy individuals (n = 16) as controls. ADAR2 was found to be consistently downregulated in t(8;21) AML samples (Figure 1C). Altogether, among 3 ADAR family genes, ADAR2 is the only 1 that is selectively downregulated in patients with t(8;21) and inv16 AML, suggesting a specific regulatory mechanism repressing ADAR2 expression in CBF AMLs.
ADAR2 is selectively downregulated in CBF AMLs. (A) Box plots showing the expression levels of ADAR1, ADAR2, and ADAR3 in healthy donors (n = 8) and patients with different subtypes of AML (n = 108) (∗∗∗∗P < .0001, 2-tailed Student t test) detected by microarray. The data were obtained from Gene Expression Omnibus database (GSE1159).31 The expression level of a particular gene is reflected by the intensity of hybridization of labeled mRNA to gene-specific probe sets (10-20 oligonucleotides per gene).31 The line in the middle of the box is plotted at the median. The whiskers indicate minimum to maximum. (B) Box plots showing the expression of ADAR1, ADAR2, and ADAR3 in patients with t(8;21) AML (n = 7) and control patients with non-CBF (n = 164) AML from the TCGA AML RNA-seq data sets. (C) Box plot showing the relative expression of ADAR2 in leukemic blasts isolated from patients with AML with t(8;21) (n = 11) or normal karyotype (n = 13) and CD34+ cells isolated from BM samples of healthy individuals (n = 16). The relative expression was calculated as described in supplemental Materials and Methods. Data are presented as technical triplicates from a representative experiment. ∗∗P < .01; ∗∗∗∗P < .0001 using two-tailed Student t test. chr, chromosome; ns, not significant.
ADAR2 is selectively downregulated in CBF AMLs. (A) Box plots showing the expression levels of ADAR1, ADAR2, and ADAR3 in healthy donors (n = 8) and patients with different subtypes of AML (n = 108) (∗∗∗∗P < .0001, 2-tailed Student t test) detected by microarray. The data were obtained from Gene Expression Omnibus database (GSE1159).31 The expression level of a particular gene is reflected by the intensity of hybridization of labeled mRNA to gene-specific probe sets (10-20 oligonucleotides per gene).31 The line in the middle of the box is plotted at the median. The whiskers indicate minimum to maximum. (B) Box plots showing the expression of ADAR1, ADAR2, and ADAR3 in patients with t(8;21) AML (n = 7) and control patients with non-CBF (n = 164) AML from the TCGA AML RNA-seq data sets. (C) Box plot showing the relative expression of ADAR2 in leukemic blasts isolated from patients with AML with t(8;21) (n = 11) or normal karyotype (n = 13) and CD34+ cells isolated from BM samples of healthy individuals (n = 16). The relative expression was calculated as described in supplemental Materials and Methods. Data are presented as technical triplicates from a representative experiment. ∗∗P < .01; ∗∗∗∗P < .0001 using two-tailed Student t test. chr, chromosome; ns, not significant.
RUNX1-ETO and its truncated variant AE9a demonstrate dominant-negative effects on ADAR2 transcription in t(8;21) AML
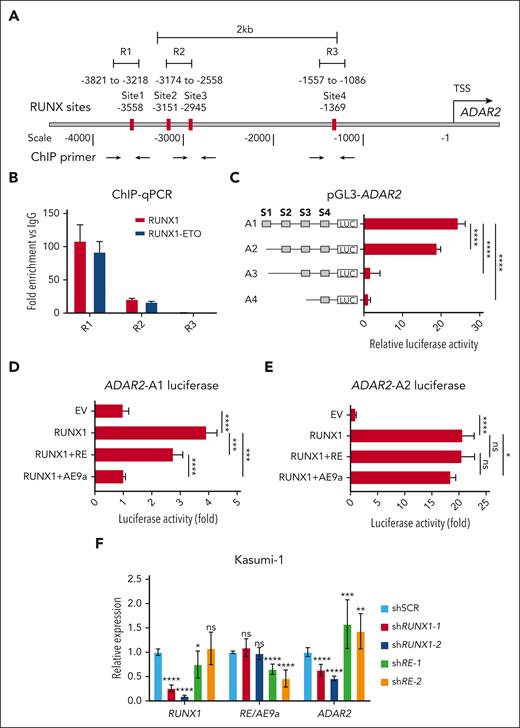
Next, we focused on the t(8;21) AML subtype and investigated the cause of ADAR2 downregulation. Because RUNX1-ETO has been reported to downregulate RUNX1 target genes through competitive DNA binding,44-52 we wondered whether ADAR2 is a RUNX1 target gene and whether such transcriptional activation can be hindered by RUNX1-ETO in a dominant-negative manner. To this end, we searched for the RUNX-binding motif (TGTGGT)53-55 within the 5000 base pair genomic DNA sequence upstream of the ADAR2 transcription start site (TSS) and found 4 putative RUNX-binding sites (Figure 2A). To test for RUNX1 and RUNX1-ETO binding to these sites, we conducted RUNX1 and RUNX1-ETO chromatin immunoprecipitation in the human t(8:21) AML cell line Kasumi-1. Notably, we used anti-RUNX1 and anti-ETO56 antibodies to differentiate binding by the 2 proteins, and binding at the putative sites was examined via chromatin immunoprecipitation–quantitative polymerase chain reaction (Figure 2A). Both RUNX1 and RUNX-ETO could bind to the RUNX-binding site in the upstream regulatory region of ADAR2, as suggested by the high enrichment of both immunoprecipitation samples within the R1 and R2 regions, when compared with the immunoglobulin G control (Figure 2B). To test the potential transcriptional activation caused by such binding, we generated luciferase reporter constructs containing the full-length sequence upstream of the ADAR2 TSS with 4 RUNX sites or its truncated derivatives (A1:-3618; A2:-3171; A3:-2956; and A4:-1520 relative to the TSS; Figure 2C). We observed a dramatic increase in the luciferase activity of the full-length sequence (A1 construct); however, such an effect was significantly abolished when a 5′ sequence containing the RUNX sites 1 and 2 was deleted (A3 construct), suggesting that the RUNX-binding sites 1 and 2 are essential for the transcriptional activation of the ADAR2 gene. Next, we cotransfected the A1 luciferase construct together with either RUNX1 alone (RUNX1) or RUNX1 and RUNX1-ETO (RUNX1 + RE) in Kasumi-1 cells (Figure 2D). Although forced expression of either RUNX1 alone or RUNX1 + RE resulted in increased luciferase activity, RUNX1-ETO attenuated RUNX1-mediated transcriptional activation of ADAR2 (Figure 2D). This repressive effect was abolished when RUNX site 1 was deleted in the A2 construct (Figure 2E). Moreover, it has been reported that alternative splicing of RUNX1-ETO could generate a truncated variant that contains an additional exon 9a (namely AML1-ETO9a or AE9a). AE9a is frequently coexpressed with RUNX1-ETO in most patients with t(8;21) AML,57 and coexpression of RUNX1-ETO and AE9a in mice could induce a rapid AML onset with more aggressive leukemic phenotypes.58 We thus queried whether AE9a demonstrates a stronger suppressive effect on RUNX1-mediated transcriptional activation of ADAR2 than on the RUNX1-ETO-mediated activation. As expected, AE9a almost completely abolished the transcriptional activation of ADAR2 driven by RUNX1, and consistently, this effect was eliminated by the deletion of site 1 (Figure 2D-E). Furthermore, we examined the changes in ADAR2 expression endogenously in Kasumi-1 cells upon knockdown or overexpression of RUNX1 and its fusion isoform, RUNX1-ETO or AE9a. Consistent with results from the luciferase reporter assays, RUNX1 could transcriptionally activate ADAR2 expression, whereas the RUNX1-ETO/AE9a isoform showed a dominant-negative effect on ADAR2 expression (Figure 2F; supplemental Figure 2A). Next, the dead Cas9 VP64-mediated activation of sites 1 or 2 significantly increased ADAR2 expression (supplemental Figure 2B-C). Together, these data suggested that RUNX1 transcriptionally activates ADAR2 by binding to the RUNX sites 1 and 2 in the distal regulatory region upstream of the ADAR2 TSS; however, in t(8,21) AML, expression of the fusion isoform RUNX1-ETO and its truncated form AE9a may outcompete RUNX1 for binding to the site 1, thereby suppressing ADAR2 transcription.
RUNX1-ETO and its truncated variant, AE9a, found in t(8:21) AML demonstrate dominant-negative effects on ADAR2 transcription. (A) Schematic diagram of the RUNX1 binding sites along the 4 kb region upstream of the TSS of the ADAR2 gene. The numbers below the double line indicate the nucleotide position with respect to the ADAR2 TSS. Black bars indicate 4 putative RUNX1 binding sites (TGTGGT) centered at positions −3558, −3151, −2945, and −1369 base pairs relative to the TSS. Black arrows indicate the locations of primers used for chromatin immunoprecipitation (ChIP) quantitative polymerase chain reaction (PCR). Notably, the primers were designed to amplify R1, R2, and R3 regions which cover site 1, sites 2 and 3, and site 4, respectively. (B) ChIP-qPCR analysis of RUNX1 or RUNX1-ETO protein binding to the indicated ADAR2 regulatory region (R1, R2, or R3) in Kasumi-1 cells. Anti-RUNX1 or anti-ETO antibodies were used to pull down RUNX1 or RUNX1-ETO, respectively. Immunoglobulin G (IgG) was used as a negative control. Notably, the anti-RUNX1 antibody recognizes the 200 to 300 amino acids of RUNX1 protein and only recognizes RUNX1, whereas the anti-ETO antibody was used for immunoprecipitation to specifically pull down RUNX1-ETO in Kasumi-1 cells, which do not express WT ETO protein. Data were presented as the mean ± standard deviation (SD) of technical triplicates from a representative experiment. (C) Luciferase activities associated with each of the indicated sequences upstream of the TSS of ADAR2. HEK293T cells were transfected with each of the indicated reporter constructs containing fragment A1, A2, A3, or A4. S1, S2, S3, and S4 refer to RUNX binding sites 1, 2, 3, and 4, respectively. Relative luciferase activity represents firefly luciferase activity normalized against the internal control Renilla luciferase, calculated as fold difference against the activity of the pGL3 EV. Data were presented as the mean ± SD of 3 independent experiments (∗∗∗∗P < .0001 using two-tailed Student t test.). (D) Luciferase activities associated with the A1 fragment in HEK293T cells (panel C) cotransfected with either RUNX1 alone or RUNX1 together with RUNX1-ETO or AE9a (RUNX1, RUNX1 + RE, or RUNX1 + AE9a, respectively). Luciferase activity was normalized to the A1 construct coexpressed with EV control and defined as Luciferase activity (fold). Data were presented as the mean ± SD of 3 independent experiments. (E) Similar to panel D, except that the A2 luciferase construct (panel C) was used. (F) Bar chart showing the relative expression of RUNX1, RUNX1-ETO/AE9a, and ADAR2 expression in Kasumi-1 cells, upon short hairpin RNA (shRNA)–mediated knockdown of RUNX1 (shRUNX1-1 and shRUNX1-2) or RUNX1-ETO/AE9a (shRE-1 and shRE-2). The relative expression was calculated as described in supplemental Materials and Methods. Data were presented as mean ± SD of 3 independent experiments (∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001 using two-tailed Student t test).
RUNX1-ETO and its truncated variant, AE9a, found in t(8:21) AML demonstrate dominant-negative effects on ADAR2 transcription. (A) Schematic diagram of the RUNX1 binding sites along the 4 kb region upstream of the TSS of the ADAR2 gene. The numbers below the double line indicate the nucleotide position with respect to the ADAR2 TSS. Black bars indicate 4 putative RUNX1 binding sites (TGTGGT) centered at positions −3558, −3151, −2945, and −1369 base pairs relative to the TSS. Black arrows indicate the locations of primers used for chromatin immunoprecipitation (ChIP) quantitative polymerase chain reaction (PCR). Notably, the primers were designed to amplify R1, R2, and R3 regions which cover site 1, sites 2 and 3, and site 4, respectively. (B) ChIP-qPCR analysis of RUNX1 or RUNX1-ETO protein binding to the indicated ADAR2 regulatory region (R1, R2, or R3) in Kasumi-1 cells. Anti-RUNX1 or anti-ETO antibodies were used to pull down RUNX1 or RUNX1-ETO, respectively. Immunoglobulin G (IgG) was used as a negative control. Notably, the anti-RUNX1 antibody recognizes the 200 to 300 amino acids of RUNX1 protein and only recognizes RUNX1, whereas the anti-ETO antibody was used for immunoprecipitation to specifically pull down RUNX1-ETO in Kasumi-1 cells, which do not express WT ETO protein. Data were presented as the mean ± standard deviation (SD) of technical triplicates from a representative experiment. (C) Luciferase activities associated with each of the indicated sequences upstream of the TSS of ADAR2. HEK293T cells were transfected with each of the indicated reporter constructs containing fragment A1, A2, A3, or A4. S1, S2, S3, and S4 refer to RUNX binding sites 1, 2, 3, and 4, respectively. Relative luciferase activity represents firefly luciferase activity normalized against the internal control Renilla luciferase, calculated as fold difference against the activity of the pGL3 EV. Data were presented as the mean ± SD of 3 independent experiments (∗∗∗∗P < .0001 using two-tailed Student t test.). (D) Luciferase activities associated with the A1 fragment in HEK293T cells (panel C) cotransfected with either RUNX1 alone or RUNX1 together with RUNX1-ETO or AE9a (RUNX1, RUNX1 + RE, or RUNX1 + AE9a, respectively). Luciferase activity was normalized to the A1 construct coexpressed with EV control and defined as Luciferase activity (fold). Data were presented as the mean ± SD of 3 independent experiments. (E) Similar to panel D, except that the A2 luciferase construct (panel C) was used. (F) Bar chart showing the relative expression of RUNX1, RUNX1-ETO/AE9a, and ADAR2 expression in Kasumi-1 cells, upon short hairpin RNA (shRNA)–mediated knockdown of RUNX1 (shRUNX1-1 and shRUNX1-2) or RUNX1-ETO/AE9a (shRE-1 and shRE-2). The relative expression was calculated as described in supplemental Materials and Methods. Data were presented as mean ± SD of 3 independent experiments (∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001 using two-tailed Student t test).
Restoration of ADAR2-regulated RNA editing inhibits t(8;21) AML cell leukemogenicity
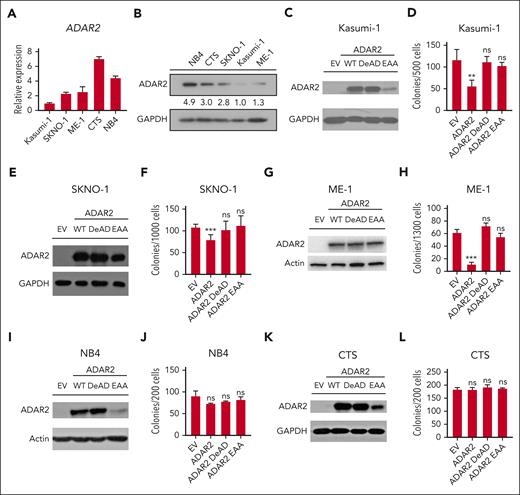
Because of the downregulation of ADAR2, it is not surprising that ADAR2-regulated RNA editome might be suppressed in t(8;21) AML. We next queried whether ADAR2 regulates the leukemogenicity of t(8:21) AML cells and, more specifically, whether such regulation is mediated by its RNA editing function. To this end, we first checked ADAR2 expression in 2 t(8:21) (Kasumi-1 and SKNO-1), 1 inv16 (ME-1), and 2 non-CBF AML cell lines (CTS and NB4), and then performed colony-formation assays using cells stably overexpressing the WT or mutant form of ADAR2 (including an RNA editing–depleted DeAD mutant59,60 and an RNA binding–depleted EAA mutant (KKXXK motif to EAXXA mutant), which lacks both RNA binding and editing capabilities60) or the empty vector (EV) control (Figure 3). The results confirmed that ADAR2 expression was specifically downregulated in Kasumi-1, SKNO-1, and ME-1 when compared with CTS and NB4 cells (Figure 3A-B). Forced expression of ADAR2 in Kasumi-1, SKNO-1, and ME-1 cells inhibited clonogenic growth dependent on the RNA editing (catalytic) function of ADAR2, whereas such inhibition was not observed in non-CBF CTS and NB4 cells (Figure 3C-L). These data indicated that the antileukemic effect of ADAR2 on CBF AML most likely requires its catalytic ability.
ADAR2 specifically represses the clonogenic growth of CBF AML cells dependent on the RNA editing capability of ADAR2. (A) Bar chart presents reverse transcription-qPCR analysis of ADAR2 expression in Kasumi-1, SKNO-1, ME-1, CTS, and NB4 cells. The relative expression was calculated as described in supplemental Materials and Methods. Data were presented as the mean ± SD of technical replicates from a representative experiment. (B) Western blot analysis of ADAR2 protein expression in Kasumi-1, SKNO-1, ME-1, CTS, and NB4 cells. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. The numbers below the lanes represent the relative protein level, which was determined by the band intensity using Photoshop CC software and normalized to the value of Kasumi-1. Western blot analysis of ADAR2 protein in Kasumi-1 (C), SKNO-1 (E), ME-1 (G), NB4 (I), and CTS (K) cells stably expressing the WT or mutant form of ADAR2 or the EV control via retroviral transduction. GAPDH or actin was used as a loading control. (D,F,H,J,L) Bar chart illustrating the number of colonies formed by the indicated cell groups described in panels C, E, G, I, and K, respectively. Data were presented as the mean ± SD of technical replicates from a representative experiment. (∗∗P < .01; ∗∗∗P < .001 using two-tailed Student t test.)
ADAR2 specifically represses the clonogenic growth of CBF AML cells dependent on the RNA editing capability of ADAR2. (A) Bar chart presents reverse transcription-qPCR analysis of ADAR2 expression in Kasumi-1, SKNO-1, ME-1, CTS, and NB4 cells. The relative expression was calculated as described in supplemental Materials and Methods. Data were presented as the mean ± SD of technical replicates from a representative experiment. (B) Western blot analysis of ADAR2 protein expression in Kasumi-1, SKNO-1, ME-1, CTS, and NB4 cells. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. The numbers below the lanes represent the relative protein level, which was determined by the band intensity using Photoshop CC software and normalized to the value of Kasumi-1. Western blot analysis of ADAR2 protein in Kasumi-1 (C), SKNO-1 (E), ME-1 (G), NB4 (I), and CTS (K) cells stably expressing the WT or mutant form of ADAR2 or the EV control via retroviral transduction. GAPDH or actin was used as a loading control. (D,F,H,J,L) Bar chart illustrating the number of colonies formed by the indicated cell groups described in panels C, E, G, I, and K, respectively. Data were presented as the mean ± SD of technical replicates from a representative experiment. (∗∗P < .01; ∗∗∗P < .001 using two-tailed Student t test.)
Analysis of AML sample editing sites suggests that ADAR2 prevents leukemogenesis mainly via altering gene function through editing-mediated protein recoding
To understand how ADAR2 prevents leukemogenesis via its RNA editing function, we went on to perform the RNA-seq analysis of Kasumi-1 cells overexpressing the WT or mutant form of ADAR2 or the EV control. We found that the WT ADAR2, but not the DeAD and EAA mutants, significantly increased the global A-to-I editing level, whereas no obvious transcriptome-wide changes in gene expression were observed in Kasumi-1 cells (supplemental Figure 3A-B). In addition, we selected 21 ADAR2-regulated editing sites using the TCGA AML RNA-seq data sets based on the following criteria: (1) ≥75% of the TCGA AML samples having a coverage of >10 reads at the inferred ADAR2 target site, and (2) the editing frequency of the site having a correlation with ADAR2 expression level (correlation coefficient > 0.1) among all the TCGA AML samples. The editing frequencies of 21 editing sites were significantly lower in t(8,21) and inv(16) AML samples than in their non-CBF AML counterparts (Figure 4A). It is known that RNA editing events, particularly those occurring in noncoding regions, may regulate gene expression by affecting RNA transport, degradation, and microRNA targeting, among other mechanisms.61-64 However, in this study, overexpression of ADAR2 had no effect on global gene expression (supplemental Figure 3B). We thus queried whether ADAR2 might regulate leukemogenesis via editing-mediated protein recoding rather than by affecting gene expression. Notably, editing-mediated protein-recoding events are known to be of high biological conservation and importance.65 With this question, we first identified 147 ADAR2-regulated protein recoding editing targets using our Kasumi-1 RNA-seq data and then performed gene ontology analysis. We found that ADAR2-regulated protein-recoding targets are functionally enriched in multiple important biological pathways, such as protein localization to organelles, RNA splicing, and DNA repair, among others (supplemental Figure 3C). Of the 172 differently edited protein-recoding sites in ADAR2 overexpressing Kasumi-1 cells, COG3 and COPA were among the top hits (Figure 4B), and both demonstrated significantly lower editing frequencies in CBF AML samples than in their non-CBF AML counterparts (Figure 4C). Because both COG3 and COPA are critical factors for protein transport between the Golgi complex and endoplasmic reticulum,66,67 it is highly possible that ADAR2 prevents leukemogenesis, at least partially, by mediating critical biological processes, such as protein localization to organelles via editing-mediated protein recoding.
ADAR2 repression of the clonogenic growth of t(8,21) AML cells is associated with RNA editing–mediated protein recoding of COPA and COG3. (A) Box plots illustrating editing frequencies of 21 ADAR2-regulated editing sites in patients with t(8;21) (n = 7), inv16 (n = 8), and control non-CBF (n = 164) AML, from the TCGA RNA-seq data sets. The box extends from the 25th to 75th percentiles. The line in the middle of the box is plotted at the median. The whiskers indicate 5% to 95% percentile. (∗∗∗P < .001; ∗∗P < .01 using two-tailed Student t test.). (B) Scatter plots showing the changes in editing frequency of each of 172 protein-recoding editing targets between Kasumi-1 cells stably overexpressing the WT ADAR2 and the EV control. Red lines indicate the cutoff values that discriminate sites showing ≥5% editing change from those with <5% change in editing frequency. (C) Box plots showing the editing frequency of COG3 and COPA in patients with CBF AML (n = 15) and those with non-CBF (n = 164) AML, from the TCGA AML RNA-seq data sets. Western blot analysis of COG3 (D) or COPA (E) protein levels in Kasumi-1 cells stably overexpressing the WT or edited COG3 (COG3-WT or COG3-I635V); or WT or edited COPA (COPA-WT or COPA-I164V); or the pMSCV-flag-puro EV control via retroviral transduction. GAPDH was used as a loading control. (F-G) Sequence chromatograms illustrating the editing levels of COG3 and COPA transcripts in the same samples as described in panels D and E, respectively. The arrowhead indicates the position of the edited nucleotide. (H-I) Bar chart illustrating the number of colonies formed by the indicated group of cells as described in panels D and E. Data are presented as the mean ± SD of technical replicates from 2 representative experiment out of 3 independent experiments. ∗∗∗P < .001; ∗∗P < .01; ∗P < .05 using two-tailed Student t test.
ADAR2 repression of the clonogenic growth of t(8,21) AML cells is associated with RNA editing–mediated protein recoding of COPA and COG3. (A) Box plots illustrating editing frequencies of 21 ADAR2-regulated editing sites in patients with t(8;21) (n = 7), inv16 (n = 8), and control non-CBF (n = 164) AML, from the TCGA RNA-seq data sets. The box extends from the 25th to 75th percentiles. The line in the middle of the box is plotted at the median. The whiskers indicate 5% to 95% percentile. (∗∗∗P < .001; ∗∗P < .01 using two-tailed Student t test.). (B) Scatter plots showing the changes in editing frequency of each of 172 protein-recoding editing targets between Kasumi-1 cells stably overexpressing the WT ADAR2 and the EV control. Red lines indicate the cutoff values that discriminate sites showing ≥5% editing change from those with <5% change in editing frequency. (C) Box plots showing the editing frequency of COG3 and COPA in patients with CBF AML (n = 15) and those with non-CBF (n = 164) AML, from the TCGA AML RNA-seq data sets. Western blot analysis of COG3 (D) or COPA (E) protein levels in Kasumi-1 cells stably overexpressing the WT or edited COG3 (COG3-WT or COG3-I635V); or WT or edited COPA (COPA-WT or COPA-I164V); or the pMSCV-flag-puro EV control via retroviral transduction. GAPDH was used as a loading control. (F-G) Sequence chromatograms illustrating the editing levels of COG3 and COPA transcripts in the same samples as described in panels D and E, respectively. The arrowhead indicates the position of the edited nucleotide. (H-I) Bar chart illustrating the number of colonies formed by the indicated group of cells as described in panels D and E. Data are presented as the mean ± SD of technical replicates from 2 representative experiment out of 3 independent experiments. ∗∗∗P < .001; ∗∗P < .01; ∗P < .05 using two-tailed Student t test.
Next, we focused on COPA and COG3, 2 known genes undergoing ADAR2-mediated protein recoding editing68,69 to understand their functional relevance to t(8,21) leukemogenesis. Of note, both edited products have been reported to have tumor suppressor functions: COG3-I635V has been shown to increase drug sensitivity, including MEK inhibitors;9,70 whereas COPA-I164V represses the oncogenic function of COPA-WT (unedited), functioning in a dominant-negative manner.68 These reports intrigued us to ask whether restoration of the edited protein variant could at least partially phenocopy ADAR2-mediated suppression of leukemogenesis of t(8;21) AML. Upon overexpression of the WT ADAR2 but not the DeAD or EAA mutant, editing frequency of COPA or COG3 transcripts reached 22.8% or 52.1%, respectively, from an undetectable level in Kasumi-1 cells (supplemental Figure 4A), indicating that COPA and COG3 are bona fide ADAR2 targets in t(8;21) AML cells. Moreover, expression of COPA or COG3 at mRNA and protein levels was not significantly influenced by the overexpression of ADAR2 (supplemental Figure 4B-F). We then studied the functional difference between the WT and edited COPA or COG3 in Kasumi-1 cells (Figure 4F-I). Notably, Kasumi-1 cells stably expressing COPA-I164V or COG3-I635V demonstrated significantly lower colony-forming ability than the EV control cells, whereas cells expressing the WT form (COPA-WT or COG3-WT) did not show any obvious difference in the colony-forming ability when compared with the control cells (Figure 4F-I). Taken together, these data suggested that in t(8,21) AML cells, downregulation of ADAR2 may promote leukemogenesis, at least partially, because of the loss of edited protein products that are of antileukemic capability (eg, COPA-I164V and COG3- I635V).
The RNA editing capability of ADAR2 is essential for its suppression of leukemogenesis in an AE9a-induced mouse AML model
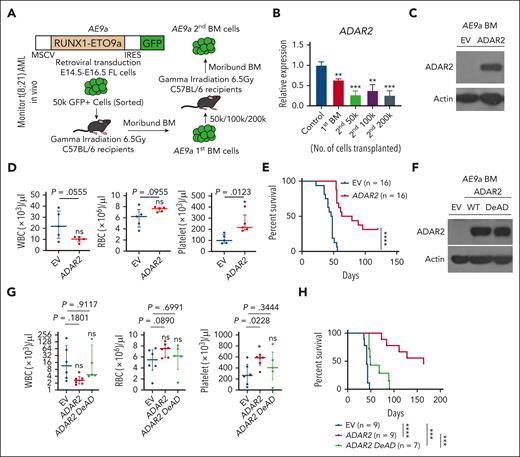
Next, we established an AE9a-induced AML mouse model, as reported previously,58 to further investigate the role of ADAR2 in vivo (Figure 5A). Notably, we found that ADAR2 expression had consistently decreased in BM cells isolated from the recipients after the first or second transplantation (Figure 5B), suggesting a negative role of ADAR2 in leukemia-initiating potential. We further facilitated the overexpression of ADAR2 in BM cells that were obtained from the recipients after the second transplantation (Figure 5C; supplemental Figure 5A-B). These ADAR2-expressing BM cells were further transplanted into sublethally (6.5 Gy) γ-irradiated C57BL/6J mice for peripheral blood (PB) collection and survival monitoring. We observed a trend of reduction in the white blood cell counts to normal ranges in the mice receiving ADAR2-expressing BM cells, accompanied by a slight increase in the number of red blood cells and platelets (Figure 5D). More importantly, stable expression of ADAR2 significantly extended the survival time of recipient mice (Figure 5E). Furthermore, we confirmed that most BM cells harvested from mice receiving ADAR2-expressing BM cells (ADAR2 group) underwent differentiation, whereas most of those from mice receiving control cells (EV group) remained undifferentiated (supplemental Figure 5C). This was in line with a significant reduction in the AE9a (green fluorescent protein [GFP])–positive cell population in the PB of mice in the ADAR2 group (supplemental Figure 5D). In conclusion, results from the t(8,21) AML mouse model provided solid evidence that the CBF fusion oncogenes can promote leukemogenesis by suppressing ADAR2 expression.
RNA editing capability of ADAR2 is essential for the suppression of leukemogenesis in an AE9a mouse model. (A) Experimental strategy for AE9a BM transplantation mouse model. (B) Bar chart showing the relative expression of ADAR2 transcripts in BM cells from moribund mice after the first transplantation, and in the second transplantation injected with different numbers of AE9a cells (50 000, 100 000, and 200 000). The relative expression was calculated as described in supplemental Materials and Methods. Data are presented as the mean ± SD of technical triplicates from a representative experiment. (C) Western blot analysis of ADAR2 protein expression in AE9a BM cells stably overexpressing ADAR2 or the MSCV-IRES-tdTOMATO EV. (D) Dot plot representing counts of white blood cells (WBCs), red blood cells (RBCs), and platelets in the PB from recipients at 28 days after transplantation of AE9a AML cells stably overexpressing AE9a or the MSCV-IRES-tdTOMATO EV. n = 5 in each group. (E) Kaplan-Meier survival curve of recipients who underwent transplantation with 50 000 AE9a AML cells stably overexpressing ADAR2 or MSCV-IRES-tdTOMATO EV. n = 16 in each group. Statistical analysis was performed using log-rank (Mantel-Cox) test. (F) Western blot analysis of ADAR2 or ADAR2 DeAD protein expression in BM cells from the same recipients the indicated groups of cells described in panel E. Actin was used as a loading control. (G) Dot plot representing counts of WBCs, RBCs, and platelets in the PB from recipients at 45 days after transplantation of AE9a AML cells stably overexpressing of ADAR2 (n = 6), ADAR2 DeAD (n = 5), or MSCV-IRES-tdTOMATO EV (n = 6). (H) Kaplan-Meier survival curve of recipients who underwent transplantation with 50 000 AE9a AML cells stably overexpressing ADAR2 (n = 9), ADAR2DeAD (n = 7), or MSCV-IRES-tdTOMATO EV (n = 9). Statistical analysis was performed using log-rank (Mantel-Cox) test. FL, full-length; IRES, internal ribosome entry site. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001 using two-tailed Student t test.
RNA editing capability of ADAR2 is essential for the suppression of leukemogenesis in an AE9a mouse model. (A) Experimental strategy for AE9a BM transplantation mouse model. (B) Bar chart showing the relative expression of ADAR2 transcripts in BM cells from moribund mice after the first transplantation, and in the second transplantation injected with different numbers of AE9a cells (50 000, 100 000, and 200 000). The relative expression was calculated as described in supplemental Materials and Methods. Data are presented as the mean ± SD of technical triplicates from a representative experiment. (C) Western blot analysis of ADAR2 protein expression in AE9a BM cells stably overexpressing ADAR2 or the MSCV-IRES-tdTOMATO EV. (D) Dot plot representing counts of white blood cells (WBCs), red blood cells (RBCs), and platelets in the PB from recipients at 28 days after transplantation of AE9a AML cells stably overexpressing AE9a or the MSCV-IRES-tdTOMATO EV. n = 5 in each group. (E) Kaplan-Meier survival curve of recipients who underwent transplantation with 50 000 AE9a AML cells stably overexpressing ADAR2 or MSCV-IRES-tdTOMATO EV. n = 16 in each group. Statistical analysis was performed using log-rank (Mantel-Cox) test. (F) Western blot analysis of ADAR2 or ADAR2 DeAD protein expression in BM cells from the same recipients the indicated groups of cells described in panel E. Actin was used as a loading control. (G) Dot plot representing counts of WBCs, RBCs, and platelets in the PB from recipients at 45 days after transplantation of AE9a AML cells stably overexpressing of ADAR2 (n = 6), ADAR2 DeAD (n = 5), or MSCV-IRES-tdTOMATO EV (n = 6). (H) Kaplan-Meier survival curve of recipients who underwent transplantation with 50 000 AE9a AML cells stably overexpressing ADAR2 (n = 9), ADAR2DeAD (n = 7), or MSCV-IRES-tdTOMATO EV (n = 9). Statistical analysis was performed using log-rank (Mantel-Cox) test. FL, full-length; IRES, internal ribosome entry site. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001 using two-tailed Student t test.
To explore whether RNA editing function of ADAR2 is required for its antileukemogenic effect in vivo, we facilitated the overexpression of WT ADAR2 or its DeAD mutant in BM cells isolated from the recipients after the second transplantation (Figure 5F; supplemental Figure 5E), followed by the transplantation into γ-irradiated mice as above described. The antileukemogenic effect of ADAR2, as manifested by altered PB counts (decreased white blood cell counts together with increased RBC and platelet counts), was not observed for the ADAR2 DeAD mutant (Figure 5G). More importantly, although all recipients receiving ADAR2 DeAD-expressing BM cells (ADAR2 DeAD group) died within 100 days, mice receiving WT ADAR2-expressing BM cells (ADAR2 group) demonstrated further extended lifespans, with 50% of recipients surviving over 150 days (Figure 5H). As expected, the DeAD mutant failed to restore BM cell differentiation, as evident from the undifferentiated PB morphology and the high percentage of AE9a (GFP)–positive cells in the BM of mice in the ADAR2 DeAD group (supplemental Figure 5F-G). Moreover, we could also detect editing of COG3 and COPA transcripts in ADAR2-expressing AE9a BM cells (supplemental Figure 6). Finally, to study whether ADAR2 may possibly affect the viability of normal BM cells, we transduced ADAR2 into normal BM cells; 99% of ADAR2 (GFP)–positive cells remained viable after 2 to 3 days of culture (supplemental Figure 7A-B). Furthermore, to study whether ADAR2 may possibly affect the differentiation of normal BM cells, we transduced ADAR2 into lineage− Sca-1+ c-Kit+cells, followed by sorting ADAR2 (GFP)–positive cells for cell differentiation. Thus, overexpression of ADAR2 had no effect on the integrity of hematopoietic stem and progenitor cells (supplemental Figure 7C-E). Taken together, these data supported that ADAR2 suppresses leukemogenesis of t(8:21) AML in vivo, which is dependent of its RNA editing function.
Discussion
A-to-I RNA editing is one of the most common posttranscriptional RNA modification processes that changes the DNA coding in the mammalian transcriptome. ADAR1 and ADAR2 are the primary RNA editing enzymes catalyzing this process.3 ADAR proteins demonstrate specific expression patterns in different tissues and environments.71,72 Although dysregulation of A-to-I RNA editing is implicated in multiple cancer types,6,8,10,12,13,73-76 roles of RNA editing in leukemia, particularly those mediated by ADAR2, remain largely unexplored.
In this study, using both t(8;21) AML cell lines and mouse models, we revealed a previously undescribed role of ADAR2-mediated RNA editing in suppressing leukemogenesis. We found that ADAR2, but not ADAR1 or ADAR3, was specifically downregulated in patients with CBF AML, as evidenced by the expression analysis of multiple cohorts of patients with AML. Mechanistically, we showed that the WT RUNX1 could transcriptionally activate ADAR2 by binding to 2 distal RUNX-binding sites ∼3 kb upstream of the TSS of the ADAR2 gene. However, RUNX1-ETO and its truncated variant, AE9a, suppressed RUNX1-driven ADAR2 transcription in a dominant-negative manner, possibly through outcompeting RUNX1 for binding to the RUNX site 1. Because ADAR2 can suppress the development and/or progression of solid tumors, such as hepatocellular carcinoma and colorectal cancer through RNA editing,68,77-80 we wondered whether loss of ADAR2 and RNA editing may specifically contribute to leukemogenesis of t(8;21) AML. This hypothesis was well supported by the results obtained from cell models as well as in vivo mouse experiments. Briefly, we showed that forced expression of ADAR2 in t(8:21) and inv16 AML cells, but not non-CBF AML cells, could suppress leukemogenesis dependent on the catalytic ability of ADAR2. Although ADAR2 had no major effect on global gene expression, we found 2 targets demonstrating the protein-recoding type of editing regulated by ADAR2, COPA, and COG3, were significantly underedited in patients with CBF AML. More importantly, their edited forms (COG3-I635V and COPA-I164V) were shown to suppress the clonogenicity of t(8,21) AML cells. However, because ADAR2 has thousands of A-to-I edits in the human transcriptome, it is unlikely that COPA and COG3 are the only 2 target genes accounting for the suppressive role of ADAR2 in t(8,21) AML leukemogenesis. In gastric cancer, one of ADAR2 substrates, PODXL (podocalyxin-like), can promote tumorigenesis by interacting with the actin-binding protein EZR to induce invasion and migration in cancer cells.7 However, upon editing by ADAR2, PODXL lost its oncogenic ability.7 Moreover, ADAR2-mediated editing of IGFBP7 inhibits the proteolytic cleavage of IGFBP7, leading to increased protein stability. In esophageal squamous cell carcinoma, downregulation of ADAR2 results in more frequent cleavage of IGFBP7 in tumors than in the corresponding normal tissues.15,81 Whether these ADAR2 substrates contribute to AML leukemogenesis will remain the subject of further investigation. In addition to the cell culture–based experiments, our in vivo results showed that transplantation recipients of AE9a+ BM cells, which were stably expressing the WT ADAR2 but not the editing-depleted DeAD mutant, demonstrated significantly prolonged survival. Editing of Cog3 and Copa transcripts was also detected in ADAR2-expressing AE9a BM cells. It is not surprising to observe that mouse Cog3 and Copa transcripts could be edited by human ADAR2 protein, given the high homology between human and mouse ADAR2.82 More importantly, editing of COG3 and COPA is biologically conserved between mice and humans.68,83,84 Notably, we further corroborated the in vivo antileukemogenic effect of ADAR2 in t(8,21) AML with 2 additional findings: firstly, the transduction of ADAR2 into AE9a BM cells led to the death of AE9a+ cells; and secondly, ADAR2 had no effect on the viability of normal BM cells and the integrity of hematopoietic stem and progenitor cells. It should be noted that recipients receiving AE9a BM cells expressing editing-deficient ADAR2 also showed a slight increase in their lifespan. This might be attributed to the nonediting functions of ADAR2. For example, ADAR2 may regulate microRNA maturation through interaction with Dicer,85 or alternatively, ADAR2 may protect its target mRNAs from destabilizing proteins.58
How changes in RNA processing steps, such as splicing, polyadenylation, RNA modifications, and editing, contribute to AML remained unknown until recent years. For example, the leukemogenic role of fat mass and obesity–associated protein through reducing N6-methyladenosine (m6A) RNA modification was first reported in 2017.86 Since then, a number of studies from different groups have further confirmed the biological importance of m6A modification in normal hematopoiesis and AML and highlighted the therapeutic potential of targeting m6A RNA modification in AML.87-94 In line with these studies, we further emphasized the importance of RNA metabolism in AML. Here, we showed that forced expression of edited isoforms, such as COG3-I635V, could inhibit the colony-forming of t(8:21) AML cells, suggesting a novel therapeutic strategy for restoring edited RNAs in AML cells. Future studies using single-cell RNA-seq analysis would be essential for identifying transcriptome-wide ADAR2 targets in different cell populations. Functional importance of potential disease-associated editing targets will need to be further verified in cell lines and preclinical models, such as organoids derived from patients with AML and AML mouse models.
Although ADAR genes are frequently dysregulated in multiple cancer types, the regulatory mechanisms underlying such dysregulation remain elusive. From our study, RUNX1 was identified as a novel transcriptional factor of ADAR2; however, the existence of RUNX1-ETO and AE9a demonstrated dominant-negative effects on ADAR2 transcription, leading to downregulation of ADAR2 in t(8,21) AML. It has been reported that ADAR2 self-editing at its proximal 3′ acceptor site converts an intronic AA-to-AI dinucleotide, giving rise to an alternatively spliced transcript with a 47-nucleotide frameshift insertion in its coding sequence.18,95 This forms a negative autoregulatory feedback loop to maintain proper A-to-I editing levels in cells. However, from our RNA-seq analysis of ADAR2-overexpressing Kasumi-1 cells, we detected neither self-editing nor alternative splicing of ADAR2. Therefore, ADAR2 downregulation is most likely to be modulated through transcriptional regulation in human t(8,21) AML cells. It is known that ADAR2 is also dysregulated in gastric cancer, hepatocellular carcinoma, esophageal squamous cell carcinoma, and glioblastoma.6,7,13,15,16,18,96-99 Because both RUNX1 and ADAR2 are ubiquitously expressed in multiple tissue types, it is possible that such regulatory mechanisms are also present in other tissues. In contrast, besides t(8;21) AML, ADAR2 was also significantly downregulated in inv(16) CBF leukemia, which is associated with the formation of CBFβ-MYH11 fusion gene. The DNA-binding activity of CBFα is stabilized by dimerizing with CBFβ. As the CBFβ-MYH11 fusion protein has a stronger binding affinity to RUNX1 than CBFβ, it therefore sequesters RUNX proteins from their target genes.100-103 This may explain why ADAR2 was also downregulated in patients with inv(16) AML. However, the precise regulatory mechanism remains to be investigated. It might also be worth exploring other non-CBF-associated mechanisms contributing to the downregulation of ADAR2 in CBF leukemia. For example, self-editing and changes in chromatin status, such as histone modifications and chromatin interactions, have been linked to hematopoiesis.104,105 Wang et al reported that changes in chromatin accessibility regulate the transcription of the m6A demethylase gene ALKBH5, thereby promoting leukemogenesis via a KDM4C-ALKBH5-AXL signaling axis.106 In summary, our findings shed new light on future research on the causes and functional consequences of ADAR2 dysregulation in multiple diseases, including cancer, as well as the underlying mechanisms (editing-dependent or -independent) leading to its biological implications.
Acknowledgments
The authors thank Michelle Mok Meng Huang for assistance with flow cytometry experiments and Wright-Giemsa stain, Sudhakar Jha for providing the pMSCV flag puro plasmid, and Dong-Er Zhang for advice regarding AML ETO9a.
This work was supported by the Singapore Ministry of Health’s National Medical Research Council (NMRC), the Singapore National Research Foundation, the Singapore Ministry of Education (MOE) under its Research Centres of Excellence initiative, and the National Institutes of Health with the following details: Singapore Translational Research Investigator Award, National Institutes of Health grant (P01 HL131477 (D.G.T.), MOE Tier 2 grants (MOE2019 T2-1-083 and MOE2019 T2-2-008), NMRC Clinician Scientist-Individual Research grants (project IDs: MOH 000214 and MOH-001092-00), and the Health and Biomedical Sciences Industry Alignment Fund Pre-Positioning (H20C6a0034) (L.C.), NMRC Open Fund Young Individual Research grant (MOH OFYIRG20nov-0011) and MOE Research Scholarship Block grant (N 171 000 019 001) (Q.Z.), as well as MOE Tier 3 grant (MOE2014-T3-1-006) (D.G.T. and L.C.).
Authorship
Contribution: D.G.T. and L.C. conceived and cosupervised the study with Q.Z., M.G., and T.H.M.C.; Q.Z. designed the experiments; M.G. and T.H.M.C. performed all experiments with input from Q.Z., Y.S., and V.H.E.N.; H.Y., O.A., and Y.L. conducted all bioinformatics analyses; Z.H.T., V.H.E.N., and Z.Q.T. assisted with mouse experiments; P.S.P. and J.A.P. conducted colony-forming unit assay in inv(16) AML cells; E.I., M.M., and M.O. provided the samples from patients with t(8,21) AML; M.G.M.O., W.Y.J., L.P.K., E.C., L.K.T., and W.J.C. provided the samples from patients with the normal karyotype and t(8;21) AML; Y.G., W.W., B.T.H.K., C.M.C., and M.J.F. provided the CD34+ bone marrow samples; and D.G.T., L.C., Q.Z., and M.G. wrote the manuscript, and T.H.M.C. and X.C. provided the input for it.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Daniel G. Tenen, Cancer Science Institute of Singapore, National University of Singapore, 14 Medical Drive, Singapore 117599, Singapore; e-mail: daniel.tenen@nus.edu.sg; and Leilei Chen, Cancer Science Institute of Singapore, National University of Singapore, 14 Medical Drive, Singapore 117599, Singapore; e-mail: polly_chen@nus.edu.sg.
References
Author notes
∗M.G., T.H.M.C., and Q.Z. contributed equally to this study.
Sequencing data reported in this article have deposited in the Gene Expression Omnibus database (accession number GSE220488).
Qualified researchers may request access to individual patient level data through the data request platform at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE220488 (enter token ulmhguwohpqplen).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal