Key Points
FOXP1 directly represses multiple proapoptotic genes in primary mature human B cells and DLBCL cell lines.
FOXP1 cooperates with NF-κB signaling to promote expansion of primary mature human B cells by inhibition of caspase-dependent apoptosis.
Abstract
The forkhead transcription factor FOXP1 is involved in B-cell development and function and is generally regarded as an oncogene in activated B-cell–like subtype of diffuse large B-cell lymphoma (DLBCL) and mucosa-associated lymphoid tissue lymphoma, lymphomas relying on constitutive nuclear factor κB (NF-κB) activity for survival. However, the mechanism underlying its putative oncogenic activity has not been established. By gene expression microarray, upon overexpression or silencing of FOXP1 in primary human B cells and DLBCL cell lines, combined with chromatin immunoprecipitation followed by next-generation sequencing, we established that FOXP1 directly represses a set of 7 proapoptotic genes. Low expression of these genes, encoding the BH3-only proteins BIK and Harakiri, the p53-regulatory proteins TP63, RASSF6, and TP53INP1, and AIM2 and EAF2, is associated with poor survival in DLBCL patients. In line with these findings, we demonstrated that FOXP1 promotes the expansion of primary mature human B cells by inhibiting caspase-dependent apoptosis, without affecting B-cell proliferation. Furthermore, FOXP1 is dependent upon, and cooperates with, NF-κB signaling to promote B-cell expansion and survival. Taken together, our data indicate that, through direct repression of proapoptotic genes, (aberrant) expression of FOXP1 complements (constitutive) NF-κB activity to promote B-cell survival and can thereby contribute to B-cell homeostasis and lymphomagenesis.
Introduction
The forkhead transcription factor FOXP1 plays an important role in a wide diversity of biological processes, including T-cell development and differentiation1,2 and B-cell development and function.3-5 Furthermore, FOXP1 has long been recognized as a potential oncogene in various types of B-cell non-Hodgkin lymphomas; however, its mode of oncogenic action is largely unknown.6,7
In diffuse large B-cell lymphoma (DLBCL) and mucosa-associated lymphoid tissue (MALT) lymphoma, aberrantly high expression of FOXP1 is associated with poor prognosis and FOXP1-positive MALT lymphomas were shown to be at risk of transforming into aggressive DLBCLs.8,9 This overexpression of FOXP1 can be caused by a t(3;14)(p14;q32) translocation, involving FOXP1 and IgH loci, which has recurrently been observed in MALT lymphoma and activated B-cell–like (ABC) DLBCL.10-13 FOXP1 expression is also frequently upregulated in ABC-DLBCL as a result of trisomy 3 or more restricted focal amplifications,14 whereas aberrant Myc expression in transformed gastric MALT lymphomas leads to upregulation of FOXP1 due to repression of the FOXP1 targeting miRNA 34a.15 Furthermore, expression levels of FOXP1 can be used as a discriminator between the ABC and germinal center (GC) subtypes of DLBCL, which are distinct biological disease entities, the former having significant worse survival rates.12,13
Interestingly, the type of lymphomas in which FOXP1 is highly expressed are characterized by constitutive activation of the nuclear factor κB (NF-κB) pathway, on which they rely for survival.16 Activation of various receptors, such as the B-cell receptor (BCR), CD40, or Toll-like receptors, will lead to formation of the CARD11-BCL10-MALT1 signaling complex, which results in the activation of the NF-κB pathway.17 A large proportion of MALT lymphomas express a BCR with rheumatoid factor activity, which is continuously stimulated by autoreactive immunoglobulins, causing continuous activation of the NF-κB signaling pathway.18 Moreover, MALT lymphomas often contain recurrent translocations that affect MALT1 or BCL10, resulting in constitutive activation of the NF-κB pathway.16 In ABC-DLBCL, the NF-kB signaling pathway is constitutively active as a result of oncogenic mutations in CARD11, the adaptor protein MYD88 and/or of the BCR subunit CD79 (which causes chronic active BCR signaling), and by inactivating mutations in A20, a negative regulator of the NF-κB pathway.19-24
In the present study, we aimed to further investigate the mechanistic role of FOXP1 in human B-cell function and lymphomagenesis. We show that FOXP1 directly represses the expression of a panel of proapoptotic genes in primary human B cells and DLBCL cell lines and that overexpression of FOXP1 promotes survival and outgrowth of primary human B cells by cooperating with NF-κB pathway. Together, our study provides novel insights into the role of FOXP1 in B-cell homeostasis and establishes a new oncogenic mechanism by which aberrantly expressed FOXP1 may contribute to B-cell lymphomagenesis.
Materials and methods
Constructs
pcDNA3.1-FOXP1-myc-his encoding human FOXP1 was obtained from Daniel Simon (Harvard Medical School, Boston, MA)25 and subcloned to generate LZRS-FOXP1-IRES-YFP (see supplemental Methods available on the Blood Web site). MSCV-CA-IKK2-IRES-GFP was kindly provided by Dr J. Schuringa (University of Groningen, Groningen, The Netherlands) and LZRS-BCL6-IRES-GFP by Dr H. Spits.26
B-cell cultures, retroviral transductions, and small interfering RNA–mediated knockdown
Isolated human B cells (see supplemental Methods) were cultured on CD40L-L cells,27 with interleukin-21 (IL-21) (25 ng/mL; R&D Systems, Abingdon, UK) and IL-2 (40 U/mL; Prospec, East Brunswick, NJ), and transduced essentially as described previously.28,29 Cells were passaged and cultured either with or without CD40L-L cells 72 hours after transduction. For microarray analysis, after transduction, cells were cultured without cytokines for 3 days.
DLBCL cell lines OCI-Ly1, OCI-Ly3, OCI-Ly7, OCI-Ly10, U2932, and SUDHL-6 were obtained and cultured as described previously.30 Small interfering RNA (siRNA)-mediated knockdown was performed essentially as described previoulsy31 (see supplemental Methods). Blood was obtained in accordance with local institutional board requirements and the Declaration of Helsinki.
Microarray analysis, ChIP-seq, and qRT-PCR
Flow cytometry and Caspase-Glo 3/7 assay
For full details on labeling and/or analysis of eFluor 670, propidium iodide,35 green fluorescent protein (GFP)/yellow fluorescent protein (YFP) fluorescence, the Caspase-Glo 3/7 assay, and other methods, see supplemental Methods.
Results
FOXP1 represses the expression of multiple proapoptotic genes
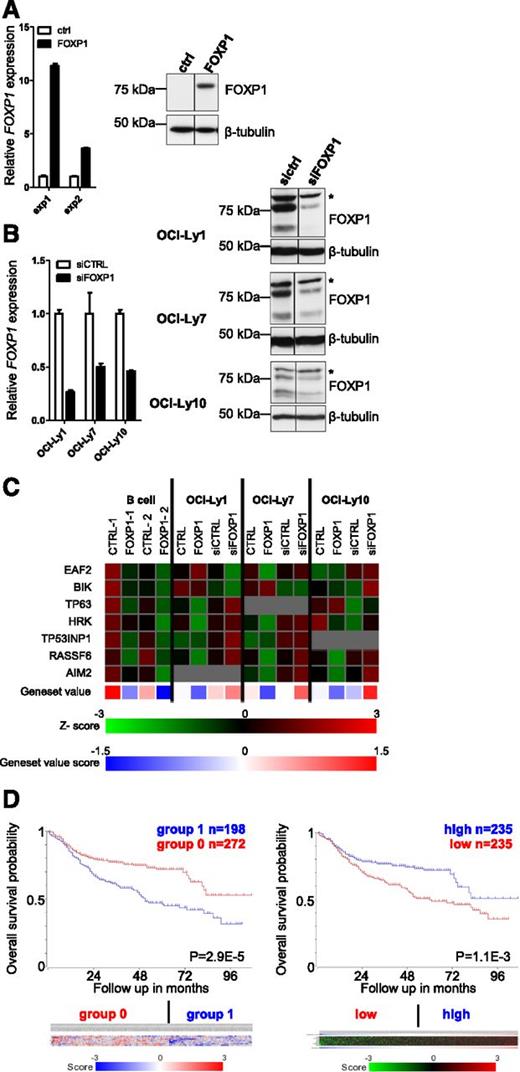
To provide insight into the potential mechanism(s) by which FOXP1 may affect B-cell function and lymphomagenesis, we performed gene expression microarray (GEM) analysis of primary human B cells retrovirally transduced with FOXP1-IRES-YFP to constitutively overexpress FOXP1 (Figure 1A). After transduction, the cells were cultured on CD40L-L cells for 3 days, YFP-positive fractions were sorted, RNA was isolated, and GEM was performed. In total, 201 genes were found to be reproducibly regulated by FOXP1 in B cells from 2 different donors (supplemental Table 1). Gene ontology (GO) analysis of these genes showed high enrichment for genes involved in induction of apoptosis (Table 1), which is of specific interest because of the key role for repression of apoptosis in (antigen-specific) differentiation and oncogenic transformation of GC B cells. Strikingly, the FOXP1- regulated genes within the various proapoptotic GO categories were all downregulated, and some of these genes, including HRK, RASSF6, and TP53INP1, were among the 15 most strongly repressed genes in either one or both of the donors.
Identification of FOXP1-regulated genes in primary human B cells and DLBCL cell lines by GEM analysis. To identify FOXP1-regulated genes, we conducted GEM analysis upon retroviral overexpression or RNA interference–mediated silencing of FOXP1 in primary B cells and DLBCL cell lines. (A-B) Expression levels of FOXP1 in primary B cells (A) and DLBCL cell lines (B) upon retroviral transduction or siRNA transfection. (A) Human primary B cells were transduced with FOXP1-IRES-YFP or ctrl-IRES-YFP and cultured with CD40L-L cells. (Left) Three days after transduction, YFP-positive fractions were sorted, RNA was isolated, and gene expression levels were analyzed by qRT-PCR. Expression levels were normalized to expression levels in empty vector–transduced cells. (Right) Three days after transduction, cell lysates were harvested and immunoblotted for FOXP1. β-Tubulin was used as a loading control. (B) DLBCL cell lines were transfected with siRNA against FOXP1. (Left) One day after nucleofection, RNA was isolated and gene expression levels were analyzed by qRT-PCR. Expression levels were normalized to expression levels in control siRNA–transduced cells. (Right) Two days after nucleofection with siRNA against FOXP1, cell lysates were harvested and immunoblotted for FOXP1. β-Tubulin was used as a loading control. Asterisk (*) indicates a nonspecific background band. (C) Expression of the proapoptotic genes that were significantly and reproducibly regulated by FOXP1 in microarray analysis of primary B cells and DLBCL cell lines. Data are presented as z-scores calculated within samples of each cell line. The gray squares indicate expression beneath the threshold value (= no expression). The lower panel shows the mean relative expression values of a gene set composed of the 7 proapoptotic genes. (D) Kaplan-Meier plots of the OS of 498 DLBCL patients treated with rituximab-CHOP therapy,37 stratified in 2 groups by expression of the 7 FOXP1-repressed proapoptotic genes. (Left) OS of DLBCL patients stratified in 2 groups by k-means clustering of the 7 genes. The 5-year OS is 72% for the patient group with overall higher (group 0; red) vs 47% in the group with overall lower (group 1; blue) expression of the 7 genes. The color bar displays the z-scores (blue = low, red = high) of each of the 7 proapoptotic genes (from top to bottom): TP63, HRK, EAF2, TP53INP1, AIM2, RASSF6, and BIK. (Right) OS of DLBCL patients stratified in 2 equal-sized groups (separated at the median) by ranking the patients according to their mean z-score of the gene set. The 5-year OS is 72% in the high-expressing (blue) vs 49% in the low-expressing (red) group. The heatmap displays the z-scores (green = low, red = high) of each of the 7 proapoptotic genes (from top to bottom): TP63, RASSF6, BIK, HRK, EAF2, TP53INP1, and AIM. The blue-red bar displays the mean z-scores (blue is low; red is high) for the gene set, according to which the patients were ranked.
Identification of FOXP1-regulated genes in primary human B cells and DLBCL cell lines by GEM analysis. To identify FOXP1-regulated genes, we conducted GEM analysis upon retroviral overexpression or RNA interference–mediated silencing of FOXP1 in primary B cells and DLBCL cell lines. (A-B) Expression levels of FOXP1 in primary B cells (A) and DLBCL cell lines (B) upon retroviral transduction or siRNA transfection. (A) Human primary B cells were transduced with FOXP1-IRES-YFP or ctrl-IRES-YFP and cultured with CD40L-L cells. (Left) Three days after transduction, YFP-positive fractions were sorted, RNA was isolated, and gene expression levels were analyzed by qRT-PCR. Expression levels were normalized to expression levels in empty vector–transduced cells. (Right) Three days after transduction, cell lysates were harvested and immunoblotted for FOXP1. β-Tubulin was used as a loading control. (B) DLBCL cell lines were transfected with siRNA against FOXP1. (Left) One day after nucleofection, RNA was isolated and gene expression levels were analyzed by qRT-PCR. Expression levels were normalized to expression levels in control siRNA–transduced cells. (Right) Two days after nucleofection with siRNA against FOXP1, cell lysates were harvested and immunoblotted for FOXP1. β-Tubulin was used as a loading control. Asterisk (*) indicates a nonspecific background band. (C) Expression of the proapoptotic genes that were significantly and reproducibly regulated by FOXP1 in microarray analysis of primary B cells and DLBCL cell lines. Data are presented as z-scores calculated within samples of each cell line. The gray squares indicate expression beneath the threshold value (= no expression). The lower panel shows the mean relative expression values of a gene set composed of the 7 proapoptotic genes. (D) Kaplan-Meier plots of the OS of 498 DLBCL patients treated with rituximab-CHOP therapy,37 stratified in 2 groups by expression of the 7 FOXP1-repressed proapoptotic genes. (Left) OS of DLBCL patients stratified in 2 groups by k-means clustering of the 7 genes. The 5-year OS is 72% for the patient group with overall higher (group 0; red) vs 47% in the group with overall lower (group 1; blue) expression of the 7 genes. The color bar displays the z-scores (blue = low, red = high) of each of the 7 proapoptotic genes (from top to bottom): TP63, HRK, EAF2, TP53INP1, AIM2, RASSF6, and BIK. (Right) OS of DLBCL patients stratified in 2 equal-sized groups (separated at the median) by ranking the patients according to their mean z-score of the gene set. The 5-year OS is 72% in the high-expressing (blue) vs 49% in the low-expressing (red) group. The heatmap displays the z-scores (green = low, red = high) of each of the 7 proapoptotic genes (from top to bottom): TP63, RASSF6, BIK, HRK, EAF2, TP53INP1, and AIM. The blue-red bar displays the mean z-scores (blue is low; red is high) for the gene set, according to which the patients were ranked.
GO term analysis of genes regulated upon FOXP1 overexpression in primary human B cells
| Biological process . | P . |
|---|---|
| Immune response | 2.76E-7 |
| Defense response | 6.38E-5 |
| Leukocyte activation | 1.52E-04 |
| Induction of apoptosis | 2.54E-04 |
| Induction of programmed cell death | 2.54E-04 |
| Response to protein stimulus | 4.12E-04 |
| Cell activation | 4.27E-4 |
| Positive regulation of apoptosis | 1.14E-3 |
| Positive regulation of programmed cell death | 1.21E-3 |
| Positive regulation of cell death | 1.25E-3 |
| Biological process . | P . |
|---|---|
| Immune response | 2.76E-7 |
| Defense response | 6.38E-5 |
| Leukocyte activation | 1.52E-04 |
| Induction of apoptosis | 2.54E-04 |
| Induction of programmed cell death | 2.54E-04 |
| Response to protein stimulus | 4.12E-04 |
| Cell activation | 4.27E-4 |
| Positive regulation of apoptosis | 1.14E-3 |
| Positive regulation of programmed cell death | 1.21E-3 |
| Positive regulation of cell death | 1.25E-3 |
GO term analysis of genes that are regulated by FOXP1 in both of 2 independent GEM studies of human peripheral blood memory B cells transduced with LZRS-IRES-FOXP1, compared with control-transduced cells. The 10 most significantly enriched GO categories are shown.
Given the putative oncogenic role of FOXP1 in DLBCL, we also investigated FOXP1-regulated gene expression upon overexpression or silencing of FOXP1 in 1 ABC-type (OCI-Ly10) and 2 GC-type (OCI-Ly1 and OCI-Ly7) DLBCL cell lines. Nucleofection of these cell lines with previously described FOXP1-targeting siRNA36 resulted in a 50% to 70% knockdown of FOXP1 expression (Figure 1B). Two days after transduction with FOXP1 or after electroporation with siRNA directed against FOXP1, RNA was isolated and GEM was performed. Interestingly, many of the proapoptotic genes repressed by FOXP1 in primary B cells were also repressed upon FOXP1 overexpression and/or induced upon FOXP1 knockdown in one or more of the cell lines (Figure 1C). Of these genes, HRK, BIK, and RASSF6, were among the 15 most strongly repressed genes in OCI-Ly1, OCI-Ly7, and/or OCI-Ly10. The microarray results for the 7 reproducibly FOXP1-repressed proapoptotic genes (ie, EAF2, BIK, TP63, HRK, TP53INP1, RASSF6, and AIM2) were validated by qRT-PCR of FOXP1-transduced primary human B cells (not shown; see, however, Figures 4C and 5C). Combining these 7 proapoptotic genes into 1 gene set clearly shows for each cell line that FOXP1 overexpression represses, and FOXP1 knockdown induces, expression of this proapoptotic gene set (Figure 1C).
Low expression of the FOXP1-repressed apoptotic genes is associated with poor survival in DLBCL patients
To investigate if the 7 apoptotic gene panel may have prognostic clinical significance in DLBCL, we analyzed the GEM data from 498 DLBCL patients treated with rituximab-CHOP37 by means of the R2 microarray analysis and visualization platform (http://r2.amc.nl). The DLBCL patients were stratified in 2 groups, either by k-means clustering of the 7 genes or after ranking the patients according to the mean z-score or the summed rank of the 7 genes (Figure 1D and supplemental Figure 1). Interestingly, irrespective of the stratification method, statistically significant differences in the probability of overall survival (OS) and progression-free survival were observed, with the patients with low expression of the 7 proapoptotic gene signature always having a worse prognosis (Figure 1D and supplemental Figure 1). Notably, in accordance with the observed FOXP1-mediated repression of these genes in ABC- and GC-type DLBCL cell lines (Figure 1C), this difference in OS (and progression-free survival; data not shown) was observed irrespective of the ABC- vs GCB-subtype classification of the patients (supplemental Figure 1). Together, our findings are completely in line with the proapoptotic function of these genes and with a potential role for their FOXP1-mediated repression in lymphomagenesis.
FOXP1 binds to the promoter regions of multiple proapoptotic genes
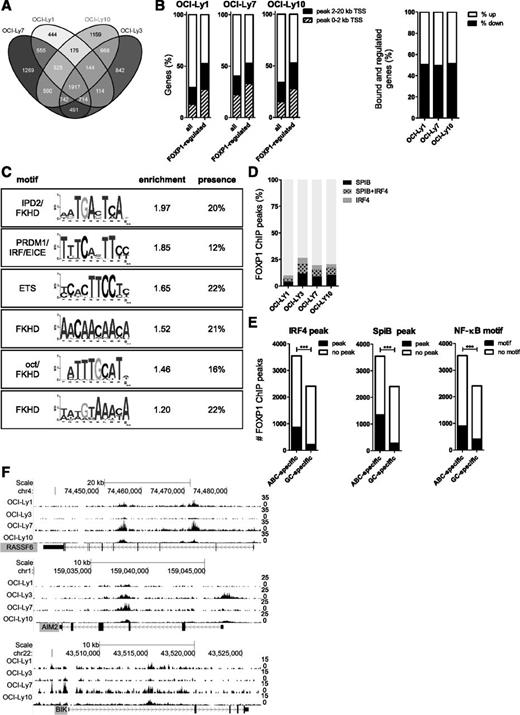
To identify genes that are directly regulated by FOXP1, we performed genome-wide ChIP-seq in 4 DLBCL cell lines, ie, OCI-Ly1, OCI-Ly3, OCI-Ly7, and OCI-Ly10. In each cell line, we identified 20 000 to 26 000 FOXP1 binding sites. These binding sites were most strongly enriched within 2 kb of the transcription start sites (TSSs) of protein-coding genes. Interestingly, however, the majority of sites were located at a larger distance from the closest TSS (supplemental Figure 2A). FOXP1 ChIP-seq peaks were found in the vicinity (within a 20-kb window around the TSS) of 4000 to 7000 genes in each cell line and approximately 2000 genes had a FOXP1 ChIP-seq peak in all 4 cell lines (Figure 2A).
Chromatin immunoprecipitation and sequencing analysis of FOXP1 target genes. To identify direct FOXP1 target genes, we conducted genome-wide mapping of the FOXP1 binding sites in 4 DLBCL cell lines, by chromatin immunoprecipitation with a FOXP1-specific antibody followed by high-throughput sequencing (ChIP-seq). (A) A Venn diagram showing the number of genes in each cell line with a FOXP1 binding peak within 20 kb of the TSS, as determined by ChIP-seq. (B) Percentage of all expressed genes or FOXP1-regulated genes (as determined by microarray analysis) in a cell line that showed a FOXP1 binding peak within 2 kb or 20 kb of the TSS, as determined by ChIP-seq (left), and the percentage of genes being either up- or downregulated, among the genes that are both regulated and bound by FOXP1 within 20 kb of the TSS (right). (C) De novo motif analysis of FOXP1 ChIP-seq peaks in OCI-Ly3 reveals the presence of several enriched motifs in the FOXP1-binding regions. Relative enrichment to control regions and percentage of peaks containing the motif are shown. (D) Overlap of IRF4 ChIP-seq peaks and SpiB ChIP-seq peaks in HBL138 with FOXP1 ChIP-seq peaks in DLBCL cell lines. (E) The number of FOXP1 ChIP-seq peaks that were found exclusively either in both ABC-DLBCL cell lines (OCI-Ly3 and OCI-Ly10) or in both GC-DLBCL cell lines (OCI-Ly1 and OCI-Ly7) and the proportion of these peaks that were also found among IRF4 or SpiB ChIP-seq peaks in HBL138 or that contained a consensus NF-κB binding site. (***P < .001 significant difference between presence of SpiB or IRF4 peaks or a NF-κB consensus motif among ABC-specific vs GC-specific FOXP1 CHIP-seq peaks as determined by χ2 test). (F) Tracks showing the locations of the FOXP1 ChIP-seq peaks in the proximity of the TSS of RASSF6, AIM2, and BIK, some of the proapoptotic genes that are downregulated by FOXP1 in primary human B cells and DLBCL cell lines.
Chromatin immunoprecipitation and sequencing analysis of FOXP1 target genes. To identify direct FOXP1 target genes, we conducted genome-wide mapping of the FOXP1 binding sites in 4 DLBCL cell lines, by chromatin immunoprecipitation with a FOXP1-specific antibody followed by high-throughput sequencing (ChIP-seq). (A) A Venn diagram showing the number of genes in each cell line with a FOXP1 binding peak within 20 kb of the TSS, as determined by ChIP-seq. (B) Percentage of all expressed genes or FOXP1-regulated genes (as determined by microarray analysis) in a cell line that showed a FOXP1 binding peak within 2 kb or 20 kb of the TSS, as determined by ChIP-seq (left), and the percentage of genes being either up- or downregulated, among the genes that are both regulated and bound by FOXP1 within 20 kb of the TSS (right). (C) De novo motif analysis of FOXP1 ChIP-seq peaks in OCI-Ly3 reveals the presence of several enriched motifs in the FOXP1-binding regions. Relative enrichment to control regions and percentage of peaks containing the motif are shown. (D) Overlap of IRF4 ChIP-seq peaks and SpiB ChIP-seq peaks in HBL138 with FOXP1 ChIP-seq peaks in DLBCL cell lines. (E) The number of FOXP1 ChIP-seq peaks that were found exclusively either in both ABC-DLBCL cell lines (OCI-Ly3 and OCI-Ly10) or in both GC-DLBCL cell lines (OCI-Ly1 and OCI-Ly7) and the proportion of these peaks that were also found among IRF4 or SpiB ChIP-seq peaks in HBL138 or that contained a consensus NF-κB binding site. (***P < .001 significant difference between presence of SpiB or IRF4 peaks or a NF-κB consensus motif among ABC-specific vs GC-specific FOXP1 CHIP-seq peaks as determined by χ2 test). (F) Tracks showing the locations of the FOXP1 ChIP-seq peaks in the proximity of the TSS of RASSF6, AIM2, and BIK, some of the proapoptotic genes that are downregulated by FOXP1 in primary human B cells and DLBCL cell lines.
About half of the genes that were regulated by FOXP1 in our microarray analysis also have a FOXP1 ChIP-seq peak within 20 kb from their TSS (Figure 2B, left panel). In accordance with previous studies on the effects of FOXP1,5,33,36 these direct FOXP1 targets included both positive and negative regulated genes (Figure 2B, right panel).
De novo motif search verified the presence of a forkhead binding motif in the majority of the peaks (Figure 2C and supplemental Figure 2B-C). Interestingly, binding motifs for various other transcription factors, including IRF and ETS motifs, were also significantly enriched among FOXP1-bound regions (Figure 2C and supplemental Figure 2B). At these regions, FOXP1 appears to directly bind to DNA via its own consensus motif, because >90% of these peaks also contain a consensus forkhead motif. Furthermore, high overlap was found between FOXP1 binding sites and previously defined binding sites for the ABC-DLBCL oncogenes IRF4 and the ETS factor SpiB (identified in HBL1)38 (Figure 2D-E). Notably, FOXP1 peaks exclusively detected in ABC-type DLBCL cell lines vs GC-type DLBCL cell lines were significantly enriched for IRF4- and SpiB-bound regions and for NF-κB consensus motifs (Figure 2E). Because high expression of IRF4 and SpiB as well as high NF-κB activity are hallmarks of the ABC- but not GC-type DLBCL, these findings suggest that FOXP1, by analogy to its family member FOXP3,39,40 might preferentially bind sites cooccupied by other transcription factors and regulate gene expression through physical or functional interaction with those factors.
In compliance with the results of our microarray study, GO analysis of the FOXP1-bound genes showed enrichment for genes involved in cell death and apoptosis across all 4 cell lines, irrespective of ABC or GCB subtype (supplemental Figure 2D). The top 10 GO terms based upon genes exclusively bound by FOXP1 in both ABC-type DLBCL cell lines are dominated by terms related to regulation of chemotaxis, immune response, and cell/lymphocyte activation, and the top 10 of both GC-type DLBCL cell lines are dominated by terms related to the regulation of cell cycle, translational initiation, and chromosome/nucleosome organization (supplemental Table 2). Importantly, all proapoptotic genes repressed by FOXP1 in primary B cells and cell lines (Figure 1C) were also bound by FOXP1 in the vicinity of their TSS in at least 2 cell lines (Figure 2F and supplemental Figure 2E-F). Combined, our GEM and ChIP-seq results clearly demonstrate that FOXP1 directly represses expression of multiple proapoptotic genes, strongly suggesting a role for FOXP1 in B-cell survival.
FOXP1 induces expansion of primary human B cells by repression of caspase-dependent apoptosis, without affecting proliferation
To address the functional consequence of FOXP1 expression, we employed a culture system that allows for manipulation of human primary B cells ex vivo. Primary B cells cultured in vitro will undergo spontaneous apoptosis; however, this cell death can be partially overcome by CD40 ligation, which results in B-cell expansion by activating multiple pathways including the NF-κB pathway.41-43 This expansion can be further enhanced by addition of cytokines such as IL-21 and IL-2.27,42 However, B cells cultured under these conditions only have a limited replication potential, and overexpression of antiapoptotic proteins or transcription factors, such as BCL6, is required to further extend their lifespan, by promoting survival or proliferation and/or by repressing differentiation.26,29
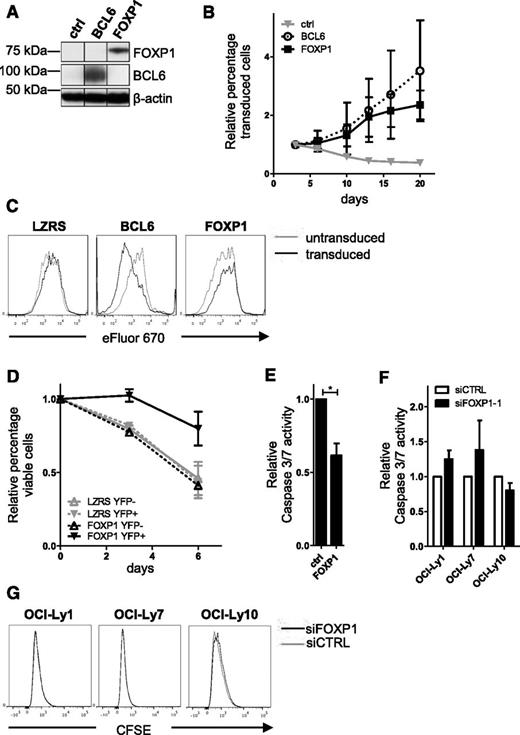
To investigate if aberrant FOXP1 expression could promote expansion of primary human B cells, we retrovirally transduced primary human B cells with FOXP1-IRES-YFP, control-IRES-YFP (as a negative control), or BCL6-IRES-GFP (as a positive control) (Figure 3A). The transduced B cells were cultured on L cells expressing CD40-ligand (CD40L-L cells), in the presence of IL-21 and IL-2. In compliance with previous studies,26,29 the percentage of GFP-positive cells in BCL6-transduced cultures rapidly increased with time (Figure 3B). Interestingly, a similar increase in the percentage of YFP-positive cells was observed in cultures transduced with FOXP1 (Figure 3B). These B-cell cultures were polyclonal as established by PCR-based GeneScan analysis of IgVH and IgVκ gene segments (data not shown).44
FOXP1 promotes expansion of primary human B cells, not by stimulating proliferation but by repressing apoptosis. Memory B cells were sorted from human peripheral blood and transduced with FOXP1-IRES-YFP, BCL6-IRES-GFP, or ctrl-IRES-YFP and cultured with CD40L-L cells, IL-21, and IL-2. (A) Representative example of FOXP1 and BCL6 overexpression in primary YFP+ or GFP+ B cells. Six days after transduction, YFP+ cells were sorted and analyzed by immunoblotting. β-Actin was used as loading control. (B) FOXP1-IRES-YFP, BCL6-IRES-GFP, and ctrl-IRES-YFP transduced B cells were continuously cultured with IL-21 and IL-2 and CD40L-L cells. The percentage of transduced cells in each culture was followed over time by FACS analysis and normalized to the percentage of transduced cells at day 3 after transduction. Mean ± standard deviation (SD) of 3 independent experiments are shown. (C) Seven days after transduction, cells were labeled with eFluor 670 and cultured for 4 days, after which the eFluor 670 intensity was determined by flow cytometry. Representative graphs of 3 independent experiments are shown. (D) A total of 6 to 7 days after transduction, live YFP-positive and YFP-negative fractions of FOXP1 and control vector single-transduced cultures were sorted. After a recovery period of 4 to 5 days, the percentage of cells in the forward-scatter/side-scatter live gate was determined by flow cytometry at 3 consecutive time points. The data were normalized to the percentage of living cells measured at the first time point. Mean ± standard error or the mean (SEM) of 2 independent experiments are shown. (E) A total of 6 to 7 days after transduction, the YFP-positive fractions of FOXP1- and control vector–transduced cultures were sorted. After a recovery period of 5 to 7 days, caspase-3/7 activity was determined by the Caspase-Glo 3/7 assay. Values were corrected for number of living cells as determined by FACS analysis. Mean ± SD of 4 independent experiments are shown (Student t test, *P < .05). (F-G) DLBCL cell lines were nucleofected with control siRNA or siRNA against FOXP1. (F) Four days after nucleofection, caspase-3/7 activity was determined by the Caspase-Glo 3/7 assay. Values were corrected for number of living cells as determined by FACS analysis. Mean ± SEM of 2 independent experiments are shown. (G) Cell lines were labeled with CFSE 1 day before nucleofection. Three days after nucleofection, CFSE intensity was determined by flow cytometry. Representative graphs of 2 independent experiments are shown. Efficient knockdown in these experiments was validated by qRT-PCR analysis.
FOXP1 promotes expansion of primary human B cells, not by stimulating proliferation but by repressing apoptosis. Memory B cells were sorted from human peripheral blood and transduced with FOXP1-IRES-YFP, BCL6-IRES-GFP, or ctrl-IRES-YFP and cultured with CD40L-L cells, IL-21, and IL-2. (A) Representative example of FOXP1 and BCL6 overexpression in primary YFP+ or GFP+ B cells. Six days after transduction, YFP+ cells were sorted and analyzed by immunoblotting. β-Actin was used as loading control. (B) FOXP1-IRES-YFP, BCL6-IRES-GFP, and ctrl-IRES-YFP transduced B cells were continuously cultured with IL-21 and IL-2 and CD40L-L cells. The percentage of transduced cells in each culture was followed over time by FACS analysis and normalized to the percentage of transduced cells at day 3 after transduction. Mean ± standard deviation (SD) of 3 independent experiments are shown. (C) Seven days after transduction, cells were labeled with eFluor 670 and cultured for 4 days, after which the eFluor 670 intensity was determined by flow cytometry. Representative graphs of 3 independent experiments are shown. (D) A total of 6 to 7 days after transduction, live YFP-positive and YFP-negative fractions of FOXP1 and control vector single-transduced cultures were sorted. After a recovery period of 4 to 5 days, the percentage of cells in the forward-scatter/side-scatter live gate was determined by flow cytometry at 3 consecutive time points. The data were normalized to the percentage of living cells measured at the first time point. Mean ± standard error or the mean (SEM) of 2 independent experiments are shown. (E) A total of 6 to 7 days after transduction, the YFP-positive fractions of FOXP1- and control vector–transduced cultures were sorted. After a recovery period of 5 to 7 days, caspase-3/7 activity was determined by the Caspase-Glo 3/7 assay. Values were corrected for number of living cells as determined by FACS analysis. Mean ± SD of 4 independent experiments are shown (Student t test, *P < .05). (F-G) DLBCL cell lines were nucleofected with control siRNA or siRNA against FOXP1. (F) Four days after nucleofection, caspase-3/7 activity was determined by the Caspase-Glo 3/7 assay. Values were corrected for number of living cells as determined by FACS analysis. Mean ± SEM of 2 independent experiments are shown. (G) Cell lines were labeled with CFSE 1 day before nucleofection. Three days after nucleofection, CFSE intensity was determined by flow cytometry. Representative graphs of 2 independent experiments are shown. Efficient knockdown in these experiments was validated by qRT-PCR analysis.
Next, we analyzed proliferation in these cultures by labeling the cells with the proliferation dye eFluor 670 and measuring dilution of the staining in the YPF+ and YFP− populations after 4 days of culturing, by fluorescence-activated cell sorter (FACS) analysis. eFluor 670 dilution was not increased in FOXP1-transduced cells (Figure 3C). In accordance, cell-cycle analysis indicated no major changes in cell-cycle distribution upon FOXP1 overexpression (supplemental Figure 3). BCL6 overexpression, on the other hand, did result in increased eFluor 670 dilution and a higher percentage of cells in S phase (Figures 3C and supplemental Figure 3). Combined, these results clearly establish that FOXP1 overexpression does not affect cell proliferations in primary human B cells cultured under these conditions.
To assess whether FOXP1 promotes B-cell survival, viable cells of the YFP-positive and YFP-negative fractions of FOXP1- or empty vector–transduced cells were sorted, and the percentage of live cells was monitored by flow cytometry during 1 week of subsequent culturing. FOXP1-transduced cells showed a clear survival benefit compared with YFP-negative– or empty vector–transduced cells (Figure 3D). To investigate whether this was mediated by inhibition of caspase-dependent apoptosis, a Caspase-Glo 3/7 assay, which measures the activity of caspase-3 and caspase-7, was performed on sorted YFP+ cells that had been cultured for 5 to 7 days after sorting. Caspase-3/7 activity was significantly reduced in cells transduced with FOXP1 (Figure 3E). Notably, in the DLBCL cell lines, siRNA-mediated silencing of FOXP1 did not affect cell proliferation, viability, or caspase-3/7 activity (Figure 3F-G and supplemental Figure 6; see also “Discussion”). Together, our results indicate that FOXP1 promotes primary B-cell survival (at least in part) by inhibiting caspase-dependent apoptosis.
FOXP1 is dependent upon NF-κB activation and cooperates with constitutive NF-κB activity for promotion of B-cell expansion and survival
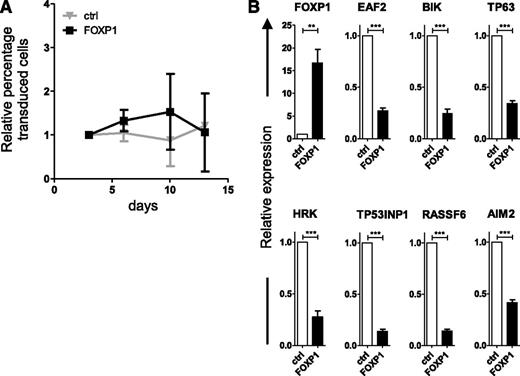
Interestingly, if FOXP1-transduced primary B cells were deprived of CD40L stimulation, the FOXP1-driven outgrowth was no longer observed (Figure 4A). Likewise, BCL6-driven outgrowth was no longer observed in the absence of CD40L. BCL6 expression resulted in a selective growth disadvantage, which could not be overcome by coexpression of FOXP1 (supplemental Figure 4). Notably, in the absence of CD40L, FOXP1 overexpression still resulted in repression of the proapoptotic genes identified by GEM and ChIP-seq (Figure 4B). Thus, aberrant expression of FOXP1 provides primary human B cells with a survival advantage; however, in order to benefit from this advantage, these cells are dependent upon signals provided by the CD40L-L cells.
FOXP1 is dependent upon CD40 stimulation for promotion of human B-cell expansion but not for repression of proapoptotic genes. Memory B cells were sorted from human peripheral blood and transduced with either FOXP1-IRES-YFP or control-IRES-YFP and cultured without CD40L-L cells as of day 3 after transduction. (A) The percentage of transduced cells in each culture was followed over time by FACS analysis and normalized to the percentage of transduced cells at day 3 after transduction. Mean ± SD of 3 independent experiments are shown. (B) Six days after transduction, YFP-positive cells were sorted. Gene expression levels of the proapoptotic genes were analyzed by qRT-PCR. Expression levels were normalized to expression levels in control-transduced cells. Mean ± SEM of six independent experiments are shown. (1-sample t test, **P < .01, ***P < .001).
FOXP1 is dependent upon CD40 stimulation for promotion of human B-cell expansion but not for repression of proapoptotic genes. Memory B cells were sorted from human peripheral blood and transduced with either FOXP1-IRES-YFP or control-IRES-YFP and cultured without CD40L-L cells as of day 3 after transduction. (A) The percentage of transduced cells in each culture was followed over time by FACS analysis and normalized to the percentage of transduced cells at day 3 after transduction. Mean ± SD of 3 independent experiments are shown. (B) Six days after transduction, YFP-positive cells were sorted. Gene expression levels of the proapoptotic genes were analyzed by qRT-PCR. Expression levels were normalized to expression levels in control-transduced cells. Mean ± SEM of six independent experiments are shown. (1-sample t test, **P < .01, ***P < .001).
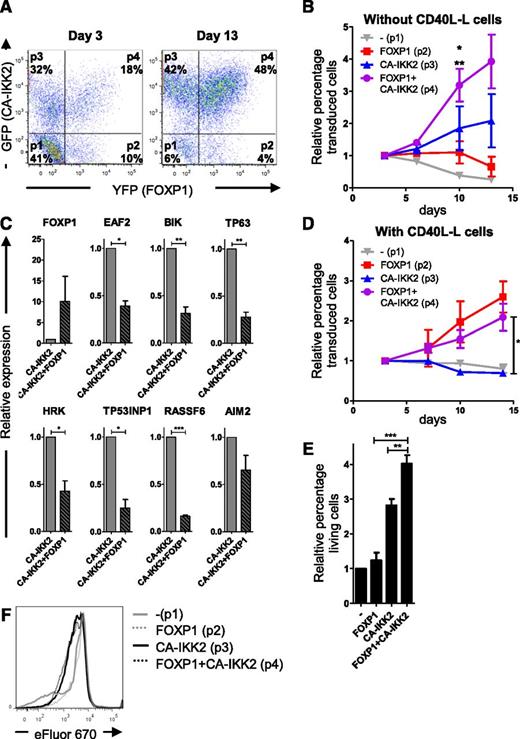
CD40 ligation activates several pathways, including the mitogen-activated protein kinase, phosphatidylinositol 3-kinase (PI3K), and NF-κB pathways.45,46 NF-κB activity plays a crucial role in normal B-cell differentiation, proliferation, and survival,47 and B-cell non-Hodgkin lymphomas in which FOXP1 is highly expressed are characterized by constitutive activation of the NF-κB pathway, which is critical for their survival.19-22 To investigate whether exclusive activation of the NF-κB pathway is sufficient to mimic the effect of CD40 activation, human primary B cells were cotransduced with FOXP1-IRES-YFP and CA-IKK2-IRES-GFP, which expresses a constitutively active mutant of IKK2 resulting in constitutive activation of the NF-κB-pathway,48 and cultured with IL-21 and IL-2. Based on YFP and GFP expression, 4 separate populations could be identified in these cultures by FACS analysis: nontransduced cells (p1), FOXP1 single-transduced cells (p2), CA-IKK2 single-transduced cells (p3), and FOXP1/CA-IKK2 double-transduced cells (p4), each typically constituting 10% to 40% of the total population (Figure 5A, left panel). In compliance with the ability of the NF-κB pathway to induce proliferation and inhibit apoptosis of in vitro cultured primary B cells,48,49 we observed a rapid increase in the percentage of CA-IKK2 single-transduced cells in time (Figure 5A [compare the left and right panels] and Figure 5B). In contrast, the proportion of the FOXP1 single-transduced population did not increase. Importantly, however, cells overexpressing both CA-IKK2 as well as FOXP1 showed a more pronounced expansion as compared with the CA-IKK2 single-transduced cells (Figure 5A-B). In line with this observation, expression of the proapoptotic FOXP1 target genes, as identified by GEM and ChIP-seq (Figures 1 and 2), was repressed in the FOXP1/CA-IKK2 double-transduced cells as compared with CA-IKK2 single-transduced cells (Figure 5C), whereas transduction with CA-IKK2 does not affect expression of these genes (supplemental Figure 5). Notably, CA-IKK2-transduced and CD40L-stimulated cells display enhanced expression of a gene signature of established NF-κB target genes (supplemental Figure 5), and when the cotransduced cells were cultured on CD40L-L cells, the growth-advantage of the CA-IKK2 single-transduced population over nontransduced cells was lost (Figure 5D), indicating that NF-κB is already optimally activated by the CD40L-L cells. In accordance, the relative percentages of the FOXP1 single- and FOXP1/CA-IKK double-transduced populations showed a comparable increase over time under these conditions (Figure 5D).
FOXP1 is dependent upon and cooperates with (constitutive) NF-κB activity to promote expansion and survival of human B cells. Memory B cells were sorted from human peripheral blood and cotransduced with FOXP1-IRES-YFP and CA-IKK2-IRES-GFP. Transduced B cells were cultured with IL-21, IL-2, and CD40L-L cells for the first 3 days and subsequently with IL-21 and IL-2, either in the absence (A-C,E-F) or presence (D) of CD40L-L cells. (A) Cells were analyzed by flow cytometry, 3 (left) and 13 (right) days after transduction. Four populations can be identified: YFP single positive (FOXP1 overexpression; p2), GFP single positive (CA-IKK2 overexpression; p3), YFP/GFP double positive (FOXP1 and CA-IKK2 overexpression; p4), and double negative (p1). (B,D) The percentages of the 4 populations within a single unsorted culture, cultured with IL-21 and IL-2 only (B), or with IL21, IL-2, and CD40L-L cells (D), were followed over time by FACS analysis and normalized to the percentage of each population at day 3 after transduction. Mean ± SEM of 3 independent experiments are shown. (B) Significant differences in relative expansion were observed at day 10 after transduction between the FOXP1+CA-IKK2 double-transduced population vs the CA-IKK2 single-transduced population (*P < .05) and the FOXP1 single-transduced population (**P < .01) by repeated-measures analysis of variance (ANOVA) followed by Bonferroni’s multiple comparison test. (D) Significant difference in relative expansion was observed between the FOXP1+CA-IKK2 double-transduced population vs the CA-IKK2 single-transduced population (*P < .05) by repeated-measures ANOVA followed by Bonferroni’s multiple comparison test. (C) Six days after transduction, GFP single-positive and GFP/YFP double-positive cells, cultured in the absence of CD40L (as in panel B), were sorted. Gene expression levels of the proapoptotic genes were analyzed by qRT-PCR. Expression levels were normalized to expression levels in CA-IKK2 single-transduced cells. Mean ± SEM of 3 independent experiments are shown. (1-sample t test, *P < .05, **P < .01, ***P < .001). (E) Six days after transduction, live cells of the 4 separate populations of cotransduced cells were sorted and cultured for 4 to 5 more days, after which the percentage of cells in the forward-scatter/side-scatter live gate was determined by flow cytometry and normalized to the percentage of living cells in the nontransduced population. Mean ± SEM of 3 independent experiments are shown (repeated-measures ANOVA followed by Bonferroni’s multiple comparison test; **P < .01, ***P < .001). (F) Seven days after transduction, cells were labeled with eFluor 670 and cultured for 4 days, after which the eFluor 670 intensity was determined by flow cytometry. Representative graphs of 2 independent experiments are shown.
FOXP1 is dependent upon and cooperates with (constitutive) NF-κB activity to promote expansion and survival of human B cells. Memory B cells were sorted from human peripheral blood and cotransduced with FOXP1-IRES-YFP and CA-IKK2-IRES-GFP. Transduced B cells were cultured with IL-21, IL-2, and CD40L-L cells for the first 3 days and subsequently with IL-21 and IL-2, either in the absence (A-C,E-F) or presence (D) of CD40L-L cells. (A) Cells were analyzed by flow cytometry, 3 (left) and 13 (right) days after transduction. Four populations can be identified: YFP single positive (FOXP1 overexpression; p2), GFP single positive (CA-IKK2 overexpression; p3), YFP/GFP double positive (FOXP1 and CA-IKK2 overexpression; p4), and double negative (p1). (B,D) The percentages of the 4 populations within a single unsorted culture, cultured with IL-21 and IL-2 only (B), or with IL21, IL-2, and CD40L-L cells (D), were followed over time by FACS analysis and normalized to the percentage of each population at day 3 after transduction. Mean ± SEM of 3 independent experiments are shown. (B) Significant differences in relative expansion were observed at day 10 after transduction between the FOXP1+CA-IKK2 double-transduced population vs the CA-IKK2 single-transduced population (*P < .05) and the FOXP1 single-transduced population (**P < .01) by repeated-measures analysis of variance (ANOVA) followed by Bonferroni’s multiple comparison test. (D) Significant difference in relative expansion was observed between the FOXP1+CA-IKK2 double-transduced population vs the CA-IKK2 single-transduced population (*P < .05) by repeated-measures ANOVA followed by Bonferroni’s multiple comparison test. (C) Six days after transduction, GFP single-positive and GFP/YFP double-positive cells, cultured in the absence of CD40L (as in panel B), were sorted. Gene expression levels of the proapoptotic genes were analyzed by qRT-PCR. Expression levels were normalized to expression levels in CA-IKK2 single-transduced cells. Mean ± SEM of 3 independent experiments are shown. (1-sample t test, *P < .05, **P < .01, ***P < .001). (E) Six days after transduction, live cells of the 4 separate populations of cotransduced cells were sorted and cultured for 4 to 5 more days, after which the percentage of cells in the forward-scatter/side-scatter live gate was determined by flow cytometry and normalized to the percentage of living cells in the nontransduced population. Mean ± SEM of 3 independent experiments are shown (repeated-measures ANOVA followed by Bonferroni’s multiple comparison test; **P < .01, ***P < .001). (F) Seven days after transduction, cells were labeled with eFluor 670 and cultured for 4 days, after which the eFluor 670 intensity was determined by flow cytometry. Representative graphs of 2 independent experiments are shown.
To establish if FOXP1 elicits its potentiating effect on NF-κB–induced B-cell expansion by a similar mechanism as observed in CD40L-stimulated cells, ie, by specifically promoting B-cell survival, viable cells of each of the 4 populations were FACS sorted and cultured for 5 days on cytokines only. Subsequently, the percentage of live cells was determined by flow cytometry (Figure 5E). Whereas the percentage of viable cells was not affected in FOXP1 single-transduced cultures, it increased in cultures transduced with CA-IKK2 only. Moreover, the percentage of viable cells further increased in cultures in which CA-IKK2 and FOXP1 were coexpressed. eFluor 670 staining of FOXP1/CA-IKK2 cotransduced cells indicated that whereas proliferation was clearly increased in the CA-IKK2 single-transduced population, confirming the role of NF-κB activation in B-cell proliferation, no further dilution of eFluor staining was observed in the CA-IKK2/FOXP1 double-transduced population (Figure 5F). Together, these results demonstrate that whereas FOXP1 has the capacity to repress expression of the proapoptotic genes independent of NF-κB activation, it is dependent upon and cooperates with (constitutive) NF-κB activity for the promotion of human primary B-cell expansion by further enhancing B-cell survival.
Discussion
In our study, GEM of primary B cells and DLBCL cell lines in which we overexpressed or silenced FOXP1, combined with ChIP-seq analysis, revealed that FOXP1 directly represses multiple genes encoding proapoptotic proteins, including BIK, HRK, EAF2, RASSF6, TP63, TP53INP1, and AIM2 (Figures 1C, 2F and supplemental Figure 2E-F). The proapoptotic BH3-only protein BIK is involved in spontaneous apoptosis of primary human GC B cells in vitro50,51 and in apoptosis of B-cell lymphoma cell lines.52,53 Interestingly, BIK has already been identified as a FOXP1 target gene in 2 other cellular systems using ChIP-seq technology.33,36 Van Boxtel et al reported that FOXP1 prevents FOXO-induced cell death in a colon carcinoma cell line by directly modulating expression of a specific subset of FOXO target genes, including BIK.33 This mode of transcriptional regulation by FOXP1, ie, through repression of FOXO target genes, has also been described in naive T cells, in which FOXP1 antagonizes FOXO1-induced expression of IL7RA.2 Because repression of FOXO1 activity through the PI3K pathway plays an important role in survival of mature resting B cells and in a subset of DLBCLs,54,55 the antiapoptotic role of FOXP1 in B cells may also be (partially) mediated through inhibition of FOXO target genes. In line with this, HRK, another proapoptotic BH3-only protein we found to be repressed by FOXP1, is also repressed by the PI3K pathway in a subset of DLBCLs, which is important for their survival.55 However, FOXP1 most likely also affects B-cell survival via other mechanisms, because the other proapoptotic FOXP1 target genes are not established FOXO targets.
The role of the other FOXP1-repressed proapoptotic genes in survival of B cells and B-cell lymphomas has not been extensively studied. However, given the established role of the tumor protein p53 in B-cell differentiation and lymphomagenesis,56 it is intriguing that 3 of the FOXP1-repressed proapoptotic genes (ie, encoding TP63, TP53INP1, and RASSF6) are tumor suppressors that share the capacity to enhance p53 activity.57-59 Apart from that, p63 expression has been correlated to DLBCL prognosis,60,61 expression of TP53INP1 is low in MALT lymphomas,62 and methylation of the RASSF6 promoter has been observed in B-cell acute lymphoid leukemia and chronic lymphoid leukemia.63,64 Although further studies are needed to determine if and how these proapoptotic genes control B-cell survival and lymphomagenesis, combined the 7 FOXP1-repressed proapoptotic genes clearly have prognostic significance for overall and progression-free survival of DLBCL patients (Figure 1D); the patients with low expression of the gene signature always have a worse prognosis, which is completely in line with their proapoptotic function and with a potential role in lymphomagenesis.
In accordance with previous literature,26,29 ectopic overexpression of BCL6 in mature B cells, cultured with CD40L-L cells and cytokines, results in selective outgrowth of transduced cells. Remarkably, whereas FOXP1-transduced cells cultured in the same conditions displayed very similar growth dynamics (Figure 3), the mechanism of action is totally different: whereas FOXP1 exclusively promotes B-cell survival, not proliferation, BCL6 overexpression enhanced cell proliferation, but not survival (Figure 3 and data not shown). The latter is in line with the results of Kuo et al, who showed that transduction of in vitro cultured human mature B cells with BCL6 did not affect B-cell survival.65 Interestingly, Craig et al recently reported that siRNA-mediated silencing of FOXP1 in DLBCL cell lines U2932, SUDHL-4, SUDHL-6, and SUDHL-7 reduced their proliferation.15 However, we did not observe impaired proliferation or survival upon knockdown of FOXP1 in various DLBCL cell lines, including U2932 and SUDHL-6, also not if combined with reduced serum conditions or NF-κB inhibition, or with the same pool of siRNAs used in the study of Craig et al (supplemental Figure 6). This may be the consequence of somewhat lower knockdown efficiencies or different type of proliferation assays used in our studies (supplemental Figure 6), although, as such, additional genetic changes during lymphomagenesis, ongoing mutation ex vivo, and/or extensive (clonal) selection may also render cell lines less dependent upon a single gene for growth and survival. Conversely, overexpression of FOXP1 in GC-DLBCL cell lines with low endogenous FOXP1 expression (supplemental Figure 7), either alone or in combination with CA-IKK2, did not have a positive effect on selective outgrowth (supplemental Figure 8); rather, we reproducibly observed a selective growth disadvantage, suggesting the high levels of FOXP1 expression may be toxic to these cells.
Activation of the NF-κB pathway in our culture system increased B-cell expansion by stimulating both B-cell proliferation and survival (Figure 5B,E-F). Simultaneous overexpression of FOXP1 further enhanced B-cell survival and reduced caspase-3/7 activity but did not affect proliferation (Figures 3 and 5). Importantly, in the absence of (constitutive) NF-κB activation, FOXP1 overexpression did not promote survival of primary B cells (Figures 4 and 5), but FOXP1 still repressed its proapoptotic target genes (Figure 4B). In accordance, FOXP1-mediated repression of these genes was observed not only in the ABC-DLBCL cell-line OCI-LY10, which carries NF-κB–activating CD79a and MyD88 gain-of-function mutations20,66 but also in the GC-DLBCL cell lines OCI-LY1 and LY7, which are not characterized by NF-κB activation (Figure 1C). This indicates that whereas repression of these genes by FOXP1 might be required for the prosurvival effect of FOXP1, their repression is not sufficient to promote survival of primary human B cells, because activation of the NF-κB pathway is required as well. Conversely, although it is well established that NF-κB activation and the subsequent upregulation of various antiapoptotic NF-κB targets plays a major role in B-cell survival,47,67,68 activation of NF-κB alone is apparently not sufficient to accomplish an optimal survival response (Figure 5). Thus, corroborated by the observation that aberrant expression of FOXP1 typically occurs in lymphomas that carry mutations resulting in constitutive NF-κB activity, our results indicate that combining NF-κB–mediated induction of antiapoptotic genes with FOXP1-mediated repression of proapoptotic genes is functionally complementary and provides a highly efficacious combination for optimal B-cell survival and lymphomagenesis.
Taken together, we have shown that FOXP1 directly represses transcription of various proapoptotic genes and cooperates with CD40 and NF-κB signaling in promoting B-cell expansion by inhibition of caspase-dependent apoptosis. Apart from providing novel insight into the role of FOXP1 in B-cell homeostasis, our results indicate that aberrant high FOXP1 expression, as a result of chromosomal translocation t(3;14), trisomy 3, or by other means, contributes to lymphomagenesis by promoting sustained B-cell survival. Aberrant expression of FOXP1 is typical for ABC-DLBCL and MALT lymphoma, which are characterized by, and are dependent upon, constitutive NF-κB activation. Therefore, from a diagnostic and therapeutic perspective, it is of major importance that the FOXP1-promoted B-cell survival and expansion is NF-κB dependent: targeted therapy of lymphoma patients with agents that result in inhibition of NF-κB activity (eg, treatment with the clinically active BTK inhibitor ibrutinib20,69,70 or inhibitors of IRAK or MALT171-73 ) will also overcome the additional survival benefit due to aberrant FOXP1 expression.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Berend Hooibrink for FACS sorting; Richard Volckman for providing help with microarray analysis; Edwin Cuppen, Ewart de Bruijn, Nico Lansu, and Wensi Hao for deep sequencing; and Sander Boymans for mapping of the sequencing data.
This study was supported by a grant from the Dutch Cancer Society.
Authorship
Contribution: M.v.K. designed the research, performed experiments, analyzed data, designed figures, and wrote the manuscript; L.J.G., M.M., and R.v.B. performed experiments, analyzed data, and designed figures; J.K. analyzed array/survival data; P.J.C. interpreted data and cosupervised part of the study; S.T.P. cosupervised the study and reviewed the manuscript; and M.S. designed the research, supervised the study, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marcel Spaargaren, Department of Pathology, Academic Medical Center, University of Amsterdam, Meibergdreef 9, 1105 AZ Amsterdam, The Netherlands; e-mail: marcel.spaargaren@amc.uva.nl.
References
Author notes
S.T.P. and M.S. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal