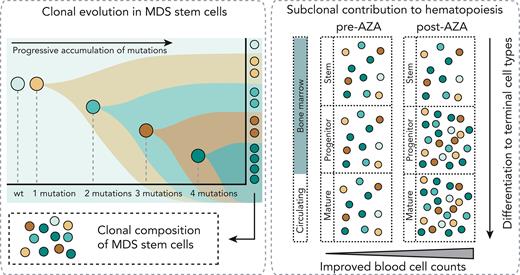
Key Points
The mutational burden in HSCs is proportionally reflected throughout hematopoietic differentiation in MDS/CMML.
Improved hematopoiesis in response to AZA therapy was associated with increased clonal output from mutant progenitors to mature cells.
Abstract
Myelodysplastic neoplasms (MDSs) and chronic myelomonocytic leukemia (CMML) are clonal disorders driven by progressively acquired somatic mutations in hematopoietic stem cells (HSCs). Hypomethylating agents (HMAs) can modify the clinical course of MDS and CMML. Clinical improvement does not require eradication of mutated cells and may be related to improved differentiation capacity of mutated HSCs. However, in patients with established disease it is unclear whether (1) HSCs with multiple mutations progress through differentiation with comparable frequency to their less mutated counterparts or (2) improvements in peripheral blood counts following HMA therapy are driven by residual wild-type HSCs or by clones with particular combinations of mutations. To address these questions, the somatic mutations of individual stem cells, progenitors (common myeloid progenitors, granulocyte monocyte progenitors, and megakaryocyte erythroid progenitors), and matched circulating hematopoietic cells (monocytes, neutrophils, and naïve B cells) in MDS and CMML were characterized via high-throughput single-cell genotyping, followed by bulk analysis in immature and mature cells before and after AZA treatment. The mutational burden was similar throughout differentiation, with even the most mutated stem and progenitor clones maintaining their capacity to differentiate to mature cell types in vivo. Increased contributions from productive mutant progenitors appear to underlie improved hematopoiesis in MDS following HMA therapy.
Introduction
Somatic mutations in hematopoietic stem cells (HSCs) are a central pathogenic event in myelodysplastic neoplasms (MDSs) and chronic myelomonocytic leukemia (CMML).1-6 Patients with high-risk disease who are ineligible for allogeneic bone marrow (BM) transplantation are treated with hypomethylating agents (HMAs), usually 5-azacytidine (AZA). AZA treatment can improve peripheral blood (PB) counts and delay progression to acute myeloid leukemia in some patients.7-10 Mutations progress in stepwise branching fashion to establish clonal structures in HSCs, but contribution of specific clones in circulating mature blood cell types is unclear (supplemental Figure 1, available on the Blood website). We and others have previously described cohorts with hematological response to AZA despite persistently high variant allele fractions (VAFs) in BM.11-13 Colonies derived from in vitro assays of stem cell function following AZA treatment showed decreased mutational complexity, suggesting a shift in hematopoiesis from clones with high to low mutational burden in response to treatment.12 However, in vitro colony-forming capacity might not correlate with in vivo hematopoietic potential, and whether mutated clones are proportionally represented in progenitor and mature cells or whether cells with fewer mutations are better able to contribute to mature cells is unknown. Furthermore, it is unclear how such contributions vary between patients who do, or do not, respond to AZA. Single-cell genotyping techniques can resolve combinations of mutations in cells.1,14-17 Index sorting and single-cell genotyping were used to characterize the haplotype composition of individual stem cells (HSCs/multipotent progenitors [MPPs] and MDS stem cells [MDS-SCs]), progenitors (common myeloid progenitors, granulocyte monocyte progenitors, and megakaryocyte erythroid progenitors), and high-turnover circulating cells (monocytes, neutrophils, and naïve B cells [nBCs]) in treatment-naïve and AZA-treated MDS and CMML. We further characterized the VAF in progenitor and mature cells before and after treatment in AZA responders and nonresponders.
Study design
Samples were collected with patient consent and institutional ethics approval. The BM was enriched for CD34+ cells and single-cell index sorted into 384-well plates (supplemental Figure 2). A multiplex polymerase chain reaction–based strategy was used to amplify mutations in single cells. Amplicons were then barcoded, followed by Illumina sequencing (supplemental Figure 3; supplemental Material). Capture sequencing was performed using a targeted panel for myeloid driver mutations (supplemental Material). Mutational calling was performed via pairwise sequence alignments using SeqAn18 and seqanpy (https://github.com/iosonofabio/seqanpy). Analysis code is available at https://github.com/julie-thoms/MDS_amplicons.
Results and discussion
Matched stem cells and progenitor cells from BM and high-turnover differentiated cells from PB from 3 patients were analyzed (Figure 1A; supplemental Table 1) and VAFs were determined in bulk samples from each cell type. Variant alleles were detected at high frequency (a 0.5 VAF indicates that essentially every diploid cell carries a mutated copy of that allele). VAF distributions were similar across BM and PB cell types in all patients with the exception that nBCs in patient #H198304 were predominantly wild-type (WT) (Figure 1B-D, left), which were confirmed using an orthogonal approach (supplemental Figure 4). Then VAFs of known mutations were determined in single cells (Figure 1B-D, right; supplemental Figure 5). Allele fractions were highly correlated between bulk- and single-cell assessments (supplemental Figure 6; Pearson r = 0.8989).
Mutational burden in matched stem and progenitor cells in the BM and differentiated cells in PB. (A) Blood differentiation hierarchy in MDS/CMML showing stem cells (healthy stem cells [HSCs/MPP] and MDS-SC), progenitors (common myeloid progenitors [CMP], granulocyte monocyte progenitors [GMP], megakaryocyte eythroid progenitors [MEP], and common lymphoid progenitors [CLP]), and differentiated mature cells (left). Those cell types colored white were not characterized in this figure. Schematic showing collection and cell sorting strategies for PB and BM (right). PB was flow sorted into neutrophils (Neut: SSChi, CD45+, immunoglobulin D–negative, CD16+, and CD66b+), monocytes (Mono: SSClo, CD45+, immunoglobulin D–negative, and CD16+), and nBC (SSClo, CD45+, IgD+, and CD27−). BM mononuclear cells (BM-MNC) were isolated on Ficoll and used directly for bulk capture sequencing. MACS-enriched CD34+ cells (BM-CD34+) were dropped into 384-well plates for amplicon sequencing, with indexing for CD38, CD123, CD45RA, CD90, and IL1RAP, and post hoc assignment of cell type (HSC/MPP: LIN−, CD34+, CD38lo, CD45RA−, CD123−, IL1RAP−; MDS-SC: LIN−, CD34+, CD38lo, [CD45RA+ or CD123+ or IL1RAP+]; CMP: LIN−, CD34+, CD38+, CD45RA−, CD123+; GMP: LIN−, CD34+, CD38+, CD45RA+, CD123+; and MEP: LIN−, CD34+, CD38+, CD45RA−, CD123−). (B-D) (i) VAFs determined by capture sequencing in bulk BM and PB cell types (bulk VAF) and corresponding VAFs determined by amplicon sequencing in single cells (single-cell VAF). VAFs refer to alleles: in diploid cells a VAF of 0.5 indicates that every cell carries a mutated allele. VAFs >0.5 can occur where there is loss of heterozygosity or in the case of X-linked genes in male patients where each cell carries only 1 copy of the allele. For single-cell VAFs, each allele was analyzed individually, and the bar graph indicates the number of cells analyzed for each mutation in each cell type. Bars show standard error of the mean. (ii) Single-cell haplotypes. Pie charts show the proportions of cells across the hematopoietic hierarchy carrying 0, 1, 2, 3, or 4 mutations in the specified alleles; the number of individual cells analyzed for each population are as indicated in square brackets. Patient #H198302 (B), patient #H198303 (C), and patient #H198304 (D) are shown respectively. Ery, erythrocyte; Meg, megakaryocyte.
Mutational burden in matched stem and progenitor cells in the BM and differentiated cells in PB. (A) Blood differentiation hierarchy in MDS/CMML showing stem cells (healthy stem cells [HSCs/MPP] and MDS-SC), progenitors (common myeloid progenitors [CMP], granulocyte monocyte progenitors [GMP], megakaryocyte eythroid progenitors [MEP], and common lymphoid progenitors [CLP]), and differentiated mature cells (left). Those cell types colored white were not characterized in this figure. Schematic showing collection and cell sorting strategies for PB and BM (right). PB was flow sorted into neutrophils (Neut: SSChi, CD45+, immunoglobulin D–negative, CD16+, and CD66b+), monocytes (Mono: SSClo, CD45+, immunoglobulin D–negative, and CD16+), and nBC (SSClo, CD45+, IgD+, and CD27−). BM mononuclear cells (BM-MNC) were isolated on Ficoll and used directly for bulk capture sequencing. MACS-enriched CD34+ cells (BM-CD34+) were dropped into 384-well plates for amplicon sequencing, with indexing for CD38, CD123, CD45RA, CD90, and IL1RAP, and post hoc assignment of cell type (HSC/MPP: LIN−, CD34+, CD38lo, CD45RA−, CD123−, IL1RAP−; MDS-SC: LIN−, CD34+, CD38lo, [CD45RA+ or CD123+ or IL1RAP+]; CMP: LIN−, CD34+, CD38+, CD45RA−, CD123+; GMP: LIN−, CD34+, CD38+, CD45RA+, CD123+; and MEP: LIN−, CD34+, CD38+, CD45RA−, CD123−). (B-D) (i) VAFs determined by capture sequencing in bulk BM and PB cell types (bulk VAF) and corresponding VAFs determined by amplicon sequencing in single cells (single-cell VAF). VAFs refer to alleles: in diploid cells a VAF of 0.5 indicates that every cell carries a mutated allele. VAFs >0.5 can occur where there is loss of heterozygosity or in the case of X-linked genes in male patients where each cell carries only 1 copy of the allele. For single-cell VAFs, each allele was analyzed individually, and the bar graph indicates the number of cells analyzed for each mutation in each cell type. Bars show standard error of the mean. (ii) Single-cell haplotypes. Pie charts show the proportions of cells across the hematopoietic hierarchy carrying 0, 1, 2, 3, or 4 mutations in the specified alleles; the number of individual cells analyzed for each population are as indicated in square brackets. Patient #H198302 (B), patient #H198303 (C), and patient #H198304 (D) are shown respectively. Ery, erythrocyte; Meg, megakaryocyte.
Stem cells with multiple mutations might contribute proportionally (neutral selection) or asymmetrically (negative clonal selection) to mature circulating cells (supplemental Figure 1B). To resolve this, BM cells from 3 patients were classified (supplemental Table 1) as healthy HSC/MPPs, MDS-SCs, common myeloid progenitors, granulocyte monocyte progenitors, or megakaryocyte erythroid progenitors using indexed fluorescence-activated cell sorting and assessed the presence/absence of known variants in BM cell types and matched PB neutrophils, monocytes, and nBCs (Figure 1B-D; supplemental Figure 7).
In patient #H198302, 4 mutations were tracked (Figure 1Bii; SRSF2, CUX1, and biallelic TET2). Most stem cells carried 2 or 3 mutations and we detected no WT HSC/MPPs. Cells across the progenitor compartment were similar and mostly highly mutated, which is a pattern that was maintained particularly in differentiated monocytes and neutrophils.
In patient #H198303, 2 mutations12 were tracked (Figure 1Cii; biallelic TET2). In the stem compartment, cells carrying biallelic TET2 mutations were frequent, but numerous cells were WT or carried a single mutant allele. Progenitors were relatively homogenous, although mutational burden differed slightly from the stem compartment. The haplotype distribution in mature cells was again like progenitor cells, with the exception that no WT neutrophils were detected.
In patient #H198304, 4 mutations were tracked (Figure 1Dii; SRSF2, RUNX1, and biallelic TET2). Most stem cells carried 2 or 3 mutations, a few healthy HSC/MPPs had no mutations detected, and ∼20% of MDS-SCs carried an additional mutated RUNX1 allele. In the progenitor compartment, all cell types were similar. In the mature compartment, myeloid populations contained cells with 1, 2, 3, or 4 mutations, with a higher proportion of cells with all 4 alleles mutated than in progenitors, suggesting that highly mutated stem cells can produce differentiated myeloid cells. Consistent with bulk analysis, few mutant cells were detected in nBCs. Additional PB analysis in this patient revealed that naïve T cells, but not natural killer (NK) cells, were also predominantly WT (supplemental Figure 4C-D), suggesting specific impairment of B- and T-lineage maturation in mutated cells.
Attrition of highly mutated cells during myeloid maturation was not observed in any patient with single-cell analysis, suggesting that highly mutated stem and progenitor cells retain some capacity to differentiate in vivo. To extend these observations and further characterize the impact of HMA therapy, we analyzed archived pre- or post-AZA samples from a second MDS cohort (supplemental Table 1) and measured VAFs in mononuclear cells and bulk sorted immature myeloid progenitors (imMyes), monocytes, and NK cells from BM (Figure 2A). The pretreatment clonal composition of mature myeloid cells (monocytes) essentially mirrored total BM and imMye in all patients (Figure 2B-C). The NK cells, which are generally believed to derive from lymphoid progenitors, shared a similar clonal composition with their myeloid counterparts, albeit with lower VAFs. In 2 patients, (#61293005 and #61293004) subclones with biallelic TP53 mutations that did not contribute to mature cells were detectable in imMye (VAF range, 0.08-0.41). Following AZA treatment, the VAFs of these noncontributing subclones diminished as patients responded to treatment.
Fidelity of mutational burden in immature and mature cells before and after 6 cycles of AZA in clinical responders and nonresponders. (A) Schematic showing AZA treatment regime and sorting strategy to assess VAF in multiple cell types before and after AZA treatment. Those cell types colored white were not characterized in this figure. (B-C) VAFs in individual cell types, pre- and post-AZA treatment in 9 patients with MDS, for the indicated variants for each patient with corresponding clinical parameters (neutrophils, ×109/L; platelets [Plts], ×109/L; hemoglobin [Hb], g/L; and blasts, % blasts in BM). VAFs refer to alleles: in diploid cells a VAF of 0.5 indicates that every cell carries a mutated allele. VAFs >0.5 can occur where there is loss of heterozygosity or in the case of X-linked genes in male patients where each cell carries only 1 copy of the allele. For each patient the International Working Group [IWG] (2006) assessment post-AZA is shown.19 Responders (B) (complete remission [CR], marrow complete remission [mCR], and hematological improvement [HI]) are indicated in purple and nonresponders (C) (all stable disease [SD]) are indicated in lime. Variant names are abbreviated as gene names; multiple occurrences of the same gene in a single patient indicate multiple variants detected. Full variant IDs are provided in the supplemental Table 2.
Fidelity of mutational burden in immature and mature cells before and after 6 cycles of AZA in clinical responders and nonresponders. (A) Schematic showing AZA treatment regime and sorting strategy to assess VAF in multiple cell types before and after AZA treatment. Those cell types colored white were not characterized in this figure. (B-C) VAFs in individual cell types, pre- and post-AZA treatment in 9 patients with MDS, for the indicated variants for each patient with corresponding clinical parameters (neutrophils, ×109/L; platelets [Plts], ×109/L; hemoglobin [Hb], g/L; and blasts, % blasts in BM). VAFs refer to alleles: in diploid cells a VAF of 0.5 indicates that every cell carries a mutated allele. VAFs >0.5 can occur where there is loss of heterozygosity or in the case of X-linked genes in male patients where each cell carries only 1 copy of the allele. For each patient the International Working Group [IWG] (2006) assessment post-AZA is shown.19 Responders (B) (complete remission [CR], marrow complete remission [mCR], and hematological improvement [HI]) are indicated in purple and nonresponders (C) (all stable disease [SD]) are indicated in lime. Variant names are abbreviated as gene names; multiple occurrences of the same gene in a single patient indicate multiple variants detected. Full variant IDs are provided in the supplemental Table 2.
Following 6 cycles of AZA therapy, VAFs in mature myeloid cells continued to reflect those in imMye, irrespective of clinical response. As in pretreatment samples, NK cells generally had lower VAFs than their myeloid counterparts, which is consistent with observations in the primary cohort that mutant HSCs contribute to both myeloid and lymphoid lineages but with a bias toward the former. Overall, highly mutated progenitor clones contributed proportionally to the mature population in all patients except for biallelic TP53 mutant clones, and clinical response to HMA occurred without substantial variations in subclonal structure.
In summary, the mutational profiles of thousands of individual stem/progenitor/mature cells plus bulk sorted cells were characterized from a total of 12 patients with MDS/CMML and found that in vivo, highly mutated stem cells contribute to productive hematopoiesis (supplemental Figure 1B, neutral selection). We observed proportional contribution from specific clones across the stem to mature trajectory and found these proportions were retained following HMA therapy. In patients who showed clinical response to HMAs, we observed modest variations in subclonal structure after treatment. Notably, several clones (all with biallelic TP53 alterations) did not contribute to mature cells and diminished after HMA treatment. However, in patients who failed to respond, there was little or no variation in subclonal structure following AZA treatment. Our data also support the hypothesis that clinical response to HMAs can and frequently does occur without substantial variations in subclonal structure, suggesting that improved circulating cell counts in responder patients are driven by increased output from mutated HSCs.
These findings are pertinent when combining cytotoxic therapies designed to eliminate mutant cells with HMAs.20 Given recent reports that the number of HSCs that contribute to blood formation in clonal hematopoiesis and elderly individuals are small,21,22 our study questions whether therapeutic principles and end points that apply in high-blast acute myeloid leukemia (clonal eradication and minimal residual disease monitoring for relapse) are appropriate in contexts where clinical improvement does not require ablation of mutant clones.
Acknowledgments
The authors thank the study participants for their generosity in providing samples and Swapna Joshi and Ameline Lim for assistance in processing the peripheral blood and bone marrow samples.
A.S.S.-K. was supported by a Swiss National Research Foundation early post doctoral mobility grant (P2LAP3_181273). J.A.I.T. was supported by the Anthony Rothe Memorial Trust. S.K.B. and P.M.K. are supported by Leukaemia and Blood Cancer New Zealand and the family of Marijana Kumerich. J.E.P. was supported by grants from the National Health and Medical Research Council of Australia (grants GNT1139787, GNT2011627, and MRF1200271), a translational program grant from the Leukemia Lymphoma Society (LLS)-Snowdome Foundation-Leukaemia Foundation (6620-21), and the Anthony Rothe Memorial Trust.
Authorship
Contribution: A.S.S.-K., J.A.I.T., R.L., P.W., A.R.-M., A.J.M., A.U., M.N.P., M.H., O.R.F., C.J.J., F.Z., and J.E.P. designed the study; A.S.S.-K., J.A.I.T., G.S.B., H.R.H., L.V., P.M.K., H.M.L., E.M.V.J., T.W.F., G.M.A., J.K., S.D., O.R.F., and C.J.J. performed the research; A.S.S.-K., J.A.I.T., K.R., P.M.K., S.K.B., E.P., C.J.J., F.Z., and J.E.P. analyzed and interpreted the data; and A.S.S.-K., J.A.I.T., C.J.J., F.Z., and J.E.P. wrote the manuscript.
Conflict-of-interest disclosure: M.N.P. received research funding and/or provision of drug for clinical trials (to institution) from AstraZeneca, BRII Biosciences, Celgene/Bristol Myers Squibb (BMS), CSL Behring, Eli Lilly, Emergent Biosciences, Gilead Pharmaceuticals, GlaxoSmithKline, Grifols, Janssen/Johnson and Johnson, Takeda, and ViiV Pharmaceuticals and has advisory roles with Celgene/BMS, Gilead Pharmaceuticals, and ViiV Pharmaceuticals. M.H. is a consultant/advisory board member at Roche, Gilead, Otsuka, Janssen, BeiGene, and Takeda. E.P is a founder, equity holder, and holds a fiduciary role in Isabl Inc. J.E.P. received research funding and/or provision of drug for clinical trials (to institution) from Celgene/BMS, Astex, Verastem Oncology and received honoraria from AbbVie as an advisory board member. The remaining authors declare no competing financial interests.
Correspondence: John E. Pimanda, School of Biomedical Sciences, Lowy Cancer Research Centre, UNSW Sydney, Sydney, NSW 2052, Australia; e-mail: jpimanda@unsw.edu.au; Fabio Zanini, School of Clinical Medicine, Lowy Cancer Research Centre, UNSW Sydney, Sydney, NSW 2052, Australia; e-mail: fabio.zanini@unsw.edu.au; Christopher J. Jolly, School of Biomedical Sciences, Lowy Cancer Research Centre, UNSW Sydney, Sydney, NSW 2052, Australia; e-mail: c.jolly@unsw.edu.au; and Omid R. Faridani, School of Biomedical Sciences, Lowy Cancer Research Centre, UNSW Sydney, Sydney, NSW 2052, Australia; e-mail: o.faridani@unsw.edu.au.
References
Author notes
∗A.S.S.-K. and J.A.I.T. contributed equally to this study.
†F.Z. and J.E.P. are joint senior authors and contributed equally to this study.
The raw data reported in this article have been deposited in the Sequence Read Archive database (accession number PRJNA798507) and https://flowrepository.org/id/FR-FCM-Z4PR.
Data are available on request from the corresponding authors; John E. Pimanda (jpimanda@unsw.edu.au), Fabio Zanini (fabio.zanini@unsw.edu.au), Christopher J. Jolly (c.jolly@unsw.edu.au), and Omid R. Faridani (o.faridani@unsw.edu.au).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![Mutational burden in matched stem and progenitor cells in the BM and differentiated cells in PB. (A) Blood differentiation hierarchy in MDS/CMML showing stem cells (healthy stem cells [HSCs/MPP] and MDS-SC), progenitors (common myeloid progenitors [CMP], granulocyte monocyte progenitors [GMP], megakaryocyte eythroid progenitors [MEP], and common lymphoid progenitors [CLP]), and differentiated mature cells (left). Those cell types colored white were not characterized in this figure. Schematic showing collection and cell sorting strategies for PB and BM (right). PB was flow sorted into neutrophils (Neut: SSChi, CD45+, immunoglobulin D–negative, CD16+, and CD66b+), monocytes (Mono: SSClo, CD45+, immunoglobulin D–negative, and CD16+), and nBC (SSClo, CD45+, IgD+, and CD27−). BM mononuclear cells (BM-MNC) were isolated on Ficoll and used directly for bulk capture sequencing. MACS-enriched CD34+ cells (BM-CD34+) were dropped into 384-well plates for amplicon sequencing, with indexing for CD38, CD123, CD45RA, CD90, and IL1RAP, and post hoc assignment of cell type (HSC/MPP: LIN−, CD34+, CD38lo, CD45RA−, CD123−, IL1RAP−; MDS-SC: LIN−, CD34+, CD38lo, [CD45RA+ or CD123+ or IL1RAP+]; CMP: LIN−, CD34+, CD38+, CD45RA−, CD123+; GMP: LIN−, CD34+, CD38+, CD45RA+, CD123+; and MEP: LIN−, CD34+, CD38+, CD45RA−, CD123−). (B-D) (i) VAFs determined by capture sequencing in bulk BM and PB cell types (bulk VAF) and corresponding VAFs determined by amplicon sequencing in single cells (single-cell VAF). VAFs refer to alleles: in diploid cells a VAF of 0.5 indicates that every cell carries a mutated allele. VAFs >0.5 can occur where there is loss of heterozygosity or in the case of X-linked genes in male patients where each cell carries only 1 copy of the allele. For single-cell VAFs, each allele was analyzed individually, and the bar graph indicates the number of cells analyzed for each mutation in each cell type. Bars show standard error of the mean. (ii) Single-cell haplotypes. Pie charts show the proportions of cells across the hematopoietic hierarchy carrying 0, 1, 2, 3, or 4 mutations in the specified alleles; the number of individual cells analyzed for each population are as indicated in square brackets. Patient #H198302 (B), patient #H198303 (C), and patient #H198304 (D) are shown respectively. Ery, erythrocyte; Meg, megakaryocyte.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/11/10.1182_blood.2022018602/4/m_blood_bld-2022-018602-gr1.jpeg?Expires=1770135309&Signature=CSnZAdohMhJdF6xYlrKLM5R1ybCuvmH3pn8zGkzvYzkzXOJkOdwki8fqjobF4jtyb2cyn8HVwYQQCtqddjmFKgQCRi0li6Q4nrCTlwZ4540aU2xVlW75omhCpQh-h9yeIwAwy9vgZUMKUlePyUG6DK1oASG-s6kchdKaagAmHL4LT0NWQ9SGyRekqUTlbnTSUEMcgjknsM8xJsOPm8wEwg6YlQzoq698WPx53NgpScoXvpLu857yc6stqr0q7ADP4nHP0Lf4nB95fv2IoEArhMUXEzs2O34Ayjd9kDN04x2pWsIQKQZ~J7Ds51Vo5JH9Io2AGu7Car28xyJ66u3qKw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fidelity of mutational burden in immature and mature cells before and after 6 cycles of AZA in clinical responders and nonresponders. (A) Schematic showing AZA treatment regime and sorting strategy to assess VAF in multiple cell types before and after AZA treatment. Those cell types colored white were not characterized in this figure. (B-C) VAFs in individual cell types, pre- and post-AZA treatment in 9 patients with MDS, for the indicated variants for each patient with corresponding clinical parameters (neutrophils, ×109/L; platelets [Plts], ×109/L; hemoglobin [Hb], g/L; and blasts, % blasts in BM). VAFs refer to alleles: in diploid cells a VAF of 0.5 indicates that every cell carries a mutated allele. VAFs >0.5 can occur where there is loss of heterozygosity or in the case of X-linked genes in male patients where each cell carries only 1 copy of the allele. For each patient the International Working Group [IWG] (2006) assessment post-AZA is shown.19 Responders (B) (complete remission [CR], marrow complete remission [mCR], and hematological improvement [HI]) are indicated in purple and nonresponders (C) (all stable disease [SD]) are indicated in lime. Variant names are abbreviated as gene names; multiple occurrences of the same gene in a single patient indicate multiple variants detected. Full variant IDs are provided in the supplemental Table 2.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/11/10.1182_blood.2022018602/4/m_blood_bld-2022-018602-gr2.jpeg?Expires=1770135309&Signature=ay-Rh~b~-ZxOIMD0Bk8SnvBmhRkdWtka-NM9bxnfFZrgM8qluIJOFYd~~OnnZZnrUPOlPfieJH8QmuPNOD5NnHYQKM5ox2ve1a8sWhq36DvrNDfQx3ms6n1ufQXfYTtz5kXd97JuOcB533JH9mM2~5aTzUiLC0DxJfqoTqZtz-VIgNxHxDQF5hUpNfc7gygFNiSqaHzuvCMf3VpDrRFy~SEdINdw~fyzaHPeB8Lv2fgQ1iqGRBUrlVcohvfryWhevatTAXbD6mH8FhvUmw-ABOUDEpm4LiNseR5DBylrRg6rw3jx1u1zq5~vvSe5hqHPoogbWYvOglfUCqPf2N4QKg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal