Key Points
Wnt pathway is frequently mutated in CLL.
Wnt pathway mutations can lead to pathway activation and enhanced CLL survival.
Abstract
One major goal of cancer genome sequencing is to identify key genes and pathways that drive tumor pathogenesis. Although many studies have identified candidate driver genes based on recurrence of mutations in individual genes, subsets of genes with nonrecurrent mutations may also be defined as putative drivers if they affect a single biological pathway. In this fashion, we previously identified Wnt signaling as significantly mutated through large-scale massively parallel DNA sequencing of chronic lymphocytic leukemia (CLL). Here, we use a novel method of biomolecule delivery, vertical silicon nanowires, to efficiently introduce small interfering RNAs into CLL cells, and interrogate the effects of 8 of 15 mutated Wnt pathway members identified across 91 CLLs. In HEK293T cells, mutations in 2 genes did not generate functional changes, 3 led to dysregulated pathway activation, and 3 led to further activation or loss of repression of pathway activation. Silencing 4 of 8 mutated genes in CLL samples harboring the mutated alleles resulted in reduced viability compared with leukemia samples with wild-type alleles. We demonstrate that somatic mutations in CLL can generate dependence on this pathway for survival. These findings support the notion that nonrecurrent mutations at different nodes of the Wnt pathway can contribute to leukemogenesis.
Introduction
The advent of massively parallel sequencing (MPS) has enabled the unprecedented ability to systematically discover key genetic alterations underlying cancer.1 In one example, we and others previously reported the results of large-scale whole-exome sequencing of chronic lymphocytic leukemia (CLL), a common adult leukemia marked by a highly variable clinical course among patients.2-5 In these studies, each of the significantly mutated genes suggested key pathways critical to CLL pathogenesis. In addition, the Wnt pathway was supported as a CLL-associated pathway because significantly more mutations in the Wnt pathway components were detected than expected, even while no single Wnt pathway member was identified as a putative CLL driver.2 These findings complement the previous observations of highly dysregulated gene expression and hypermethylation of Wnt pathway genes, as well as of the key pathway member LEF1 as a CLL risk loci identified by genome-wide association.1,6-13
The Wnt pathway is critical for the proliferation and cell fate determination of many cell types, including B cells.14 The discovery of multiple mutated Wnt pathway members motivated us to query the role of pathway member mutations in altering signaling and cell survival. A major barrier to the functional assessment of genetic alterations in CLL has been the lack of cell lines faithful to this malignancy and the poor efficiency of conventional transfection methodologies to genetically manipulate primary CLL-B cells. Herein, we used a recently developed biomolecule delivery platform based on vertical silicon nanowires (NWs)15,16 to assess the effects of gene knockdown on primary CLL-B cell survival. We demonstrate that inhibition of the Wnt pathway at different levels adversely affects CLL survival. Moreover, we observe that CLLs harboring dysregulating Wnt pathway mutations were dependent on their expression for survival. Hence, somatic mutation is a mechanism by which the Wnt pathway is modulated in CLL, and genetic characterization of the Wnt signaling can identify subsets of CLL patients with greater sensitivity to targeting of this pathway.
Methods
Human samples
Heparinized blood skin biopsies were obtained from normal donors and patients enrolled on clinical research protocols at the Dana-Farber Harvard Cancer Center approved by the Dana-Farber Harvard Cancer Center Human Subjects Protection Committee.2 Peripheral blood mononuclear cells were isolated by Ficoll/Hypaque density gradient centrifugation. Mononuclear cells were used fresh or cryopreserved with fetal bovine serum/10% dimethylsulfoxide, and were stored in vapor-phase liquid nitrogen until the time of analysis. This study was conducted in accordance with the Declaration of Helsinki.
Calculation of Wnt pathway significance in CLL
Gene expression microarray data analysis
Total RNA was isolated from immunomagnetically sorted CD19+ peripheral blood B cells and CLL cells (>95% CD19+CD5+) using TRIzol (Invitrogen), followed by column purification (RNeasy Mini Kit; Qiagen, Valencia CA). RNA samples were hybridized to Affymetrix U133A+ 2.0 arrays (Santa Cruz Biotechnology) at the Dana-Farber Cancer Institute (DFCI) Microarray Core Facility. Microarray data can be accessed at http://www.ncbi.nlm.nih.gov/geo/info/linking.html with accession number GSE31048. Details regarding the microarray data analysis can be found in the supplemental Methods.
Detection of Wnt activation
Depending on the putative function of the various Wnt pathway genes, activation of the Wnt pathway was interrogated using: (1) A plasmid-based luciferase reporter assay (SuperTOPflash, pRL-TK; gift from Xi He, Children’s Hospital Boston); (2) a reverse transcription–polymerase chain reaction (RT-PCR) assay for detection of the expression of Wnt pathway targets; or (3) a western blot–based assay for detection of phosphorylation of DVL2. Detailed information regarding these assays of Wnt activation is provided in the supplemental Methods.
Wnt target gene silencing by NWs
Silicon NWs were fabricated as before,15 placed in a 96-well flat bottom plate, coated with 3 μL of 50 to 100 μM control small-interfering RNA (siRNA) (ON-TARGETplus Negative Controls; Dharmacon, Lafayette, CO; silencer negative control siRNA; Applied Biosystems, Carlsbad, CA), or siRNAs targeting mutated Wnt pathway members (Dharmacon, Lafayette, CO; Applied Biosystems, Carlsbad, CA), or Alexa 546 labeled anti-vimentin siRNA and then air-dried under sterile conditions. The 1.2 × 104 CLL-B cells in 10 μL were introduced atop NWs and incubated at 37°C for 40 minutes, followed by addition of 100 μL of B-cell culture medium. At 48 hours, cell viability was evaluated by luminescence cell viability assay (CellTiter-Glo; Promega, Madison, WI), according to the manufacturer’s recommendation. Gene silencing was confirmed either by Taqman quantitative RT-PCR or immunofluorescence imaging of cells16 after NW-mediated siRNA delivery. In some experiments, cells were subjected to scanning electron microscope imaging, as previously described,15 24 to 48 hours after plating.
Statistical considerations
Normalized luciferase activity between normal and CLL-B cells were compared using the 2-sided Wilcoxon rank-sum test. The significance of changes to cell survival after NW delivery of siRNAs targeting Wnt pathway components as compared with normal B cells, or among CLL samples, was calculated using the 2-sided Welch t test (DVL1, CTNNB1, LEF1), the 1-tailed 1 sample mean Student t test (BCL9, DKK2), or 95% confidence interval (RYK, CSNK1E, FZD5, WNT1, WNT10A, a P value <.05 denotes the exclusion for sample with a mutation from the 95% confidence interval of the CLL without mutation group). Further details on choice of statistical test are provided in the supplemental Methods.
Results
Fourteen percent of CLL samples harbor somatic coding mutations in the Wnt pathway
We previously reported the results of DNA sequencing of 91 matched CLL/normal samples.2 These samples were collected from patients representing the broad spectrum of CLL clinical heterogeneity, based on established prognostic risk factors (ZAP70 expression; degree of somatic hypermutation in the variable region of the immunoglobulin heavy chain [IGHV] gene; presence of specific CLL-associated cytogenetic abnormalities). Of 1838 nonsynonymous coding mutations from 91 cases, we identified 15 nonsynonymous mutations in 12 unique Wnt pathway members (from 66 core members, which were selected based on established databases), present in 13 CLL samples (supplemental Table 1). Compared with the background CLL mutation rate of 0.75/Mb,2 the conserved exonic regions of the 66 Wnt pathway genes were mutated at a significantly higher rate (1.53/Mb, MutSig analysis; P = .00067).19
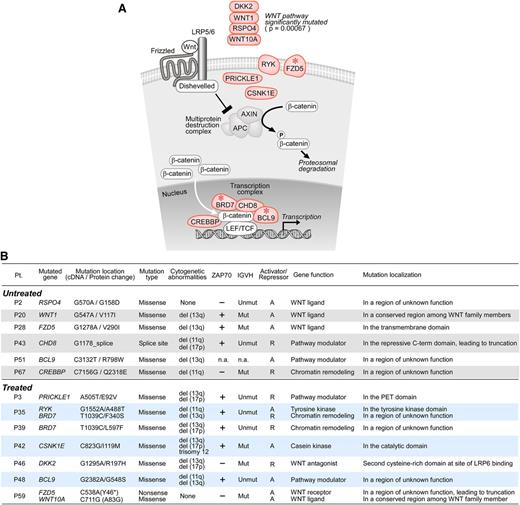
The Wnt signaling pathway contains more than 100 members.20-22 As shown in Figure 1A, in the absence of Wnt activation, β-catenin (CTNNB1) is phosphorylated by the multiprotein destruction complex and is subject to proteosomal degradation. When Wnt binds to its receptor, composed of frizzled and LRP5/6, a reaction cascade is induced, including recruitment of dishevelled (DVL1). This results in disassembly of the destruction complex, accumulation of cytoplasmic β-catenin and the subsequent nuclear translocation of β-catenin, and its binding to transcription factors such as TCF/LEF that regulate target gene transcription.
The Wnt pathway is significantly mutated in CLL (P = .00067). (A) Cellular localization of mutations in Wnt pathway components in CLL (light red). *Pathway genes mutated in more than 1 CLL sample. (B) Clinical characteristics of CLL samples harboring Wnt pathway mutations, as well as the putative function of the mutated Wnt pathway genes and their genomic localization. A, pathway activator; R, pathway repressor.
The Wnt pathway is significantly mutated in CLL (P = .00067). (A) Cellular localization of mutations in Wnt pathway components in CLL (light red). *Pathway genes mutated in more than 1 CLL sample. (B) Clinical characteristics of CLL samples harboring Wnt pathway mutations, as well as the putative function of the mutated Wnt pathway genes and their genomic localization. A, pathway activator; R, pathway repressor.
We identified 4 mutations in extracellular Wnt pathway components (WNT1, WNT10A, DKK2, RSPO4) (Figure 1A). Other identified mutated components were transmembrane receptors (FZD5, RYK), cytoplasmic factors (CSNK1E, PRICKLE1), and nuclear factors that modulate the TCF/LEF complex (CHD8, BRD7, CREBBP, BCL9). Notably, 3 genes were each mutated in the tumors of distinct patients (FZD5 [in patients P28, P59], BRD7 [P35, P39], BCL9 [P48, P51]), but not at recurrent sites. We found mutations in genes with known activating function on the Wnt signaling pathway (WNT1, WNT10A, RSPO4, CSNK1E, CREBBP, RYK, FZD5, BCL9), as well as genes with known pathway-repressive effects (DKK2, PRICKLE1, CHD8, BRD7). Targeted RNA pyrosequencing confirmed transcript expression of 5 of 5 mutated alleles (DKK2, BCL9, RYK, FZD5, and BRD7) in 4 patients (P28, P35, P46, P48) (supplemental Figure 1 and supplemental Table 2). Mutated transcripts were measured at a frequency of 25% to 50%, consistent with their heterozygous mutation status.
Mutations in the Wnt pathway were not associated with any known CLL prognostic factor. Six of 13 (46%) cases had unmutated IGHV status, and 8 of 13 (62%) were positive for ZAP70 expression (Figure 1B). Furthermore, no associations were observed between mutation and age at diagnosis, clinical stage, presence of cytogenetic abnormalities, mutation rate, or time to first therapy (supplemental Table 3). Six of 13 patients harboring mutations were chemotherapy-naïve, which suggests that pathway alteration is intrinsic to CLL rather than a result of chemotherapy exposure.
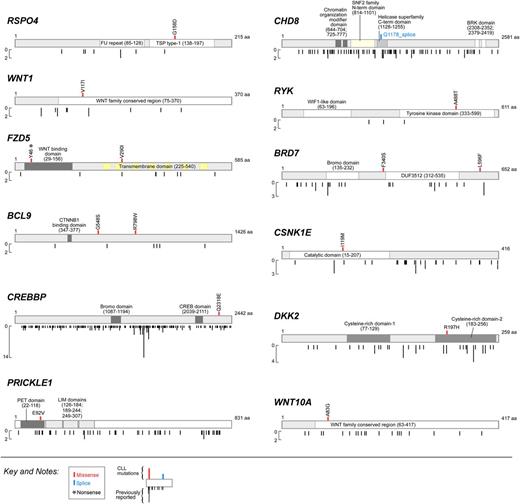
Wnt pathway mutations target evolutionarily conserved sites
With the exception of the mutations in DKK2 (p.R197H) and FZD5 (p.V290I),23 all 15 mutated Wnt pathway members localized to novel mutation sites (Figure 2, cosmic database, version 64). Furthermore, all were present at evolutionarily conserved regions, supporting a functional role for these mutations in perturbing Wnt pathway function (supplemental Figure 2). Many of the gene localizations suggested a potential mechanism through which the mutations could functionally alter pathway signaling (Figures 1B and 2). For example, we found 2 mutations in the Wnt receptor FZD5: a nonsense mutation (p.Y46*, P59) leading to protein truncation; and a missense mutation (p.V290I, P28) in the transmembrane domain. In another example, DKK2, a secreted protein that normally represses Wnt pathway activation, was mutated in P46 at a known critical point-of-contact with LRP5/6, its coreceptor.24 Two Wnt pathway-activating kinase mutations were found in regions critical for kinase function (ie, RYK [p.A488T, P35]) within the tyrosine kinase domain, and CSNK1E (p.I119M, P42) within the catalytic domain. Mutations in Wnt pathway ligands, RSPO4, WNT1, and WNT10A, were also identified in regions with potential functional interruption of ligands. PRICKLE1 was mutated in P3 in the PET domain (p.E92V), required for its protein–protein interactions.25 Finally, in P43, the g.T3543A mutation in the negative Wnt pathway regulator CHD8 is predicted to generate a truncated form lacking its repressive C-terminal helicase domain.26 Other pathway mutations (BCL9, BRD7, and CREBBP) occurred in regions of unknown gene function.
Significantly mutated Wnt pathway genes. Type (missense, splice site, nonsense) and localization of mutations in the 12 unique Wnt pathway genes identified in CLL cases (top) compared with previously reported mutations in the literature or within the COSMIC database (version 64) (bottom).
Significantly mutated Wnt pathway genes. Type (missense, splice site, nonsense) and localization of mutations in the 12 unique Wnt pathway genes identified in CLL cases (top) compared with previously reported mutations in the literature or within the COSMIC database (version 64) (bottom).
The Wnt pathway is transcriptionally and functionally hyperactivated in CLL
The Wnt pathway has been reported as dysregulated in CLL.6-9 We confirmed this finding through the comparison of a large gene expression dataset of CLL-B cells (n = 179), collected from predominantly chemotherapy-naïve patients and of normal CD19+ B cells (n = 24) (Figure 3A).27 This dataset included gene expression microarray data from 70 of the 91 MPS-characterized samples, of which 12 had Wnt pathway mutations. We examined the expression of an expanded set of Wnt pathway members and well-studied targets (supplemental Tables 4 and 5 for the gene list).20-22
Wnt pathway mutations do not contribute to the extent of dysregulated gene expression of Wnt pathway. (A) Expression profiles (from Affymetrix U133Plus2 arrays) of 60 Wnt pathway members that are significantly differentially expressed in 179 CLL-B cells compared with 24 normal CD19+ B cells (according to FDR corrected permutation test P values assessing significance of Student t test scores, FDR ≤0.05, with Student t test score ranging from −13.6-30.9). All tumor samples were comprised of >95% tumor purity. Genes were visualized in GENE-E. Wnt pathway genes downregulated in CLL are shown in the heatmap (top); Wnt pathway genes upregulated in CLL are shown (bottom). From these 60 genes, a “Wnt score” was calculated as a statistical measure of differential expression between the 37 known Wnt activators in the set (labeled in orange, in the column [right] of the heatmap) vs the 23 known repressors (labeled in black, separate column [right] of the heatmap). We generated a Student t test score for each CLL sample by comparing the differential expression of activators that are upregulated to repressors that are downregulated in each sample according to FDR-corrected permutation test P values assessing significance of Student t test scores, with FDR ≤0.05. Wnt scores ranged from −2.62 (blue) to 2.91 (red). Compared with normal B cells, overall, CLL cells demonstrate downregulation of Wnt pathway repressors and upregulation of Wnt pathway activators. (Bottom) CLL sample characteristics and whether samples also underwent whole-exome (WES) or whole-genome sequencing (WGS) (black-positive; white-negative). The rank of genes represented within the top 9% of genes differentially expressed array-wide is noted (in parentheses next to the gene names). Expression levels are log2 transformed and mean-centered for each gene for visualization. Induced levels are represented in red, repressed levels in blue, and no change is represented in white, in which levels are saturated at −0.5 and +0.5. (B) Unsupervised hierarchical clustering of Wnt pathway gene expression profiles from 12 mutated (with arrow) and 58 unmutated CLL-B cells (without arrow), performed using Pearson linear correlation with average linkage. (Right) Activators and repressors are shown (orange and black, respectively). (Upper panel) Wnt pathway genes and (lower panel) Wnt target genes, curated from the literature. A supervised analysis between the Wnt pathway mutated vs wild-type samples is presented in supplemental Figure 4. (C) The Wnt signaling pathway is chronically active in CLL-B cells. Normal and CLL-B cells were co-transfected with SuperTOPflash and pRL-TK constructs to measure endogenous TCF/LEF activity. CLL-B cells have fivefold greater normalized luciferase activity compared with normal B cells (n = 5; P < .01, Wilcoxon test, 2-tailed).
Wnt pathway mutations do not contribute to the extent of dysregulated gene expression of Wnt pathway. (A) Expression profiles (from Affymetrix U133Plus2 arrays) of 60 Wnt pathway members that are significantly differentially expressed in 179 CLL-B cells compared with 24 normal CD19+ B cells (according to FDR corrected permutation test P values assessing significance of Student t test scores, FDR ≤0.05, with Student t test score ranging from −13.6-30.9). All tumor samples were comprised of >95% tumor purity. Genes were visualized in GENE-E. Wnt pathway genes downregulated in CLL are shown in the heatmap (top); Wnt pathway genes upregulated in CLL are shown (bottom). From these 60 genes, a “Wnt score” was calculated as a statistical measure of differential expression between the 37 known Wnt activators in the set (labeled in orange, in the column [right] of the heatmap) vs the 23 known repressors (labeled in black, separate column [right] of the heatmap). We generated a Student t test score for each CLL sample by comparing the differential expression of activators that are upregulated to repressors that are downregulated in each sample according to FDR-corrected permutation test P values assessing significance of Student t test scores, with FDR ≤0.05. Wnt scores ranged from −2.62 (blue) to 2.91 (red). Compared with normal B cells, overall, CLL cells demonstrate downregulation of Wnt pathway repressors and upregulation of Wnt pathway activators. (Bottom) CLL sample characteristics and whether samples also underwent whole-exome (WES) or whole-genome sequencing (WGS) (black-positive; white-negative). The rank of genes represented within the top 9% of genes differentially expressed array-wide is noted (in parentheses next to the gene names). Expression levels are log2 transformed and mean-centered for each gene for visualization. Induced levels are represented in red, repressed levels in blue, and no change is represented in white, in which levels are saturated at −0.5 and +0.5. (B) Unsupervised hierarchical clustering of Wnt pathway gene expression profiles from 12 mutated (with arrow) and 58 unmutated CLL-B cells (without arrow), performed using Pearson linear correlation with average linkage. (Right) Activators and repressors are shown (orange and black, respectively). (Upper panel) Wnt pathway genes and (lower panel) Wnt target genes, curated from the literature. A supervised analysis between the Wnt pathway mutated vs wild-type samples is presented in supplemental Figure 4. (C) The Wnt signaling pathway is chronically active in CLL-B cells. Normal and CLL-B cells were co-transfected with SuperTOPflash and pRL-TK constructs to measure endogenous TCF/LEF activity. CLL-B cells have fivefold greater normalized luciferase activity compared with normal B cells (n = 5; P < .01, Wilcoxon test, 2-tailed).
Consistent with previous reports,6,8,9 we found LEF1, a canonical target of the Wnt pathway, to be the most significantly differentially overexpressed messenger RNA in CLL compared with normal B cells (ranked first of 20 765 features, BH-FDR ≤0.05; supplemental Table 4). We confirmed LEF1 overexpression in CLL cells at the protein level (supplemental Figure 3). In addition, other Wnt pathway members ROR1, TCF4, WNT3, FZD3, WNT10A, SMAD2, WNT5B, CALCOCO1, and HBP1 ranked within the top 9% of most differentially expressed messenger RNAs (ranked from 11th to 1681st feature). In total, 60 of 132 Wnt pathway components were expressed at altered levels in CLL compared with normal CD19+ B cells (Benjamini-Hochberg false discovery rate [BH-FDR] ≤0.05, supplemental Table 4).
To assess the aggregate change in the levels of Wnt pathway transcripts per sample, we devised a “Wnt score” as a measure of differential expression between 37 known Wnt activators and 23 known repressors (false discovery rate [FDR] ≤0.05; see supplemental Methods). We applied this test for those genes with significant change in expression in CLL-B cells vs normal B cells (Figure 3A). Most CLL samples scored positive, reflecting higher expression of activators vs repressors. Notably, activators were upregulated (eg, WNT3, TCF4, ROR1, LEF1) and repressors were downregulated (eg, SOX4, HBP1, NKD1) in CLL-B compared with normal B cells (P = .05; hypergeometric test).
To confirm dysregulated Wnt pathway activity in CLL-B cells, we examined endogenous Wnt pathway activation. We used the well-characterized TCF/LEF-dependent reporter to detect endogenous Wnt pathway activity in CLL-B cells, in which we used DNA nucleofection to deliver reporter constructs. As with other reports, using this methodology in CLL,8,9 we detected constitutive activation of the Wnt pathway in 5 of 5 CLL-B cells (Figure 3C) compared with CD19+ B cells from 4 healthy volunteers (P < .01; Wilcoxon rank-sum test).
As we previously reported, no association between known CLL prognostic factors and LEF1 expression was found.16 Likewise, we did not discover somatic mutations to contribute to Wnt pathway dysregulation through gene expression (ie, the Wnt score). Using unsupervised hierarchical clustering to compare the gene expression pattern of Wnt pathway members in the 70 CLL cases, we detected several distinct subclusters, reflecting the heterogeneity of the disease (Figure 3B). The 12 samples with Wnt mutations were spread evenly across the clusters, suggesting that the Wnt pathway gene expression dysregulation was independent of the mutation of Wnt pathway genes. In further support that these are highly heterogeneous samples, supervised hierarchical clustering of the samples with mutated vs wild-type Wnt pathway members did not reveal significant expression differences (supplemental Figure 4). We speculate that mutations may lead directly to pathway dysregulation without requiring coherent changes in expression of pathway components.
Silencing with vertical silicon NWsto efficiently examine the impact of Wnt pathway members on CLL-B cell survival
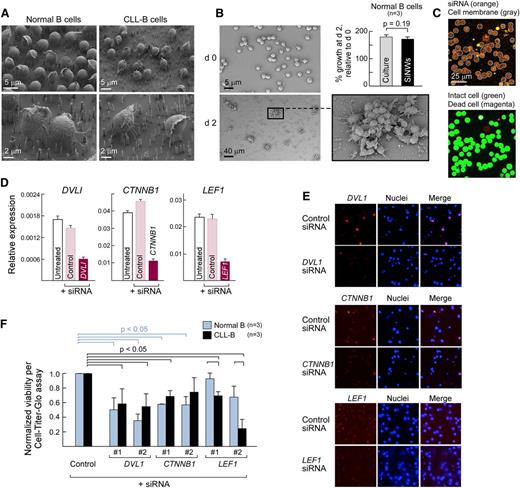
We sought to more closely examine the contribution of specific Wnt pathway components to CLL-B cell survival by manipulating the Wnt pathway of primary B cells and CLL-B cells via transfection. However, we commonly detected poor and variable viability of primary CLL-B cells within 24 hours after nucleofection (median viability, 5%; range, 1% to 10%).
As an alternative approach, NWs are capable of delivering surface-coated biomolecules directly into the cell cytosol through penetration of the cellular membrane, and they have been successfully piloted in difficult-to-manipulate primary human cells, including normal and CLL-B cells (Figure 4A).15,16 We observed equivalent growth of normal CD19+ B cells after stimulation with IL4 and CD40L,28 whether in vitro or atop NWs (Student t test; P = .19), and hence NW exposure does not apparently interfere with normal cellular functions (Figure 4B). Importantly, this approach could efficiently deliver fluorescently labeled siRNAs into human normal and malignant B cells (>90% delivery, Figure 4C, top panel) without compromising cellular viability (>95% survival, lower panel).
The expression of core Wnt pathway components is required for CLL survival. (A) Scanning electron micrographs (SEMs) of normal CD19+ B (left panels) and CLL-B cells (right panels) atop NWs taken 24 hours after plating. (B) Normal CD19+ B cells can grow and divide on NWs. Normal CD19+ B cells isolated from peripheral blood of healthy adult volunteers were stimulated with IL4 (2 ng/mL) and CD40L (0.1 mg/mLl)28 either in vitro (“culture”) or atop NWs (“SiNWs”) for 48 hours. Cell proliferation was measured using an adenosine triphosphate (ATP)-dependent Cell-Titer Glo assay. Cell growth rate was calculated based on the measurement of ATP amount after 48 hours of stimulation, normalized relative to the day 0 value. (Inset) Scanning electron microscope image showing that proliferating B cells on NWs grow in clusters. (C) Confocal scanning images of Alexa Fluor 546-labeled human anti-vimentin siRNA delivered into CLL-B cells. (Upper panel) siRNA delivery is calculated by manually counting the number of cells that have higher levels of fluorescent siRNAs compared with untransfected controls (not shown here). Alexa Fluor 546-labeled siRNA is shown (orange), whereas cell membranes are shown outlined (gray). (Lower panel) Viability was calculated as a percentage of the number of live cells in the total cells using a live–dead cell staining method. Intact cells (stained with Calcein-AM) are shown (green), whereas the nuclei of dead cells are shown in magenta (stained with EthD-1). (D) Core Wnt pathway components can be silenced in HEK293T using siRNA delivery. Gene expression of Wnt pathway members DVL1, CTNNB1, and LEF1 (relative to glyceraldehyde-3-phosphate dehydrogenase expression) were analyzed by quantitative Taqman RT-PCR using complementary DNA derived from HEK293T cells that were either untransfected (“untreated,” white bars) or transfected for 48 hours with control nontargeting siRNA (“control,” pink bars) or siRNA specific for DVL1, CTNNB1, or LEF1 (dark red bars). (E) Efficient knockdown of protein expression of Wnt pathway members in normal CD19+ and CLL-B cells via NW-mediated siRNA delivery. Representative images of target protein expression, detected by immunofluorescence 48 hours after siRNA delivery using gene-specific antibodies against the target proteins are shown. (F) Median decrease in cell survival (measured by CellTiter Glo) 48 hours after NW-mediated delivery of siRNAs against LEF1, DVL, and CTNNB1 in normal CD19+ (n = 3) and CLL-B (n = 3) compared with silencing using nontargeting siRNA controls (2 different siRNAs per target gene). Percentage of cell survival was normalized to ATP amount at day 0.
The expression of core Wnt pathway components is required for CLL survival. (A) Scanning electron micrographs (SEMs) of normal CD19+ B (left panels) and CLL-B cells (right panels) atop NWs taken 24 hours after plating. (B) Normal CD19+ B cells can grow and divide on NWs. Normal CD19+ B cells isolated from peripheral blood of healthy adult volunteers were stimulated with IL4 (2 ng/mL) and CD40L (0.1 mg/mLl)28 either in vitro (“culture”) or atop NWs (“SiNWs”) for 48 hours. Cell proliferation was measured using an adenosine triphosphate (ATP)-dependent Cell-Titer Glo assay. Cell growth rate was calculated based on the measurement of ATP amount after 48 hours of stimulation, normalized relative to the day 0 value. (Inset) Scanning electron microscope image showing that proliferating B cells on NWs grow in clusters. (C) Confocal scanning images of Alexa Fluor 546-labeled human anti-vimentin siRNA delivered into CLL-B cells. (Upper panel) siRNA delivery is calculated by manually counting the number of cells that have higher levels of fluorescent siRNAs compared with untransfected controls (not shown here). Alexa Fluor 546-labeled siRNA is shown (orange), whereas cell membranes are shown outlined (gray). (Lower panel) Viability was calculated as a percentage of the number of live cells in the total cells using a live–dead cell staining method. Intact cells (stained with Calcein-AM) are shown (green), whereas the nuclei of dead cells are shown in magenta (stained with EthD-1). (D) Core Wnt pathway components can be silenced in HEK293T using siRNA delivery. Gene expression of Wnt pathway members DVL1, CTNNB1, and LEF1 (relative to glyceraldehyde-3-phosphate dehydrogenase expression) were analyzed by quantitative Taqman RT-PCR using complementary DNA derived from HEK293T cells that were either untransfected (“untreated,” white bars) or transfected for 48 hours with control nontargeting siRNA (“control,” pink bars) or siRNA specific for DVL1, CTNNB1, or LEF1 (dark red bars). (E) Efficient knockdown of protein expression of Wnt pathway members in normal CD19+ and CLL-B cells via NW-mediated siRNA delivery. Representative images of target protein expression, detected by immunofluorescence 48 hours after siRNA delivery using gene-specific antibodies against the target proteins are shown. (F) Median decrease in cell survival (measured by CellTiter Glo) 48 hours after NW-mediated delivery of siRNAs against LEF1, DVL, and CTNNB1 in normal CD19+ (n = 3) and CLL-B (n = 3) compared with silencing using nontargeting siRNA controls (2 different siRNAs per target gene). Percentage of cell survival was normalized to ATP amount at day 0.
Therefore, we used the NW delivery platform to silence the core Wnt pathway members DVL1, CTNNB1, and LEF1 in normal and CLL-B cells. All of these genes are highly expressed in CLL-B cells, with DVL1 and LEF1 both significantly, differentially expressed compared with normal B cells (supplemental Table 4). Gene-specific silencing after NW-mediated siRNA delivery was confirmed at the transcript level in HEK293T cells (Figure 4D), and at the protein level via immunofluorescence staining of target genes in CLL-B cells (Figure 4E). Consistent with the key role of the Wnt pathway in both normal and malignant B cells, we observed reduced cell survival of normal CD19+ and CLL-B cells after silencing of DVL1 and CTNNB1 compared with the nontargeting control (P < .05). We also silenced LEF1, the terminal transcriptional activator of β−catenin/Wnt signaling and the most differentially expressed gene between normal and CLL-B cells (Figure 3A).8,9 In agreement with prior reports, LEF1 silencing resulted in reduced cell survival of CLL-B cells, but not normal B cells (P = .01). Altogether, perturbation of key nodes at different levels of the Wnt pathway can impact CLL-B cell survival.
Patterns of Wnt signaling affected by Wnt pathway mutation in HEK293T cells
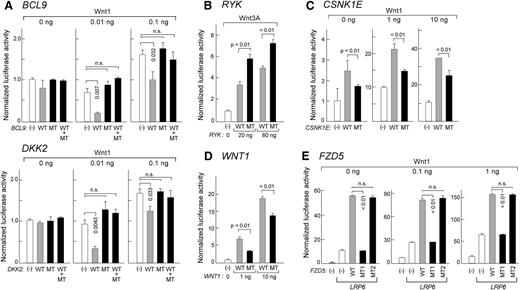
Before testing the role of Wnt pathway mutations in the survival of primary CLL-B cells, we first tested the effects of overexpressing mutated and wild-type alleles on Wnt pathway activation in easily transfectable HEK293T cells using a TCF/LEF-dependent luciferase reporter and through the examination of Wnt pathway target gene expression. We selected 8 gene mutations for 7 Wnt pathway members with well-defined roles in this signaling pathway, whereas some of the other mutations were in genes involved in additional biologic processes or pathways (supplemental Table 6), for which we generated paired wild-type and mutant expression constructs (WNT1, FZD5, BCL9, RYK, CSNK1E, DKK2, and WNT10A) (supplemental Figures 5 and 6).
We observed several distinct patterns of response after gene overexpression. First, 2 of 8 mutations (BCL9 and DKK2) demonstrated marked loss of repression of Wnt pathway signaling compared with wild-type alleles (Figure 5A). Loss of repression by mutated DKK2 was observed across a 500-fold range of gene dosages (supplemental Figure 5). In an analogous fashion, mutant DKK2 protein abolished the repressive effects of wild-type DKK2 on normal B cells after incubation of these cells with wild-type or mutant DKK2 protein (supplemental Figure 7B). For both BCL9 and DKK2, expression of a mixture of wild-type and mutated alleles in HEK293T cells eliminated the repressive effects of wild-type protein, suggesting a dominant effect of the mutation (P < .01).
Heterozygous mutations alter Wnt pathway activities in HEK293T cells. (A,C,E) HEK293T cells were cotransfected with Wnt1 expression plasmid (amounts indicated), wild-type (WT), or mutant (MT), or equal amounts of WT and MT plasmids, along with the reporter plasmids SuperTOPflash and pRL-TK. Forty-eight hours after transfection, luciferase activity was measured from 3 independent experiments. All WT, MT, or WT/MT plasmids were introduced at 0.1 ng, 0.1 ng, 50 ng, and 5 ng for BCL9, DKK2, CSNK1E, and FZD5, respectively. LRP6 plasmid (10 ng) was also included in the FZD5 mutation characterization. MT1: Y46*; MT2: V290I. Downstream Wnt pathway targets were also assessed for mutated DKK2 by gene expression (see supplemental Figure 7 and supplemental Methods). (B) HEK293T cells were cotransfected with 20 or 80 ng of WT or MT RYK along with reporter plasmids. At 24 hours after the transfection, recombinant Wnt3a was added (25 ng/mL final concentration) and incubated for 24 hours before luciferase activity was measured. Detection of phosphorylation of downstream target DVL2 was assessed by western blot analysis (see supplemental Figure 7 and supplemental Methods). (D) HEK293T cells were cotransfected with either WT or MT WNT1 along with the reporter plasmids. Luciferase activity was measured from three independent experiments 48 hours after transfection. For more details on the conditions of Wnt activation, please see supplemental Methods.
Heterozygous mutations alter Wnt pathway activities in HEK293T cells. (A,C,E) HEK293T cells were cotransfected with Wnt1 expression plasmid (amounts indicated), wild-type (WT), or mutant (MT), or equal amounts of WT and MT plasmids, along with the reporter plasmids SuperTOPflash and pRL-TK. Forty-eight hours after transfection, luciferase activity was measured from 3 independent experiments. All WT, MT, or WT/MT plasmids were introduced at 0.1 ng, 0.1 ng, 50 ng, and 5 ng for BCL9, DKK2, CSNK1E, and FZD5, respectively. LRP6 plasmid (10 ng) was also included in the FZD5 mutation characterization. MT1: Y46*; MT2: V290I. Downstream Wnt pathway targets were also assessed for mutated DKK2 by gene expression (see supplemental Figure 7 and supplemental Methods). (B) HEK293T cells were cotransfected with 20 or 80 ng of WT or MT RYK along with reporter plasmids. At 24 hours after the transfection, recombinant Wnt3a was added (25 ng/mL final concentration) and incubated for 24 hours before luciferase activity was measured. Detection of phosphorylation of downstream target DVL2 was assessed by western blot analysis (see supplemental Figure 7 and supplemental Methods). (D) HEK293T cells were cotransfected with either WT or MT WNT1 along with the reporter plasmids. Luciferase activity was measured from three independent experiments 48 hours after transfection. For more details on the conditions of Wnt activation, please see supplemental Methods.
Second, 1 mutated pathway member demonstrated augmented activating function (RYK, p.A488T) (P < .01). Consistent with this result, a downstream target of RYK, DVL2 protein and its active phosphorylated form were expressed at higher levels in the mutant RYK-expressing HEK293T cells (supplemental Figure 7C).
Third, we observed 3 mutations in known Wnt pathway activators (CSNK1E, WNT1, FZD5) that led to loss of functional pathway activation. Compared with wild-type alleles of CSNK1E and WNT1, expression of the mutated alleles in HEK293T cells resulted in dampened Wnt signaling (Figure 5C-D). This pattern was also observed in 1 of 2 FZD5 mutations (p.Y46*) that resulted in premature truncation of the protein (“MT1”) (Figure 5E). Because the Wnt receptor is composed of frizzled and LRP5/6 protein, we cotransfected LRP6 plasmid to synergize pathway activation with FZD5. As predicted, expression of truncated FZD5 (MT1) resulted in complete loss of Wnt-activating function (Figure 5E).
In contrast to MT1, a second missense mutation in FZD5 (p.V290I, “MT2”) did not change Wnt pathway activity compared with its wild-type counterpart (Figure 5E). Likewise, no significant difference in Wnt pathway activity was observed between expression of WNT10A wild-type and mutant constructs (supplemental Figure 6D).
Expression of mutated Wnt pathway alleles may be required for survival of CLL cells harboring mutations in these genes
To confirm a functional role for Wnt pathway mutations in CLL samples, we directly examined whether their expression contributed to cell survival in primary samples harboring any of the 8 mutated alleles of these genes. Wherever possible, the ability to efficiently silence these genes by NW-mediated siRNA delivery was confirmed at the protein level by immunofluorescence staining (50% to 80% reduction) (Figure 6A,C-D; supplemental Figure 7A). NWs were used to deliver gene-specific siRNAs into: (1) normal CD19+ B cells (n = 2-4); (2) CLL samples without Wnt pathway mutations (determined through MPS [n = 5-7]); and (3) CLL samples harboring Wnt pathway mutations. For these experiments, measurement of cell viability was normalized to the nontargeting control siRNA.
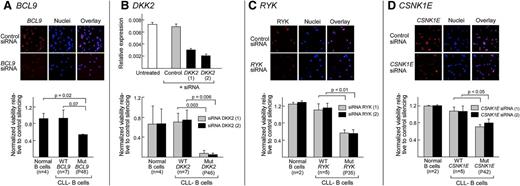
Increased dependence on Wnt signaling by CLL samples harboring Wnt pathway mutations. Silencing of BCL9, RYK, and CSNK1E protein in normal CD19+ B cells 48 hours after NW-mediated delivery of gene-specific siRNAs compared with nontargeting control siRNAs (“control”) was confirmed by gene-specific immunofluorescence staining (red) and visualized by confocal microscopy (A,C,D) (top panels). Nuclei were probed with DAPI (blue). Protein level silencing efficiency was estimated using Image J software. Per gene, 2 different targeting siRNAs were tested and the representative results are shown. (B) (Top) Silencing of DKK2 in HEK293T cells using gene-specific siRNAs detected by quantitative Taqman RT-PCR of complementary DNA derived from HEK293T cells that were either untreated (white bar) or treated with control nontargeting siRNA (“control”, gray bars) or with siRNA specific for DKK2 (black bars). (A-D) (Lower panels) Cell survival rate was normalized to nontargeting control in normal B cells (n = 4 for BCL9 and DKK2; n = 2 for RYK and CSNK1E), the CLL B cells with either mutated BCL9 (P48), DKK2 (P46), RYK (P35), or CSNK1E (P42) (all n = 1) or CLL-B samples without Wnt pathway mutations (n = 7 for BCL9 and DKK2; n = 5 for RYK and CSNK1E), using the Cell-Titer Glo assay 48 hours after NW-mediated siRNA delivery. Three replicates per independent sample were performed.
Increased dependence on Wnt signaling by CLL samples harboring Wnt pathway mutations. Silencing of BCL9, RYK, and CSNK1E protein in normal CD19+ B cells 48 hours after NW-mediated delivery of gene-specific siRNAs compared with nontargeting control siRNAs (“control”) was confirmed by gene-specific immunofluorescence staining (red) and visualized by confocal microscopy (A,C,D) (top panels). Nuclei were probed with DAPI (blue). Protein level silencing efficiency was estimated using Image J software. Per gene, 2 different targeting siRNAs were tested and the representative results are shown. (B) (Top) Silencing of DKK2 in HEK293T cells using gene-specific siRNAs detected by quantitative Taqman RT-PCR of complementary DNA derived from HEK293T cells that were either untreated (white bar) or treated with control nontargeting siRNA (“control”, gray bars) or with siRNA specific for DKK2 (black bars). (A-D) (Lower panels) Cell survival rate was normalized to nontargeting control in normal B cells (n = 4 for BCL9 and DKK2; n = 2 for RYK and CSNK1E), the CLL B cells with either mutated BCL9 (P48), DKK2 (P46), RYK (P35), or CSNK1E (P42) (all n = 1) or CLL-B samples without Wnt pathway mutations (n = 7 for BCL9 and DKK2; n = 5 for RYK and CSNK1E), using the Cell-Titer Glo assay 48 hours after NW-mediated siRNA delivery. Three replicates per independent sample were performed.
As expected from neutral or loss of function variants, silencing of mutated WNT1, WNT10A, or FZD5 (MT1, MT2) did not generate significant changes in cell viability compared with wild-type CLL samples (supplemental Figure 7A-B).
On the other hand, silencing of pathway activating mutations led to reduced cell viability (Figure 6). For example, the BCL9-mutated (P48) CLL sample was more dependent on BCL9 expression for survival than normal B cells (P = .02) or CLL samples without Wnt pathway mutations (P = .07). Likewise, silencing of DKK2 in DKK2-mutated (P46) CLL-B cells, and RYK in RYK-mutated (P < .05) CLL-B cells led to greater cell death than gene silencing in normal B cells, or CLL samples without Wnt pathway mutations (P = .003 and P < .05, respectively). Surprisingly, decreased CLL cell viability was also observed with silencing of mutated CSNK1E, which demonstrated loss-of-function in HEK293T cells (P < .05; Figure 6D).
Discussion
A central goal of cancer genome sequencing is to uncover the genes and pathways responsible for tumor initiation and progression. Although the statistically significant recurrence of mutations provides a straightforward criterion to reveal candidate driver mutations, this strategy fails to reveal the presence of “driver pathways,” wherein the pathway as a whole is recurrently mutated, but lacking in specific, recurrent mutations. To further understand the genetic alterations that generate and sustain tumors, nonrecurrent mutations that contribute to oncogenesis via these driver pathways must also be identified and investigated.
Our study directly addresses this issue by integrating sequencing and functional studies to implicate previously uncharacterized Wnt pathway members in the pathogenesis of CLL. Based on DNA sequencing of a series of CLL samples, we identified 15 novel mutations in 13 distinct genes of the Wnt pathway, representing 14% of CLL patients; and selected 8 for functional characterization, focusing on those with well-defined central roles in this signaling pathway (see supplemental Table 6). Among these, we observed several distinct patterns from HEK293T cells into which the wild-type and/or mutant alleles were introduced. First, 3 mutations (in DKK2, BCL9, RYK), indeed, led to Wnt pathway activation. These mutations either led to loss of repression or augmented activation, suggesting a gain-of-function of these genes. Second, mutations in 3 Wnt pathway activators (CSNK1E, WNT1, FZD5-MT1) resulted in reduced or loss of pathway activation. Finally, 2 mutations (in WNT10A, FZD5 MT2) lacked any functional effects on the pathway.
Further functional evaluation in which we directly silenced the expression of mutated genes in primary CLL cells with or without these mutations revealed 2 key findings. First, we observed that CLL samples harboring putative gain-of-function mutations (established in the HEK293T system, DKK2, BCL9, RYK) exhibited greater dependency on Wnt pathway signaling (“addiction”). These results support a driving role of these mutations in CLL samples harboring this class of genetic alteration. Second, cell-lineage context likely plays a role in the directionality of functional effects of the mutations and underscores the complexity of Wnt signaling circuits that likely impact CLL cell survival. We observed that silencing of mutated CSNK1E, 1 of the 5 non–gain-of-function mutations (again, established in the HEK293T system) led to decreased CLL viability, whereas the other 4 had no impact on CLL survival, as predicted. CSNK1E is a member of the noncanonical Wnt pathway, which was recently characterized to play a significant role in B-lymphocyte migration in CLL.29 Hence, mutation in this gene may have only observable effects within a B-cell context. As for the other 4 Wnt pathway mutations with no effects on CLL survival after knockdown, the absence of the signal could be attributed to the idea that these are bona fide “passenger mutations,” or that they have effects on other pathways that affect CLL, but were not measureable solely based on the cell viability readout on which we focused. Alternatively, the CLL cells were tested ex vivo and may not have been in an environment that was permissive to responding to gene silencing.
Despite these caveats, our studies suggest a rational approach for functional evaluation of gene pathway mutations. Extensive Wnt pathway dysregulation has been previously noted in CLL, and is linked in part to changes in DNA methylation.11-13,30 LEF1 overexpression has been reported in the premalignant form of CLL, monoclonal B-cell lymphocytosis,8 and is thought to drive CLL. Because expression of Wnt pathway members appears to be crucial for CLL survival and our data demonstrate that altered gene expression is indistinguishable between samples with and without mutations, we reason that somatic mutation is another layer of regulation affecting CLL function. The higher dependency of CLL cells on the expression of a mutated gene highlights somatic mutation as a mechanism for control of this critical signaling pathway. Of note, frequent mutations in Wnt pathway members have not been reported for myeloma31 or for diffuse large B-cell lymphoma.32 Hence, somatic mutation in the Wnt pathway, affecting different nodes along this pathway to promote survival of CLL cells, is a potentially distinguishing feature of CLL compared with other B-cell malignancies.
It is also likely that Wnt pathway mutations synergize with other pathways. Kaucká et al33 recently reported that the planar cell polarity pathway, a noncanonical Wnt pathway, drives CLL pathogenesis by regulating B-lymphocyte migration.29 Other investigators have shown LEF1 to inhibit CYLD, which leads to dysregulation of tumor necrosis factor-induced necroptotic signaling, thereby providing a link between survival of CLL cells and active Wnt signaling. At the same time, a large-scale genome-wide association study identified multiple risk loci for CLL that included BCL2, LEF1, and other genes,1 again highlighting the role of Wnt pathway dysregulation as a genetic factor associated with susceptibility for CLL. Altogether, these recent studies lend support to the idea that Wnt pathway mutations could potentially synergize with other pathways to modulate CLL cell survival.
From a more pragmatic perspective, patients with greater sensitivity to Wnt pathway inhibition may be potentially identifiable through relatively straightforward genetic characterization of Wnt pathway mutations. This concept is attractive because increasing numbers of inhibitors that target different components of the Wnt pathway are being developed.9,34-36 A recent study used cancer cell line profiling to identify small-molecule sensitivities and uncovered that activating mutations in CTNNB1 or mutations in members of its destruction complex (AXIN1, CSNK1A1) correlated well with sensitivity to the small molecule compound navitoclax. These results demonstrate the feasibility of developing novel drugs matched to patients by their cancer genotype and lineage.37 In general, heterogeneous clinical response to conventional chemotherapy for CLL has been the rule, with associated toxicities and lack of consistent responses. Hence, identification of subgroups of patients based on their molecular characteristics may reveal genetic dependencies, which have the potential to provide more fruitful and less toxic approaches for effective therapeutic targeting when coupled with small molecule screening.
There is an Inside Blood Commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We acknowledge all members of the Broad Institute’s Biological Samples Platform, Genetic Analysis Platform, and Genome Sequencing Platform, without whom this work would not have been possible (National Human Genome Research Institute, U54HG003067). The authors thank Wolfram Goessling and Ruben Carrasco for insightful discussions, Bethany Tesar, Stacey Fernandes, and Rutendo Gambe for their excellent technical assistance.
This work was supported by a National Institutes of Health Director's Pioneer Award (5DP1OD003893-03) and National Institutes of Health Centers of Excellence in Genomic Science Award (1P50HG006193-01) (H.P.); a National Institutes of Health Director's Pioneer Award, the Howard Hughes Medical Institute, and a fellowship with the Merkin Foundation for Stem Cell Research at the Broad Institute (A.R.); the Fund for Scientific Research – Flanders (FWO Vlaanderen), Belgium (N.P.); the National Institutes of Health, National Cancer Institute (K23 CA115682), the Melton and Rosenbach Funds, a Scholar of the American Society of Hematology, and a Scholar in Clinical Research of the Leukemia and Lymphoma Society (J.R.B.); and by the Blavatnik Family Foundation, an Innovative Research grant (SU2C/AACR) and the Damon-Runyon Cancer Research Foundation (CI-38-07) (C.J.W.).
Authorship
Contribution: L.W. designed and performed research, collected data, and wrote the initial draft of the manuscript; L.W. and C.J.W. analyzed and interpreted data; A.K.S., R.D., J.T.G., W.Z., J.W., and Q.L.S. performed experiments; M.L., P.S., C.S., and S.A.S. collected mutation data; N.P. performed gene expression analysis; N.R.G. and A.R.V. collected clinical data; K.E.S. and D.N. performed statistical analysis; B.T.M. and X.H. provided reagents; X.H., E.L., N.H., A.R., G.G., J.R.B., and H.P. contributed to the interpretation of results; C.J.W. planned the study, organized the research, and wrote the manuscript; and all of the authors edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Catherine J. Wu, Dana-Farber Cancer Institute, 450 Brookline Ave, Dana 540B, Boston, MA 02215; e-mail: cwu@partners.org.

![Figure 3. Wnt pathway mutations do not contribute to the extent of dysregulated gene expression of Wnt pathway. (A) Expression profiles (from Affymetrix U133Plus2 arrays) of 60 Wnt pathway members that are significantly differentially expressed in 179 CLL-B cells compared with 24 normal CD19+ B cells (according to FDR corrected permutation test P values assessing significance of Student t test scores, FDR ≤0.05, with Student t test score ranging from −13.6-30.9). All tumor samples were comprised of >95% tumor purity. Genes were visualized in GENE-E. Wnt pathway genes downregulated in CLL are shown in the heatmap (top); Wnt pathway genes upregulated in CLL are shown (bottom). From these 60 genes, a “Wnt score” was calculated as a statistical measure of differential expression between the 37 known Wnt activators in the set (labeled in orange, in the column [right] of the heatmap) vs the 23 known repressors (labeled in black, separate column [right] of the heatmap). We generated a Student t test score for each CLL sample by comparing the differential expression of activators that are upregulated to repressors that are downregulated in each sample according to FDR-corrected permutation test P values assessing significance of Student t test scores, with FDR ≤0.05. Wnt scores ranged from −2.62 (blue) to 2.91 (red). Compared with normal B cells, overall, CLL cells demonstrate downregulation of Wnt pathway repressors and upregulation of Wnt pathway activators. (Bottom) CLL sample characteristics and whether samples also underwent whole-exome (WES) or whole-genome sequencing (WGS) (black-positive; white-negative). The rank of genes represented within the top 9% of genes differentially expressed array-wide is noted (in parentheses next to the gene names). Expression levels are log2 transformed and mean-centered for each gene for visualization. Induced levels are represented in red, repressed levels in blue, and no change is represented in white, in which levels are saturated at −0.5 and +0.5. (B) Unsupervised hierarchical clustering of Wnt pathway gene expression profiles from 12 mutated (with arrow) and 58 unmutated CLL-B cells (without arrow), performed using Pearson linear correlation with average linkage. (Right) Activators and repressors are shown (orange and black, respectively). (Upper panel) Wnt pathway genes and (lower panel) Wnt target genes, curated from the literature. A supervised analysis between the Wnt pathway mutated vs wild-type samples is presented in supplemental Figure 4. (C) The Wnt signaling pathway is chronically active in CLL-B cells. Normal and CLL-B cells were co-transfected with SuperTOPflash and pRL-TK constructs to measure endogenous TCF/LEF activity. CLL-B cells have fivefold greater normalized luciferase activity compared with normal B cells (n = 5; P < .01, Wilcoxon test, 2-tailed).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/7/10.1182_blood-2014-01-552067/5/m_1089f3.jpeg?Expires=1769837380&Signature=5OzkGraaglw8tpnLtaNP88oVinfLHqiqk9hsldGERNWuHlDaWvsB3LRBqOCdPojK0t3bTtL8aOH0fVeIKQUcz5Tmtq1MPLWKRCkkp5ag~AiC3mj4CFx-sqngbxElzWCBET0DUI8olgbGcpeHloaAjyuDkAWjM5UBAq1XM6wFkEbF4prW1F43BNaAHibPikPgDwFMvKr46C3hb7RKThvmH9Bfkl8F8kRQXlE4wJfBiIacuS9CRs1ZN-9~6mZcz8a8ZIjvk62Jt33gk7J46Yn6E1RJpX5DKecitsF7q32z19lxNASu2LQAKhVGqpc1xrYxrpoH1Ir2K2e4fkWz1Ei1hQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal