Key Points
RAS pathway mutations are prevalent in relapsed childhood ALL, and KRAS mutations are associated with a poorer overall survival.
RAS pathway mutations confer sensitivity to mitogen-activated protein kinase kinase inhibitors.
Abstract
For most children who relapse with acute lymphoblastic leukemia (ALL), the prognosis is poor, and there is a need for novel therapies to improve outcome. We screened samples from children with B-lineage ALL entered into the ALL-REZ BFM 2002 clinical trial (www.clinicaltrials.gov, #NCT00114348) for somatic mutations activating the Ras pathway (KRAS, NRAS, FLT3, and PTPN11) and showed mutation to be highly prevalent (76 from 206). Clinically, they were associated with high-risk features including early relapse, central nervous system (CNS) involvement, and specifically for NRAS/KRAS mutations, chemoresistance. KRAS mutations were associated with a reduced overall survival. Mutation screening of the matched diagnostic samples found many to be wild type (WT); however, by using more sensitive allelic-specific assays, low-level mutated subpopulations were found in many cases, suggesting that they survived up-front therapy and subsequently emerged at relapse. Preclinical evaluation of the mitogen-activated protein kinase kinase 1/2 inhibitor selumetinib (AZD6244, ARRY-142886) showed significant differential sensitivity in Ras pathway–mutated ALL compared with WT cells both in vitro and in an orthotopic xenograft model engrafted with primary ALL; in the latter, reduced RAS-mutated CNS leukemia. Given these data, clinical evaluation of selumetinib may be warranted for Ras pathway–mutated relapsed ALL.
Introduction
Acute lymphoblastic leukemia (ALL) is the most common childhood cancer, and over the past 50 years, cure rates have consistently increased and are now approaching 90%.1 This success is attributable to the introduction and gradual intensification of combination chemotherapy, along with improvements in treatment stratification and supportive clinical care. However, for most children who relapse, the outlook is poor, and relapsed ALL is still a major cause of death in children with malignancy.2-4 Therefore, novel therapies are urgently needed.
ALL is characterized by a number of recurrent genetic abnormalities: most commonly numerical and structural chromosomal changes such as high hyperdiploidy and the ETV6-RUNX1 fusion,5 inactivation of transcription factors critical in lymphocyte development such as PAX5,6 and as we have previously shown, somatic mutation of genes that impact on the Ras/Raf/mitogen-activated protein kinase kinase (MEK)/extracellular signal-regulated kinase (ERK) signaling cascade.7 The cascade is activated in response to a variety of extracellular stimuli and transduces signals from the cell surface to nuclear and cytosolic targets and as such plays a pivotal role in a variety of cellular processes including proliferation, apoptosis, and differentiation.8 It is dysregulated in ALL by genetic alteration of an array of genes including upstream signaling molecules, such as the receptor tyrosine kinase FLT3, integral components of the pathway such as NRAS and KRAS, as well as regulators such as the phosphatase PTPN11 and the more recently described ubiquitin ligase c-CBL.9 In our cohort of children at diagnosis of ALL, mutations in these genes were found at a frequency of 35%, and the significance of pathway dysregulation in leukemogenesis was emphasized by the observation that mutations were invariably found in a mutually exclusive manner.7
Ras pathway mutations are prevalent at diagnosis of ALL and are found at high incidence in high hyperdiploidy, a group characterized by an excellent prognosis, but a similar incidence has also been reported in a “high-risk” ALL cohort and, specifically, hypodiploid ALL.7,10-15 In addition, we and others have anecdotal reports of gain of Ras pathway mutations at relapse; thus, the incidence of Ras pathway mutations in relapsed ALL may be significant.7,16 Importantly, the RAS effector RAF/MEK/ERK pathway is therapeutically tractable and may provide new, targeted therapies needed for relapsed ALL. Therefore, in this study, we have investigated Ras pathway mutations in a large cohort of relapsed ALL treated on the ALL-REZ BFM 2002 protocol and evaluated the MEK1/2 inhibitor selumetinib (AZD6244, ARRY-142886) in preclinical models of Ras pathway–positive ALL.
Materials and methods
Patients
Children included in the study were all B-cell precursor ALL at first relapse with microscopic bone marrow involvement at relapse diagnosis and treated according to the protocol ALL-REZ BFM 2002 trial (www.clinicaltrials.gov, #NCT00114348) (n = 206). Cytogenetic analysis was carried out on newly diagnosed bone marrow samples using standard G banding with specific chromosomal abnormalities, including ETV6-RUNX1, BCR-ABL1, and MLL rearrangements, characterized by standard polymerase chain reaction (PCR) and fluorescence in situ hybridization (FISH) methodologies.17 High hyperdiploidy was identified by cytogenetic analyses, FISH, or Multiplex Ligation-dependent Probe Amplification kits P007, P095 (MRC Holland, Amsterdam, Netherlands) along with the flow cytometric DNA index. Patients with intrachromosomal amplification of chromosome 21 were identified either by FISH or Multiplex Ligation-dependent Probe Amplification–kit P327 (MRC Holland) and FISH.17 Minimal residual disease analyses were performed using the European standardized PCR method for clonal antigen receptor rearrangements.18 In terms of clinical characteristics, the final study cohort presented here (n = 206) was representative of the total cohort of patients entered into the trial (n = 329) and met the same inclusion criteria (supplemental Table 1, available on the Blood Web site).The study was approved by the local ethics committee. Written informed consent was obtained from patients or guardians in accordance with the Declaration of Helsinki.
Mutation, Ras pathway activation, and preclinical evaluation of selumetinib were performed on bone marrow samples from children presenting with ALL in the northern region of England between March 2001 and September 2012 with most entered into UKALL2003 clinical trial (www.clinicaltrials.gov, #NCT00222612; see supplemental Table 2 for details). Samples were enriched for mononuclear cells using Ficoll density gradient centrifugation and washed in phosphate-buffered saline. The resulting cells were either stored as pellets at −80°C for future protein and DNA analysis or cryopreserved in 10% dimethylsulfoxide (DMSO)/fetal bovine serum (FBS) for in vivo studies.
Mutational screening
Allele-specific PCR
Taq mismatch amplification assay (TaqMAMA) assays for common NRAS and KRAS mutations were established following the method of Archambeault et al.19 Cell lines or patient samples with known mutations were used to generate standard curves, and sensitivity was determined using known wild-type (WT) samples. Bone marrow samples from children presenting with ALL were screened for low-level KRAS mutations (supplemental Table 3).
Western blotting
Whole cell lysates were prepared, and standard western blotting performed with antibodies against p–extracellular-signal regulated kinase (ERK) (Santa Cruz), Bim, and cleaved poly ADP ribose polymerase (Parp; Cell Signaling). Blots were stripped and reprobed for ERK2 (Santa Cruz) and tubulin (Sigma), which acted as loading controls.
Selumetinib
Selumetinib was initially purchased from Selleck Chemicals (supplied by Stratech, United Kingdom) and latterly was gifted from Astra Zeneca. Single-use aliquots of stock solution (20 mM) were prepared in DMSO and stored at −20°C.
Cell culture
Cell lines were obtained from European Collection of Cell Cultures or American Tissue Culture Collection and were grown in RPMI 1640 (PAA Laboratories) supplemented with 10% FBS. PreB697, Nalm6, and Molt4 all have NRAS mutations; CEM has a KRAS mutation; and Reh, Jurkat, and Raji are WT.
Cell viability assays
Primary ALL cells or cell lines were suspended in RPMI 1640 plus 10% to 15% FBS and plated out in triplicate in 96-well plates. Cells were treated with 0.1 nM to 100 µM selumetinib (plus DMSO as a control vehicle [CV]). Following a 96-hour drug exposure, cytotoxicity was assessed using CellTiter 96 Aqueous One kit (Promega). The resulting absorbances were averaged and expressed as a percent of the CV. Survival curves were plotted, and growth inhibitory (GI) 50 values calculated using GraphPad Prism software.
Xenograft studies
Primagrafts were generated using primary ALL cells injected intrafemorally into NOD SCID γ null mice and confirmed to have the same RAS mutation status as primary cells. Details of the primary samples are used are given in supplemental Table 4. For drug efficacy studies, primagraft cells were injected, and mice were monitored for engraftment every 3 to 4 weeks by tail vein bleed. Briefly, blood was red cell lysed and analyzed by flow cytometry on a BD FACSCanto II, using anti-human CD10, CD34, and CD19 and anti-mouse CD45 antibodies. Once the level of human leukemia cells reached >1% of total cells, mice were randomized into control and treatment groups. The treatment group received 25 or 100 mg/kg selumetinib, and the control group received vehicle only (CV; 0.5% hydroxypropyl methyl cellulose, 0.1% polysorbate 80) by oral gavage, twice daily throughout the week and once daily at weekends, for a total of 30 doses. Tumor burden was assessed at various time points during treatment. For pharmacodynamics studies, engrafted mice were dosed with selumetinib twice daily for 72 hours when peripheral blood leukemia load was high, and after euthanasia, the spleens were removed and assessed by flow cytometry to confirm that they contained >90% human blasts. They were then analyzed by western blotting for levels of p-ERK, ERK2, Bim, cleaved Parp, and tubulin and for annexin-V staining by flow cytometry.
Murine brain histology
Murine heads were stripped of soft tissues and decalcified in Hilleman and Lee EDTA solution (5.5% EDTA in 10% formalin) for 2 to 3 weeks, then trimmed and put in fresh EDTA for 3 to 4 days. Samples were then processed on a Tissue-Tek VIP processor using a routine overnight 17.5-hour cycle. Following paraffin wax embedding, 2.5-µm sections were cut onto slides coated with poly-l-silane. Sections were then stained with Gills hematoxylin and Putts eosin (both made in-house).
Statistical analyses
Statistical calculations were performed using SPSS software (version 18.0.1 SPSS, Chicago, IL) and STATA software (version 9.0, StataCorp, TX). Differences between groups (Ras pathway mutation vs WT) for all clinical and biological parameters were tested with Pearson’s χ2 or Fisher’s exact test and Mann-Whitney U or Kruskal-Wallis test. The probability of event-free survival and overall survival (pOS) were assessed by Kaplan-Meier analyses and the log-rank test. Probability of event-free survival or pOS was estimated from relapse diagnosis (start of relapse treatment) to a subsequent event such as death in remission, second relapse, or secondary malignancy or until any death, respectively. For multivariate analysis of survival probability, the Cox regression was performed using Wald stepwise forward testing. The likelihood-ratio test was used for comparison of different models. A P value lower than .05 was considered statistically significant.
Results
Ras pathway mutations are highly prevalent in relapsed ALL and are associated with high-risk features
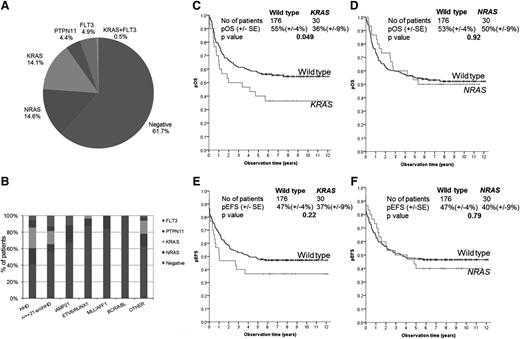
Leukemic DNA from 206 children with relapsed ALL was screened for mutations in key exons of NRAS, KRAS, FLT3, and PTPN11 by DHPLC. Mutations were found in 78 patients, giving an incidence of 37.9% (95% CI, 31.5-44.7) and were made up of NRAS (n = 30 patients), KRAS (n = 30), FLT3 (n = 10), and PTPN11 (n = 9) and are shown in Figure 1A and supplemental Table 5. Mutations were usually mutually exclusive in that only 2 patients (1%) had >1 mutation: 1 with 2 NRAS mutations (G12D and G13D) and another with a KRAS mutation (G13D), found together with FLT3 (D835H). Most mutations are well recognized, although somatically acquired insertion/deletion mutations in PTPN11 have not been previously documented.20 Using remission material as a source of constitutive DNA, PTPN11 and noncanonical KRAS mutations, including A18D and V14I, were confirmed as somatic. Importantly, in the majority (>90%) of patients, mutations were readily apparent after Sanger sequencing, indicating that they were present in the major clone among the relapsed cell population.
Ras pathway mutations in relapsed ALL and their association with cytogenetics and event-free survival. Pie chart showing the proportion of patients with Ras pathway mutations (A). Histogram of Ras pathway mutations in relation to cytogenetic subgroups (B). Kaplan Meier overall and event-free survival curves of KRAS mutants (C and E) and NRAS mutants (D and F) compared with WT.
Ras pathway mutations in relapsed ALL and their association with cytogenetics and event-free survival. Pie chart showing the proportion of patients with Ras pathway mutations (A). Histogram of Ras pathway mutations in relation to cytogenetic subgroups (B). Kaplan Meier overall and event-free survival curves of KRAS mutants (C and E) and NRAS mutants (D and F) compared with WT.
Ras pathway mutations were correlated with key clinical and biological features (Table 1 and supplemental Table 6). As previously reported in diagnostic cohorts, the frequency of Ras pathway mutations varied across the principal cytogenetic subgroups, with the highest frequency in the high hyperdiploid group (58.5%) and the lowest incidence in the ETV6-RUNX1 subgroup (12.9%) (P < .01) (Figure 1B). There was a significant relationship between Ras pathway mutations and time point of relapse, with mutation-positive patients having a higher proportion of early relapses (P = .011), defined as after 18 but <30 months after diagnosis, and this was particularly significant for NRAS/KRAS-mutated patients (P = .001) with a higher proportion relapsing on therapy (41.7% vs 25.3%; P = .02) and a median time to relapse of 2.34 years compared with 2.94 for WT patients (P = .0079). Consistent with these observations, NRAS/KRAS-mutated patients were more chemoresistant and were less likely to obtain a cytological response after the first course of therapy, and this was more marked for KRAS mutations, 22.2% compared with 46.8% for WT (P = .023). Children with a KRAS mutation had a significantly poorer 10-year pOS, 36% compared with 55% for WT (log-rank test P = .049, Figure 1C). In contrast, pOS was not different between patients with NRAS mutated (50%, standard error ± 9%) and WT (50%, standard error ± 4%) (Figure 1D). There were no differences in event-free survival (Figure 1E-F). The final model of multivariate analysis (model 2, supplemental Table 7) included time point of relapse and IKZF1 deletion and found KRAS mutation not to be independently predictive of overall survival (hazard ratio 0.99; 95% CI, 0.58-1.67). In addition, there was an overrepresentation of NRAS/KRAS-positive patients with CNS involvement, 23.3% compared with 10.3% for the mutation-negative group (P = .014), and this was more notable for NRAS mutants (26.7% vs 11.9%; P = .032). There was also an association with gender in that of the 30 KRAS mutations identified; 24 (80%) were present in males (P = .022).
Ras pathway mutations in relapsed B-lineage ALL and clinical characteristics
| Parameter/category . | Total cohort . | All RAS pathway genes . | NRAS and KRAS . | NRAS . | KRAS . | PTPN11 . | FLT3 . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n . | % . | n . | % . | P . | n . | % . | P . | n . | % . | P . | n . | % . | P . | n . | % . | P . | n . | % . | P . | |
| Gender | ||||||||||||||||||||
| Total | 206 | 100.0 | 78 | 100 | 60 | 30 | 30 | 9 | 10 | |||||||||||
| Male | 126 | 61.2 | 53 | 68.0 | .12 | 42 | 70.0 | .095 | 18 | 60.0 | .89 | 24 | 80.0 | .022 | 6 | 66.7 | .73 | 6 | 60.0 | 1.00 |
| Female | 80 | 38.8 | 25 | 32.0 | 18 | 30.0 | 12 | 40.0 | 6 | 20.0 | 3 | 33.3 | 4 | 40.0 | ||||||
| Age at relapse diagnosis | ||||||||||||||||||||
| <5 y | 29 | 14.1 | 14 | 18.0 | .46 | 11 | 18.3 | .52 | 5 | 16.7 | .89 | 6 | 20.0 | .57 | 2 | 22.2 | .72 | 2 | 20.0 | .82 |
| 5-10 y | 89 | 43.2 | 32 | 41.0 | 24 | 40.0 | 13 | 43.3 | 11 | 36.7 | 4 | 44.4 | 4 | 40.0 | ||||||
| ≥10 y | 88 | 42.7 | 32 | 41.0 | 25 | 41.7 | 12 | 40.0 | 13 | 43.3 | 3 | 33.3 | 4 | 40.0 | ||||||
| Time point of relapse | ||||||||||||||||||||
| Very early | 44 | 21.4 | 18 | 23.0 | .011 | 16 | 26.7 | .001 | 6 | 20.0 | .017 | 10 | 33.3 | .039 | 1 | 11.1 | .72 | 1 | 10.0 | .83 |
| Early | 43 | 20.9 | 24 | 30.8 | 21 | 35.0 | 12 | 40.0 | 9 | 30.0 | 1 | 11.1 | 1 | 20.0 | ||||||
| Late | 119 | 57.8 | 36 | 46.2 | 23 | 38.3 | 12 | 40.0 | 11 | 36.7 | 7 | 77.8 | 7 | 70.0 | ||||||
| Relapse stage | ||||||||||||||||||||
| On therapy | 62 | 30.1 | 28 | 35.9 | .16 | 25 | 41.7 | .02 | 13 | 43.3 | .087 | 12 | 40.0 | .20 | 2 | 22.2 | .73 | 1 | 10.0 | .29 |
| Off therapy | 144 | 69.9 | 50 | 64.1 | 35 | 58.3 | 17 | 56.7 | 18 | 60.0 | 7 | 77.8 | 9 | 90.0 | ||||||
| Site of relapse | ||||||||||||||||||||
| BM isolated | 163 | 79.1 | 59 | 75.6 | .34 | 44 | 73.3 | .19 | 20 | 66.7 | .069 | 24 | 80.0 | .90 | 7 | 77.8 | 1.00 | 8 | 80.0 | 1.00 |
| BM combined | 61 | 29.6 | 19 | 24.4 | 16 | 26.7 | 10 | 33.3 | 6 | 20.0 | 2 | 22.2 | 2 | 20.0 | ||||||
| CNS involvement | ||||||||||||||||||||
| No | 177 | 85.9 | 63 | 80.8 | .097 | 46 | 76.7 | .014 | 22 | 73.3 | .032 | 24 | 80.0 | .31 | 8 | 88.9 | 1.00 | 9 | 90.0 | 1.00 |
| Yes | 29 | 14.1 | 15 | 19.2 | 14 | 23.3 | 8 | 26.7 | 6 | 20.0 | 1 | 11.1 | 1 | 10.0 | ||||||
| Strategic group | ||||||||||||||||||||
| S2 | 131 | 63.6 | 44 | 56.4 | 30 | 50.0 | .024 | 16 | 53.3 | .15 | 14 | 46.7 | .11 | 7 | 77.8 | .88 | 8 | 80.0 | .72 | |
| S3 | 31 | 15.0 | 16 | 20.5 | 14 | 23.3 | 8 | 26.7 | 6 | 20.0 | 1 | 11.1 | 1 | 10.0 | ||||||
| S4 | 44 | 21.4 | 18 | 23.1 | 16 | 26.7 | 6 | 20 | 10 | 33.3 | 1 | 11.1 | 1 | 10.0 | ||||||
| Cytological response | ||||||||||||||||||||
| After first course | 86 | 41.7 | 26 | 34.7 | .079 | 18 | 31.6 | .031 | 12 | 40.0 | .26 | 6 | 22.2 | .023 | 4 | 44.4 | .83 | 4 | 40.0 | .70 |
| After second course | 56 | 27.2 | 27 | 36.0 | 19 | 33.3 | 9 | 30 | 10 | 37.0 | 4 | 44.4 | 5 | 50.0 | ||||||
| After third course | 17 | 8.3 | 8 | 10.7 | 8 | 14.0 | 2 | 6.7 | 6 | 22.2 | 0 | 0 | 0 | 0 | ||||||
| After fourth course | 6 | 2.9 | 4 | 5.3 | 4 | 7.0 | 3 | 10.0 | 1 | 3.7 | 0 | 0 | 0 | 0 | ||||||
| Nonresponse (no CR) | 33 | 16.0 | 10 | 13.3 | 8 | 14.0 | 4 | 13.3 | 4 | 14.8 | 1 | 11.1 | 1 | 10.0 | ||||||
| Unknown | 8 | 3 | 3 | 0 | 0 | |||||||||||||||
| Outcome | ||||||||||||||||||||
| CCR | 94 | 45.6 | 33 | 42.3 | .40 | 23 | 38.3 | .32 | 12 | 40 | .63 | 11 | 37 | .31 | 5 | 56 | 1.00 | 6 | 60 | .85 |
| Second relapse | 55 | 26.7 | 27 | 34.6 | 22 | 36.7 | 12 | 40 | 10 | 33 | 3 | 33 | 2 | 20 | ||||||
| TRD | 15 | 7.3 | 5 | 6.4 | 4 | 6.7 | 2 | 7 | 2 | 7 | 0 | 0 | 1 | 10 | ||||||
| Secondary malignancy | 2 | 9.7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| Nonresponse | 33 | 16.0 | 10 | 12.8 | 8 | 13.3 | 4 | 13 | 4 | 13 | 1 | 11 | 1 | 10 | ||||||
| Induction death | 7 | 3.4 | 3 | 3.9 | 3 | 5.0 | 0 | 0 | 3 | 10 | 0 | 0 | 0 | 0 | ||||||
| ETV6-RUNX1 | ||||||||||||||||||||
| Positive | 31 | 15.2 | 4 | 5.1 | .001 | 4 | 6.7 | .032 | 3 | 10.0 | .58 | 1 | 3.3 | .055 | 0 | 0 | .19 | 0 | 0 | .17 |
| Negative | 173 | 83.8 | 74 | 94.9 | 56 | 93.3 | 27 | 90.0 | 29 | 96.7 | 9 | 100 | 10 | 100 | ||||||
| Unknown | 2 | 0 | 0 | 0 | ||||||||||||||||
| High hyperdiploid | ||||||||||||||||||||
| Positive | 41 | 19.9 | 24 | 30.8 | .002 | 18 | 30.0 | .020 | 8 | 26.7 | .32 | 10 | 33.3 | .046 | 4 | 44.4 | .08 | 2 | 20 | 1.00 |
| Negative | 165 | 80.1 | 54 | 69.2 | 42 | 70.0 | 22 | 73.3 | 20 | 66.7 | 5 | 55.6 | 8 | 80 | ||||||
| TP53 alteration | ||||||||||||||||||||
| Positive | 23 | 11.5 | 7 | 9.2 | .43 | 6 | 10.3 | .74 | 3 | 10.3 | 1.00 | 3 | 10.34 | 1.00 | 1 | 11.1 | 1.00 | 0 | 0 | .61 |
| Negative | 177 | 88.5 | 69 | 90.8 | 52 | 89.7 | 26 | 89.7 | 26 | 89.7 | 8 | 88.9 | 10 | 100 | ||||||
| Unknown | 6 | |||||||||||||||||||
| IKZF1 deletion | ||||||||||||||||||||
| Positive | 65 | 32.5 | 28 | 36.8 | .310 | 20 | 34.5 | .70 | 11 | 37.9 | .52 | 9 | 31.0 | 1.00 | 4 | 44.4 | .48 | 4 | 40 | .73 |
| Negative | 135 | 67.5 | 48 | 63.2 | 38 | 65.5 | 18 | 62.1 | 20 | 69.0 | 5 | 55.6 | 6 | 60 | ||||||
| Unknown | 6 | |||||||||||||||||||
| Parameter/category . | Total cohort . | All RAS pathway genes . | NRAS and KRAS . | NRAS . | KRAS . | PTPN11 . | FLT3 . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n . | % . | n . | % . | P . | n . | % . | P . | n . | % . | P . | n . | % . | P . | n . | % . | P . | n . | % . | P . | |
| Gender | ||||||||||||||||||||
| Total | 206 | 100.0 | 78 | 100 | 60 | 30 | 30 | 9 | 10 | |||||||||||
| Male | 126 | 61.2 | 53 | 68.0 | .12 | 42 | 70.0 | .095 | 18 | 60.0 | .89 | 24 | 80.0 | .022 | 6 | 66.7 | .73 | 6 | 60.0 | 1.00 |
| Female | 80 | 38.8 | 25 | 32.0 | 18 | 30.0 | 12 | 40.0 | 6 | 20.0 | 3 | 33.3 | 4 | 40.0 | ||||||
| Age at relapse diagnosis | ||||||||||||||||||||
| <5 y | 29 | 14.1 | 14 | 18.0 | .46 | 11 | 18.3 | .52 | 5 | 16.7 | .89 | 6 | 20.0 | .57 | 2 | 22.2 | .72 | 2 | 20.0 | .82 |
| 5-10 y | 89 | 43.2 | 32 | 41.0 | 24 | 40.0 | 13 | 43.3 | 11 | 36.7 | 4 | 44.4 | 4 | 40.0 | ||||||
| ≥10 y | 88 | 42.7 | 32 | 41.0 | 25 | 41.7 | 12 | 40.0 | 13 | 43.3 | 3 | 33.3 | 4 | 40.0 | ||||||
| Time point of relapse | ||||||||||||||||||||
| Very early | 44 | 21.4 | 18 | 23.0 | .011 | 16 | 26.7 | .001 | 6 | 20.0 | .017 | 10 | 33.3 | .039 | 1 | 11.1 | .72 | 1 | 10.0 | .83 |
| Early | 43 | 20.9 | 24 | 30.8 | 21 | 35.0 | 12 | 40.0 | 9 | 30.0 | 1 | 11.1 | 1 | 20.0 | ||||||
| Late | 119 | 57.8 | 36 | 46.2 | 23 | 38.3 | 12 | 40.0 | 11 | 36.7 | 7 | 77.8 | 7 | 70.0 | ||||||
| Relapse stage | ||||||||||||||||||||
| On therapy | 62 | 30.1 | 28 | 35.9 | .16 | 25 | 41.7 | .02 | 13 | 43.3 | .087 | 12 | 40.0 | .20 | 2 | 22.2 | .73 | 1 | 10.0 | .29 |
| Off therapy | 144 | 69.9 | 50 | 64.1 | 35 | 58.3 | 17 | 56.7 | 18 | 60.0 | 7 | 77.8 | 9 | 90.0 | ||||||
| Site of relapse | ||||||||||||||||||||
| BM isolated | 163 | 79.1 | 59 | 75.6 | .34 | 44 | 73.3 | .19 | 20 | 66.7 | .069 | 24 | 80.0 | .90 | 7 | 77.8 | 1.00 | 8 | 80.0 | 1.00 |
| BM combined | 61 | 29.6 | 19 | 24.4 | 16 | 26.7 | 10 | 33.3 | 6 | 20.0 | 2 | 22.2 | 2 | 20.0 | ||||||
| CNS involvement | ||||||||||||||||||||
| No | 177 | 85.9 | 63 | 80.8 | .097 | 46 | 76.7 | .014 | 22 | 73.3 | .032 | 24 | 80.0 | .31 | 8 | 88.9 | 1.00 | 9 | 90.0 | 1.00 |
| Yes | 29 | 14.1 | 15 | 19.2 | 14 | 23.3 | 8 | 26.7 | 6 | 20.0 | 1 | 11.1 | 1 | 10.0 | ||||||
| Strategic group | ||||||||||||||||||||
| S2 | 131 | 63.6 | 44 | 56.4 | 30 | 50.0 | .024 | 16 | 53.3 | .15 | 14 | 46.7 | .11 | 7 | 77.8 | .88 | 8 | 80.0 | .72 | |
| S3 | 31 | 15.0 | 16 | 20.5 | 14 | 23.3 | 8 | 26.7 | 6 | 20.0 | 1 | 11.1 | 1 | 10.0 | ||||||
| S4 | 44 | 21.4 | 18 | 23.1 | 16 | 26.7 | 6 | 20 | 10 | 33.3 | 1 | 11.1 | 1 | 10.0 | ||||||
| Cytological response | ||||||||||||||||||||
| After first course | 86 | 41.7 | 26 | 34.7 | .079 | 18 | 31.6 | .031 | 12 | 40.0 | .26 | 6 | 22.2 | .023 | 4 | 44.4 | .83 | 4 | 40.0 | .70 |
| After second course | 56 | 27.2 | 27 | 36.0 | 19 | 33.3 | 9 | 30 | 10 | 37.0 | 4 | 44.4 | 5 | 50.0 | ||||||
| After third course | 17 | 8.3 | 8 | 10.7 | 8 | 14.0 | 2 | 6.7 | 6 | 22.2 | 0 | 0 | 0 | 0 | ||||||
| After fourth course | 6 | 2.9 | 4 | 5.3 | 4 | 7.0 | 3 | 10.0 | 1 | 3.7 | 0 | 0 | 0 | 0 | ||||||
| Nonresponse (no CR) | 33 | 16.0 | 10 | 13.3 | 8 | 14.0 | 4 | 13.3 | 4 | 14.8 | 1 | 11.1 | 1 | 10.0 | ||||||
| Unknown | 8 | 3 | 3 | 0 | 0 | |||||||||||||||
| Outcome | ||||||||||||||||||||
| CCR | 94 | 45.6 | 33 | 42.3 | .40 | 23 | 38.3 | .32 | 12 | 40 | .63 | 11 | 37 | .31 | 5 | 56 | 1.00 | 6 | 60 | .85 |
| Second relapse | 55 | 26.7 | 27 | 34.6 | 22 | 36.7 | 12 | 40 | 10 | 33 | 3 | 33 | 2 | 20 | ||||||
| TRD | 15 | 7.3 | 5 | 6.4 | 4 | 6.7 | 2 | 7 | 2 | 7 | 0 | 0 | 1 | 10 | ||||||
| Secondary malignancy | 2 | 9.7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| Nonresponse | 33 | 16.0 | 10 | 12.8 | 8 | 13.3 | 4 | 13 | 4 | 13 | 1 | 11 | 1 | 10 | ||||||
| Induction death | 7 | 3.4 | 3 | 3.9 | 3 | 5.0 | 0 | 0 | 3 | 10 | 0 | 0 | 0 | 0 | ||||||
| ETV6-RUNX1 | ||||||||||||||||||||
| Positive | 31 | 15.2 | 4 | 5.1 | .001 | 4 | 6.7 | .032 | 3 | 10.0 | .58 | 1 | 3.3 | .055 | 0 | 0 | .19 | 0 | 0 | .17 |
| Negative | 173 | 83.8 | 74 | 94.9 | 56 | 93.3 | 27 | 90.0 | 29 | 96.7 | 9 | 100 | 10 | 100 | ||||||
| Unknown | 2 | 0 | 0 | 0 | ||||||||||||||||
| High hyperdiploid | ||||||||||||||||||||
| Positive | 41 | 19.9 | 24 | 30.8 | .002 | 18 | 30.0 | .020 | 8 | 26.7 | .32 | 10 | 33.3 | .046 | 4 | 44.4 | .08 | 2 | 20 | 1.00 |
| Negative | 165 | 80.1 | 54 | 69.2 | 42 | 70.0 | 22 | 73.3 | 20 | 66.7 | 5 | 55.6 | 8 | 80 | ||||||
| TP53 alteration | ||||||||||||||||||||
| Positive | 23 | 11.5 | 7 | 9.2 | .43 | 6 | 10.3 | .74 | 3 | 10.3 | 1.00 | 3 | 10.34 | 1.00 | 1 | 11.1 | 1.00 | 0 | 0 | .61 |
| Negative | 177 | 88.5 | 69 | 90.8 | 52 | 89.7 | 26 | 89.7 | 26 | 89.7 | 8 | 88.9 | 10 | 100 | ||||||
| Unknown | 6 | |||||||||||||||||||
| IKZF1 deletion | ||||||||||||||||||||
| Positive | 65 | 32.5 | 28 | 36.8 | .310 | 20 | 34.5 | .70 | 11 | 37.9 | .52 | 9 | 31.0 | 1.00 | 4 | 44.4 | .48 | 4 | 40 | .73 |
| Negative | 135 | 67.5 | 48 | 63.2 | 38 | 65.5 | 18 | 62.1 | 20 | 69.0 | 5 | 55.6 | 6 | 60 | ||||||
| Unknown | 6 | |||||||||||||||||||
P values are derived from comparisons of mutated vs nonmutated group. In terms of time point of relapse, very early is <18 mo from initial diagnosis, early is between 18 mo after initial diagnosis and 6 mo after regular completion of initial treatment, and late is >6 mo after regular completion of initial treatment. P values (< .05) are in bold font.
BM, bone marrow; CCR, continuous complete remission; CNS, central nervous system; CR, complete remission; TRD, treatment-related death.
Ras pathway mutations are often “acquired” at relapse but are evident at low level at diagnosis in many patients
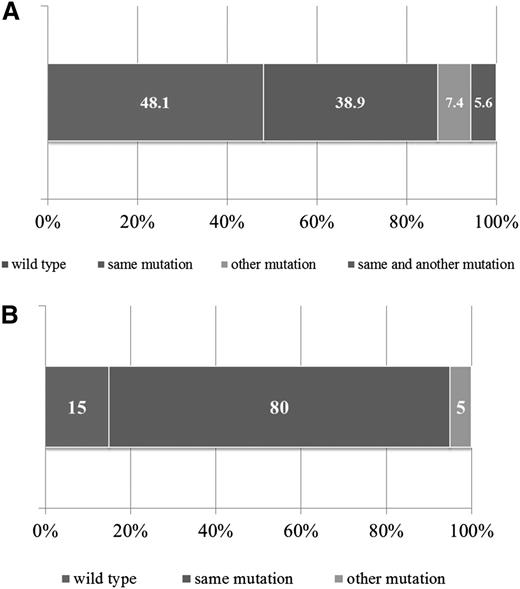
For 47 patients, matched diagnostic samples were available to determine the presence of Ras pathway mutations and their role in disease evolution. Additional nontrial samples were also included (n = 7). Mutations included NRAS (n = 21), KRAS (n = 25), PTPN11 (n = 5), and FLT3 (n = 3). In 21 of 54 patients (38.9%), the mutation found at relapse was present at diagnosis, and in 3 patients (5.6%), the same mutation was present alongside an additional one (Figure 2A). Four of the 54 patients (7.4%) showed loss of the original diagnostic mutation but gain of an alternative Ras pathway mutation at relapse. Strikingly, in 26 of the 54 patients (48.1%), the diagnostic sample was WT, suggesting acquisition of the mutation during treatment or persistence of a low-level leukemic population present in the diagnostic sample but undetected by DHPLC/Sanger sequencing. To discriminate these possibilities, a sensitive allele-specific real-time quantitative PCR (TaqMAMA) was performed for the most common NRAS and KRAS mutations, and the diagnostic samples were screened for the relevant relapse mutation. Low-level mutations, ranging from 1.00E-01 (10%) down to 5.00E-04 (0.05%) were apparent in the diagnostic samples in 5 of 6 patients (83.3%) with KRAS and 3 of 8 patients (37.5%) with NRAS mutations (supplemental Table 8), a nonstatistically significant difference (P = .12, Fisher’s exact test). There were no significant associations in the clinical or demographic features in patients with gain or persisting Ras pathway mutations (supplemental Table 9). To investigate whether mutations were also lost at relapse, mutational analyses of NRAS and KRAS were performed in paired samples available from the high-risk relapsed cohort (patients from the strategic group S3/S4) and again showed evidence of gain of mutations at relapse but also loss, at a similar frequency (supplemental Figure 1).
Backtracking and forward tracking in Ras pathway mutation–positive samples. Bar graph showing mutation status of matched diagnostic samples in those patients with Ras pathway mutations at relapse (A). Bar graph showing mutation status of Ras pathway–positive relapse samples at second relapse (B).
Backtracking and forward tracking in Ras pathway mutation–positive samples. Bar graph showing mutation status of matched diagnostic samples in those patients with Ras pathway mutations at relapse (A). Bar graph showing mutation status of Ras pathway–positive relapse samples at second relapse (B).
To assess the incidence of low-level KRAS-mutated subpopulations in newly diagnosed ALL, we accessed material from patients who were negative for RAS pathway mutations by DHPLC and screened for the most common KRAS mutations (G12D, G13D, and G12V) by TaqMAMA (n = 111). The sensitivity of these assays was between 1 × 10−3 and 5 × 10−4. Low-level KRAS mutations were relatively common and were identified in 25 of the 111 samples (22.5%) with a mean level of 2.62E-2 (standard deviation, 0.086), and a similar incidence was shown in high hyperdiploidy, ETV6-RUNX1, and “other” cytogenetic groups, 27.2%, 24.0%, and 20.3%, respectively (P = .7, Fisher’s exact test) (supplemental Figure 2). In 2 patients with low-level mutations at diagnosis, follow-up samples with high levels of minimal residual disease (MRD; >0.1%) were available allowing KRAS-mutated cells to be related to leukemic cell numbers. In one, there was clear enrichment of the 2 KRAS-mutated subpopulations during treatment, with G12D- and G12V-mutated cells found in 8.39% (standard error of the mean [SEM], 2.09) and 2.42% (SEM, 1.36) of diagnostic leukemic cells, which increased significantly to 98.3% (SEM, 20.5) and 12.93% (SEM, 1.07), respectively, by the end of induction when MRD was 3.72% (supplemental Table 10). Further samples taken at weeks 14 and 41 were MRD, G12D, and G12V negative (data not shown).
Given the association with high-risk disease, 26 Ras pathway–mutated patients in the ALL relapse cohort went on to have a second relapse, and in 20 of these, material was available for mutational screening. In 80% of patients (16 of 20), the Ras pathway mutation identified at first relapse was also present at second relapse; in 15% (3 of 20), it was no longer detectable; and in 1 case, there was loss and gain of another Ras pathway mutation (Figure 2B). Sequencing chromatogram peak heights and mutation-specific allele-specific PCR showed mutations to be dominant at both first and second relapse in most patients (supplemental Table 11). Thus, in the vast majority of patients, Ras pathway mutations persist at second relapse and are the dominant clone.
Ras pathway–mutated ALL cells show differential sensitivity to the MEK inhibitor selumetinib
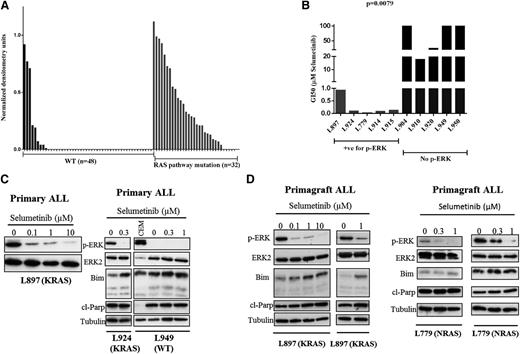
In other malignancies, Ras pathway–activating mutations are not always associated with constitutive activation of the RAS-RAF-MEK-ERK pathways; thus, we correlated pathway activation as assessed by p-ERK levels with mutation status in a large cohort of B-ALL patients (n = 80, see Figure 3A). The vast majority of Ras pathway–mutated ALL cases had constitutive activation of the pathway (27 of 32; 84.3%), and few WT leukemias were p-ERK positive (9 of 48; 18.7%) (P < .0001, Mann-Whitney U test). Three of the latter harbored chromosomal translocations involving 11q23, which are known to activate the Ras pathway; however, mutations in other genes, such as NF1, which were not screened here, may also play a role.21,22 In order to determine if Ras pathway activation is associated with differential sensitivity to MEK inhibition, we prospectively assessed the response of ALL blasts in vitro to selumetinib, a potent MEK inhibitor (Figure 3B). GI50 values were significantly lower for p-ERK/mutation-positive patient samples (n = 5, mean 250 nM, range 18-918 nM) compared with those that were negative (n = 5, mean 68 µM, range 17.8 µM to >100 µM; P = .0079, unpaired t test). Sensitivity was observed for a range of Ras pathway mutations including KRAS, G12D and G13D, NRAS, Q61R, as well as 1 patient with both a CBL and FLT3 mutation, which we have previously reported.9 Modest sensitivity was also observed in p-ERK–positive/mutation-negative patients (supplemental Figure 3). Exposure to the drug in both primary ALL cells and primagraft-derived material was associated with dose-dependent inhibition of p-ERK levels and in mutation-positive ALL cells was associated with induction of cleaved Parp and, in some cases, proapoptotic Bim (Figure 3C-D). Taken together, these data show that ALL blasts harboring a mutation activating the Ras pathway invariably have constitutive ERK activation and are differentially sensitive to the MEK inhibitor selumetinib. This differential sensitivity was also seen in ALL cell lines, but GI50 values were an order of magnitude more than primary cells (supplemental Figure 4).
Selumetinib is active in Ras pathway–mutated ALL cells in vitro and is associated with reduced levels of p-ERK and induction of Bim and cleaved Parp. Histogram showing densitometry values of p-ERK levels relative to ERK as assessed by western analyses for both WT (black bars) and Ras pathway–mutated samples (gray bars) (A). Bar chart of GI50 values as assessed by 3-(4,5 dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl-2-(4-sulfophenyl)-2H-tetrazolium assay after dosing with selumetinib for both Ras pathway–positive/p-ERK–positive samples (red bars, n = 5) and those negative for Ras pathway mutation and p-ERK (blue bars, n = 5) (B). Western analyses of ALL cell lysates from patients L897 (KRAS), L924 (KRAS), and L949 (WT) after treatment with a range of concentrations of selumetinib. Blots were probed for p-ERK, ERK2, Bim, cleaved Parp, and α-tubulin. In the case of L949, a positive control for p-ERK expression (CCRF-CEM cells) was included (C). Similar analyses of spleen cells from NOD SCID γ null mice engrafted with patient ALL cells as a source of primary-derived material (D). Primagrafts from duplicate mice implanted with blasts from patients L897 (KRAS) and L779 (NRAS) were treated with varying concentrations of selumetinib for 24 hours and processed for western analysis.
Selumetinib is active in Ras pathway–mutated ALL cells in vitro and is associated with reduced levels of p-ERK and induction of Bim and cleaved Parp. Histogram showing densitometry values of p-ERK levels relative to ERK as assessed by western analyses for both WT (black bars) and Ras pathway–mutated samples (gray bars) (A). Bar chart of GI50 values as assessed by 3-(4,5 dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl-2-(4-sulfophenyl)-2H-tetrazolium assay after dosing with selumetinib for both Ras pathway–positive/p-ERK–positive samples (red bars, n = 5) and those negative for Ras pathway mutation and p-ERK (blue bars, n = 5) (B). Western analyses of ALL cell lysates from patients L897 (KRAS), L924 (KRAS), and L949 (WT) after treatment with a range of concentrations of selumetinib. Blots were probed for p-ERK, ERK2, Bim, cleaved Parp, and α-tubulin. In the case of L949, a positive control for p-ERK expression (CCRF-CEM cells) was included (C). Similar analyses of spleen cells from NOD SCID γ null mice engrafted with patient ALL cells as a source of primary-derived material (D). Primagrafts from duplicate mice implanted with blasts from patients L897 (KRAS) and L779 (NRAS) were treated with varying concentrations of selumetinib for 24 hours and processed for western analysis.
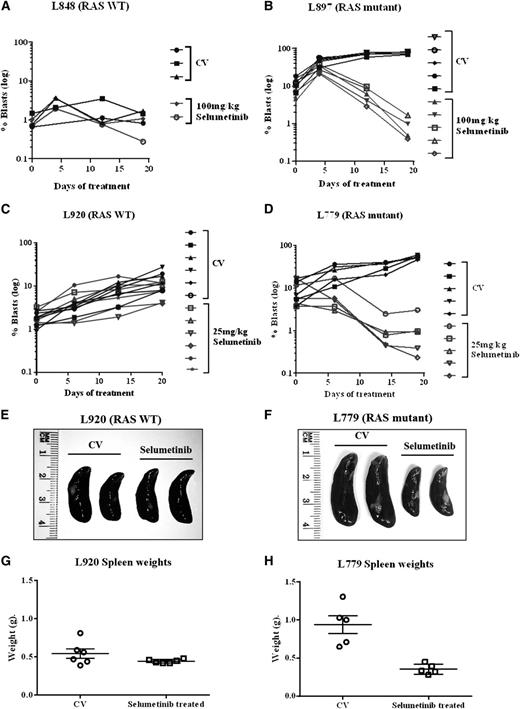
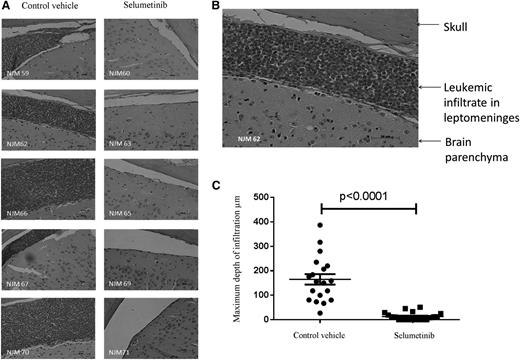
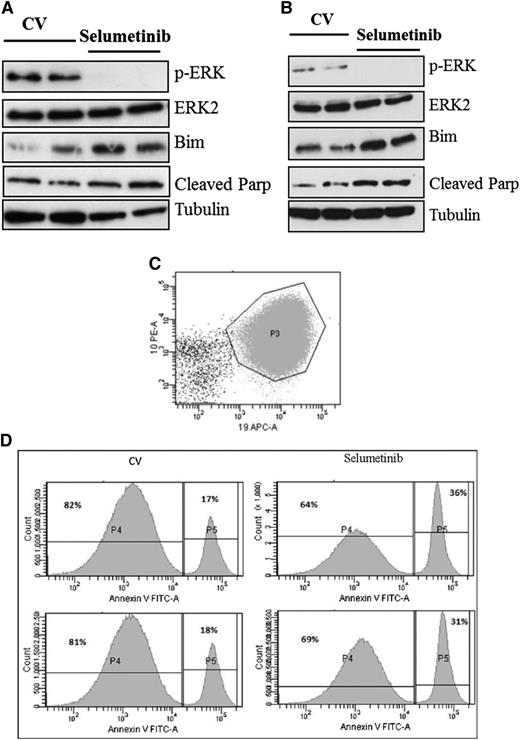
Following on from these promising in vitro experiments, we next assessed the activity of the MEK inhibitor selumetinib in vivo, using an orthotopic mouse model with primagrafts derived from primary ALL cells with known Ras pathway mutation status. After intrafemoral injection and leukemia engraftment, mice were randomized to receive either selumetinib (100 mg/kg twice daily) or CV, and levels of circulating blasts were monitored during treatment by flow cytometry. There was a dramatic reduction in circulating leukemia cell numbers in the mice implanted with the Ras mutant ALL cells (L897 KRAS, G12D) treated with selumetinib, with a mean fold decrease of 10 after 30 drug doses compared with a mean fold increase of 7 for CV and levels at the end of treatment ranging from 0.39% to 1.67% for drug treated compared with 70% to 84% for CV. There was minimal effect of selumetinib in the WT group (Figure 4A-B). The selumetinib activity on Ras pathway–mutated ALL was replicated in a second Ras-mutated primagraft (L779, NRAS, Q61R) at lower dosing (25 mg/kg twice daily), with a mean fold reduction in peripheral leukemic cell numbers of 8.5 for drug compared with 5.7-fold increase for CV, and mice euthanized at the end of the study showed a significant decrease in spleen size, mean 0.35 g compared with 0.89 g, respectively (P < .001, Student t test). Similar dosing in a second WT primagraft showed no difference in peripheral blood leukemic cell numbers or spleen sizes (P = .15) (Figure 4C-H). Histologic analysis of postmortem brains from mice engrafted with L779 (NRAS, Q61R) showed evidence of extensive meningeal leukemic infiltration in mice treated with CV, which was significantly reduced in mice treated with selumetinib (P < .001, Mann-Whitney U test) (Figure 5A-C). Pharmacodynamic assessment in spleens of mice engrafted with both the NRAS and KRAS mutant primagrafts showed absence of p-ERK and increased levels of the proapoptotic Bim as well and cleaved Parp (Figure 6A-B). In addition, flow cytometric analyses of L897-engrafted mice spleens showed a doubling of the numbers of annexin-V–positive leukemic cells in selumetinib-treated mice compared with CV (Figure 6C-D).
Selumetinib is active in Ras pathway–mutated ALL cells in an orthotopic primagraft model. Graphs showing log of the percent circulating leukemic cells in mice implanted with RAS WT (A,C) or Ras pathway mutant patient blasts (B,D). Mice were treated with either CV or selumetinib twice daily (100 mg/kg for B and 25 mg/kg for D) and the percent of circulating blasts quantified by flow cytometry during and at the end of treatment. Photographs of spleens (E,F) and graphs of spleen weights (G,H) after 30 drug doses of selumetinib in WT primagrafts (E,G) and RAS mutant primagrafts are shown (F,H).
Selumetinib is active in Ras pathway–mutated ALL cells in an orthotopic primagraft model. Graphs showing log of the percent circulating leukemic cells in mice implanted with RAS WT (A,C) or Ras pathway mutant patient blasts (B,D). Mice were treated with either CV or selumetinib twice daily (100 mg/kg for B and 25 mg/kg for D) and the percent of circulating blasts quantified by flow cytometry during and at the end of treatment. Photographs of spleens (E,F) and graphs of spleen weights (G,H) after 30 drug doses of selumetinib in WT primagrafts (E,G) and RAS mutant primagrafts are shown (F,H).
Selumetinib eradicates CNS leukemia in an orthotopic primagraft model. Photomicrographs of brain sections stained with hematoxylin and eosin from mice engrafted with Ras pathway mutant ALL cells (L779) after treatment with selumetinib or CV (×20 objective) (A). Photomicrograph of 1 section from CV-treated mouse after increased magnification (×40) (B). Dot plot of the depth of leukemic infiltrate into the leptomeninges in CV- vs selumetinib-treated mice (C).
Selumetinib eradicates CNS leukemia in an orthotopic primagraft model. Photomicrographs of brain sections stained with hematoxylin and eosin from mice engrafted with Ras pathway mutant ALL cells (L779) after treatment with selumetinib or CV (×20 objective) (A). Photomicrograph of 1 section from CV-treated mouse after increased magnification (×40) (B). Dot plot of the depth of leukemic infiltrate into the leptomeninges in CV- vs selumetinib-treated mice (C).
Pharmacodynamic analyses after selumetinib dosing in vivo shows prolonged inhibition of p-ERK and induction of apoptosis. Western analyses of spleen cells from mice engrafted with Ras pathway mutant ALL cells after 72 hours of 100 mg/kg (A, L897, KRAS) or 25 mg/kg BID (B, L779, NRAS). Flow cytometric analyses of the spleen harvests from L897 primagrafts stained with antibodies to mouse CD45, human CD19, CD10, and CD34, along with annexin V. Dot plot showing CD10 and CD19 expression of cells gated as lymphoid by light scatter and then for mouse CD45 negativity (C). CD10+CD19+ cells are then gated and displayed as histograms showing annexin-V fluorescence (D).
Pharmacodynamic analyses after selumetinib dosing in vivo shows prolonged inhibition of p-ERK and induction of apoptosis. Western analyses of spleen cells from mice engrafted with Ras pathway mutant ALL cells after 72 hours of 100 mg/kg (A, L897, KRAS) or 25 mg/kg BID (B, L779, NRAS). Flow cytometric analyses of the spleen harvests from L897 primagrafts stained with antibodies to mouse CD45, human CD19, CD10, and CD34, along with annexin V. Dot plot showing CD10 and CD19 expression of cells gated as lymphoid by light scatter and then for mouse CD45 negativity (C). CD10+CD19+ cells are then gated and displayed as histograms showing annexin-V fluorescence (D).
Discussion
This study identifies Ras pathway mutations as a highly prevalent genetic abnormality in relapsed ALL, affecting almost 40% of children. Mutations were invariably mutually exclusive, suggesting that mutations that activate the pathway preclude the necessity for a second activating Ras pathway “hit” and emphasizing the importance of the Ras pathway in ALL pathogenesis and progression. Clinically, mutations were more likely to be present in children with high-risk features such as early relapse (all mutations), on treatment relapse, and CNS involvement (NRAS/KRAS mutations). Patients with NRAS/KRAS mutations were more likely to have chemoresistant disease, as evidenced by reduced cytological remission rates and for KRAS mutations, a reduced overall survival. These data are consistent with in vitro studies showing that activation of the Ras pathway in hemapoietic cells can increase resistance to key drugs used in ALL therapy, including glucocorticoids and anthracyclines.23,24
Large studies found no prognostic significance of NRAS and KRAS mutation in ALL at diagnosis, which is surprising given the association with clinical parameters at relapse shown here.15 However, mutations in the Ras pathway are prevalent in very good risk cytogenetic group, high hyperdiploidy, but also in very poor risk groups, such as hypodiploidy and a group defined as “high risk,” which may neutralize any effect on prognosis when a patient cohort is analyzed as a whole.7,10-14,16,25 Within specific cytogenetic groups, a small study in hyperdiploid ALL showed no influence of Ras pathway mutation on prognosis, but in MLL rearranged infant ALL, NRAS/ KRAS mutations were associated with an extremely poor outcome.10,26 Mutational screening studies of large upfront ALL trials will define whether Ras pathway status has prognostic relevance within these various subgroups and if it can enhance current risk stratification strategies.
In this large relapsed cohort, we show that Ras pathway mutations appear to be acquired at relapse in almost half of Ras pathway–positive patients. Backtracking analyses using allele-specific PCR showed evidence of low-level mutated subclones in more than half of matched diagnostic samples, more so for KRAS mutations, suggesting that these mutated cells have evaded up-front therapy and “driven” relapse. These data highlight a significant role of Ras signaling in conferring a more chemoresistant phenotype and are supported by the dramatic enrichment of KRAS-mutated cells in 1 case during induction chemotherapy, with a 10% RAS-mutated leukemia burden at diagnosis increasing to 100% of the MRD. A study of relapse-associated copy number alterations also demonstrated the presence of the relapse clone as a minor subpopulation at diagnosis, but unlike this study, the affected genes were diverse, with no apparent common pathway.27 However, data from this study and elsewhere show that Ras pathway mutations present at diagnosis can also be lost at relapse and low-level KRAS mutations are common in a diagnostic cohort of long-term survivors.28,29 Thus, additional cooperating factors must influence survival of these low-level mutated clones during treatment and contribute to Ras pathway–mutated leukemia progression and relapse.30 Although transgenic models suggest that oncogenic Ras can initiate leukemia and may be a primary event (reviewed in Ward et al31 ), we show that this is a common secondary genetic event during leukemogenesis that may or may not confer a survival advantage during ALL therapy.
For those Ras-positive relapse patients who were negative in backtracking, it not clear whether mutated subclones are present but below the limit of detection of our assays or whether the mutation arises de novo, induced by the prolonged barrage of genotoxic chemotherapeutic agents during up-front therapy. Our recent report, describing TP53 mutations in the same relapse cohort, also found acquisition of mutations at relapse, but in contrast to the Ras data here, all appeared to be de novo mutations.32 A comparison of the specific NRAS/KRAS mutations of 2 previously published diagnostic ALL cohorts7,15 with the relapse cohort described here, shows similarities in the frequency of KRAS/NRAS mutations and codon position but a higher proportion of transversion mutations at relapse (data not shown). This may be indicative of mutagen exposure.
The high incidence of Ras pathway mutations at relapse and their presence in the major clone of relapsed leukemic cells and association with high-risk disease prompted us to investigate potential novel therapies that exploit pathway activation. We have comprehensively shown significant cytotoxic activity of selumetinib in Ras pathway–mutated ALL cells, in vitro, ex vivo, and in in vivo models. The differential cytotoxicity in Ras pathway–mutated ALL cells was associated with induction of the proapoptotic Bim and an increase in downstream apoptotic markers including cleaved PARP and annexin V. This differential effect may translate into an effective therapeutic window in the clinic. Importantly, given the association of NRAS/KRAS mutations and the presence of CNS disease at relapse, MEK inhibitor treatment significantly reduced CNS disease burden in our xenograft model. Selumetinib is a potent, selective, allosteric inhibitor of MEK1/2 that has reached phase 2 clinical trials in adult melanoma, colon cancer, and non–small cell lung cancer; has a favorable toxicity profile; and has demonstrated antitumor activity in some patients.33,34 It is also in a phase 1 trial for 2 pathway-activated pediatric malignancies, neurofibromatosis and pilocytic astrocytoma.
In summary, we show that Ras pathway mutations are a common genetic abnormality in relapsed ALL and are associated with high-risk features and poor prognosis. Mutations are often selected for or acquired during treatment and thus predominate in the relapsed leukemic clone and usually persist in cases of second relapse. Targeted MEK inhibition with selumetinib shows excellent activity in RAS-mutated ALL both in vitro and in vivo and may offer clinical benefit for a substantial proportion of children with relapsed ALL. Given our findings, clinical trials of selumetinib in Ras pathway–positive relapsed patients may be warranted.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Gabriele Körner, Madlen Pfau, Chris Huggins, and Rob Stewart for excellent technical assistance and Huw Thomas for useful discussions; Julia Alten and Udo zur Stadt, who provided paired samples from initial diagnosis of the ALL-REZ BFM 2002 relapse cohort; and the Newcastle Haematology Biobank for United Kingdom samples.
This work was supported by the Leukaemia and Lymphoma Research Fund (project grant 11007) (J.I.), the North of England Children’s Cancer Research Fund, Cancer Research UK (grant C27943/A12788), the German Foundation for Childhood Cancer, the German José Carreras Leukemia Foundation, and KINDerLEBEN Berlin. C. Halsey is funded by the Kay Kendall Leukaemia Fund (project grant KKL454).
Authorship
Contribution: J.I., C.E., and A.v.S. conceived and gained funding for the study; all authors performed research and analyzed and/or interpreted data; J.I., L.M., and C.E. drafted the manuscript; and all authors critically appraised and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Julie Irving, Northern Institute for Cancer Research, Newcastle University, Paul O'Gorman Building, Framlington Place, Newcastle upon Tyne, Tyne and Wear, NE2 4HH, United Kingdom; e-mail: j.a.e.irving@ncl.ac.uk.
References
Author notes
J.I. and E.M. contributed equally to this study.