To the editor:
Our understanding of acute lymphoblastic leukemia (ALL) has expanded tremendously in the past few years because of large-scale genomic studies.1,2 ALL is characterized by a relatively small number of genomic alterations that disproportionally affect hematopoietic transcriptional factors.1,3 ALL that arises from the T-cell lineage (T-ALL) vs B-cell lineage has distinctive immunophenotypes and unique genomic abnormalities.4,5 However, the vast majority of genome sequencing and subsequent mechanistic studies of ALL singularly focus on coding regions and the role of noncoding genomic aberrations in leukemogenesis remains mostly uncharacterized with few exceptions.6,7 There is growing evidence implicating noncoding genetic variation in human diseases, either by modulating transcriptional regulatory elements of protein-coding genes or by altering noncoding RNAs themselves.8-11 Somatically acquired promoter and/or enhancer mutations have also been identified that profoundly change oncogene expression and drive tumorigenesis (eg, TAL1 enhancer mutation in T-ALL).6 In this study, we systematically searched for functional noncoding genomic alterations in T-ALL by whole-genome and transcriptome sequencing.
Whole-genome sequencing was performed in germ line and tumor samples from 31 Chinese children with T-ALL, 30 of whom were also profiled by transcriptome sequencing (see supplemental Materials and methods, available on the Blood Web site). On average, we observed 6.4 coding mutations per patient, with 9.9 and 3.2 copy number losses and gains, respectively. Consistent with previous reports,12 coding mutations were most frequently observed in NOTCH1, followed by FBXW7, USP7, and PTEN (supplemental Figure 1; supplemental Table 1).
The majority of sequence mutations arose in noncoding region of the genome (median, 979 per patient [range, 157 to 3143]; supplemental Table 2). We used 3 parallel approaches to systematically identify potential functional noncoding mutations: (1) “hotspot analysis” to identify tight clusters of mutations in small focal regions of the DNA (21 bp), (2) “regional recurrence analysis” to identify annotated regulatory regions (eg, promoter) with significant enrichment of noncoding mutations, and (3) “transcription factor analysis” to identify mutations that potentially result in gain or loss of transcription factor binding sites. Remarkably, 3 known T-ALL oncogenes, LMO1, LMO2, and TAL1, were among loci with the most significant enrichment for noncoding mutations (Figure 1A). When tested for their effects on the transcription of adjacent genes, only the TAL1 and LMO1 mutations were significantly associated with TAL1 and LMO1 expression, respectively, in support of cis-regulating effects (Figure 1B-C; supplemental Figures 2 and 3). At the LMO1 locus, 3 patients (9.7%) showed an identical point mutation (chr11:8289481G>A) proximal to the transcription start site (supplemental Figure 4; supplemental Table 3) that resulted in the gain of a canonical MYB binding site (AACGG), with ∼120-fold increase in LMO1 transcription in an allele-specific fashion and no effect on other adjacent genes (supplemental Figures 5 and 6). Recurring LMO2 intronic mutations were identified in 4 patients, creating a potential binding site of the ETS factors (CATCC). Within the same intron, a fifth patient had concomitant short deletion and point mutation; however, these LMO2 noncoding mutations were not associated with LMO2 gene expression or alternative splicing (supplemental Figure 3). Of 15 cases with TAL1 overexpression, 11 were positive for the STIL-TAL1 fusion (Figure 1C) and 4 had a somatic insertion that created an MYB-mediated super enhancer (supplemental Figure 7), consistent with recent reports.6,13
Genome-wide analysis and comprehensively characterization of somatic non-coding regulatory mutations in T-ALL. (A) Hotspot analysis identified loci with significant enrichment of recurrent noncoding mutations (single nucleotide variants and/or short insertion/deletions). Statistical significance (−log10 [P value], y-axis) is plotted against the respective chromosomal position of each mutation hotspot (x-axis). Dashed horizontal line, genome-wide significance threshold (P = 5 × 10−8); open circles, mutations significantly associated with expression of adjacent genes (TAL1, LMO1); dots below y = 1, sites with only 1 mutation. (B) Association of LMO1 enhancer mutation (11:8289481G>A) with LMO1 overexpression in the discovery cohort. FPKM, fragments per kilobase of transcript per million mapped reads. (C) Patients with TAL1 enhancer mutations (Mut) had the highest TAL1 expression compared with those with an intrachromosomal rearrangement (STIL-TAL1) or those with a wild-type (WT) genotype. (D) Among a panel of T-ALL cell lines, Jurkat cells were identified as having the LMO1 enhancer mutation and LMO1 overexpression. Gray and blue rectangles, T-ALL cases or cell lines with or without the LMO1 enhancer mutation, respectively. (E) All LMO1 enhancer mutation events were completely conserved across patients and created an MYB binding site. (F) In Jurkat cells, binding of MYB, CREBBP, and RUNX1 was detected by ChIP-seq, with the peak precisely superimposing the LMO1 enhancer mutation (red arrow). RNA polymerase II (RNAPII) binding, DNase I hypersensitivity, and H3K27 acetylation confirmed active transcription at this locus. ChIP-seq peak calling was performed using the MACS2 algorithm (enrichment P < 10−7). (G) Further analysis of the ChIP-seq reads of each allele indicates that transcription factor binding and transcription activation were specific to the mutant allele. ChIP-seq and DNase I hypersensitivity signals (y-axis) were normalized to reads per million reads sequenced in each sample. Statistical significance of the differences was estimated by using 2-sided Mann-Whitney-Wilcoxon test (B-C).
Genome-wide analysis and comprehensively characterization of somatic non-coding regulatory mutations in T-ALL. (A) Hotspot analysis identified loci with significant enrichment of recurrent noncoding mutations (single nucleotide variants and/or short insertion/deletions). Statistical significance (−log10 [P value], y-axis) is plotted against the respective chromosomal position of each mutation hotspot (x-axis). Dashed horizontal line, genome-wide significance threshold (P = 5 × 10−8); open circles, mutations significantly associated with expression of adjacent genes (TAL1, LMO1); dots below y = 1, sites with only 1 mutation. (B) Association of LMO1 enhancer mutation (11:8289481G>A) with LMO1 overexpression in the discovery cohort. FPKM, fragments per kilobase of transcript per million mapped reads. (C) Patients with TAL1 enhancer mutations (Mut) had the highest TAL1 expression compared with those with an intrachromosomal rearrangement (STIL-TAL1) or those with a wild-type (WT) genotype. (D) Among a panel of T-ALL cell lines, Jurkat cells were identified as having the LMO1 enhancer mutation and LMO1 overexpression. Gray and blue rectangles, T-ALL cases or cell lines with or without the LMO1 enhancer mutation, respectively. (E) All LMO1 enhancer mutation events were completely conserved across patients and created an MYB binding site. (F) In Jurkat cells, binding of MYB, CREBBP, and RUNX1 was detected by ChIP-seq, with the peak precisely superimposing the LMO1 enhancer mutation (red arrow). RNA polymerase II (RNAPII) binding, DNase I hypersensitivity, and H3K27 acetylation confirmed active transcription at this locus. ChIP-seq peak calling was performed using the MACS2 algorithm (enrichment P < 10−7). (G) Further analysis of the ChIP-seq reads of each allele indicates that transcription factor binding and transcription activation were specific to the mutant allele. ChIP-seq and DNase I hypersensitivity signals (y-axis) were normalized to reads per million reads sequenced in each sample. Statistical significance of the differences was estimated by using 2-sided Mann-Whitney-Wilcoxon test (B-C).
In the independent validation cohort of 26 children with T-ALL, LMO1 was minimally expressed in all but 3 patients, 2 of whom had the chr11:8289481 G>A enhancer mutation (supplemental Figures 8A and 9A). In the third patient, we identified an intrachromosomal inversion event that juxtaposed the active promoter of the MED17 gene with coding sequence of LMO1, leading to constitutive expression of LMO1 (supplemental Figures 8B-E, 9B, and 10). The LMO2 noncoding mutation was observed in 3 patients, but again was not significantly associated with LMO2 expression (supplemental Figure 11).
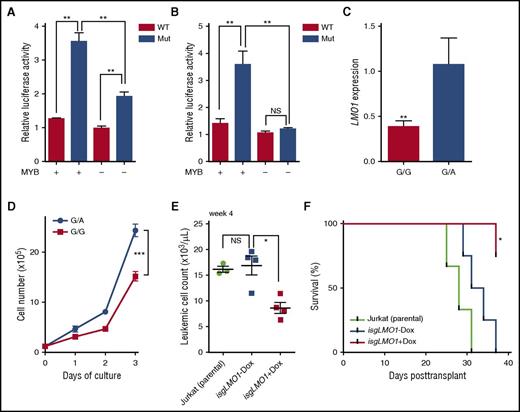
Screening a panel of T-ALL cell lines, we identified an LMO1 enhancer mutation in the Jurkat cells that also showed dramatic overexpression of LMO1 (Figure 1D-E; supplemental Figure 4). In normal human T (CD3+, CD4+, or CD8+) or T-ALL cells without the LMO1 enhancer mutation, there was a minimal H3K27me3, H3K4me3, and H3K27ac mark at this locus, suggesting an inactive chromatin state. In contrast, Jurkat cells showed a marked increase in H3K27ac, with the enhancer mutation situated precisely at the valley of 2 chromatin immunoprecipitation sequencing (ChIP-seq) peaks and DNase hypersensitivity (Figure 1F; supplemental Figures 12 and 13), suggesting transcription regulator occupancy and open chromatin structure related to the enhancer mutation. To test this hypothesis, we examined transcription factors previously implicated in MYB-mediated super enhancer activation in T-ALL (eg, MYB, RUNX1, GATA3, TAL1, CREBBP).6,14 As shown in Figure 1F, MYB binding was evident and superimposed the LMO1 enhancer mutation in Jurkat cells, along with enrichment of CREBBP, RUNX1, and RNA polymerase II. In fact, these transcription regulators were readily expressed in T-ALL patients and almost exclusively bound to the mutant allele, indicating that a transcription regulatory complex was formed as the result of the LMO1 enhancer mutation (Figure 1G; supplemental Figures 14 and 15). We also directly evaluated the effects of this LMO1 enhancer mutation on reporter gene transcription in vitro. In Jurkat cells, luciferase activity increased 2.8-fold when driven by the mutant LMO1 sequence compared with that from the wild-type sequence, but was significantly attenuated when MYB expression was downregulated by short interfering RNA-mediated knockdown (Figure 2A). In HEK293T cells, a nonhematopoietic cell line in which MYB is not expressed,15 LMO1 enhancer mutation activated luciferase transcription only when MYB was ectopically expressed, indicating that the activity of this LMO1 enhancer was specifically regulated by MYB (Figure 2B). To establish the causal relationship between the LMO1 enhancer mutation and LMO1 activation, we also converted the mutant allele at this site to the wild-type allele in Jurkat cells (ie, from G/A genotype to G/G genotype), using CRISPR/Cas9 technology. Engineered Jurkat cells with a wild-type genotype showed significantly lower LMO1 expression and a marked decrease in leukemia cell proliferation in vitro, compared with the parental cell line with LMO1 enhancer mutation (Figure 2C-D). To evaluate the effects of LMO1 enhancer mutation on leukemia progression in vivo, Jurkat cells with inducible CRISPR/Cas9 targeting LMO1 enhancer (isgLMO1) were implanted in immunodeficient mice, which were then subjected to a regular or doxycycline-supplemented diet. isgLMO1 leukemia progressed rapidly in vivo without doxycycline compared with parental Jurkat cells (Figure 2E). In contrast, there was a significantly lower disease burden and prolonged leukemia-free survival in mice bearing isgLMO1 Jurkat cells upon doxycycline treatment (Figure 2F).
LMO1 enhancer mutation analyses for transcription activity in vitro and leukemia progression in vivo. (A) Jurkat cells were transfected with a reporter gene construct with WT sequence or LMO1 enhancer mutation, and transcription activation was quantified by luciferase activity relative to Renilla. LMO1 mutant enhancer activity was significantly diminished upon downregulation of MYB by short interfering RNA (siRNA). (B) In HEK293T cells, LMO1 enhancer mutation drove luciferase transcription only when MYB was ectopically expressed. (C) LMO1 overexpression was abrogated upon conversion of the LMO1 mutant allele to the WT allele in Jurkat cells, using CRISPR/Cas9 genomic engineering. (D) Engineered Jurkat cells with WT genotype at the LMO1 enhancer locus also showed a significantly lower rate of proliferation compared with parental Jurkat cells. (E) Jurkat cells were transduced with lentiviral vectors encoding constitutive Cas9 and isgLMO1. Parental Jurkat or CRISPR/Cas9-engineered cells were injected into sublethally irradiated NOD.Cg-PrkdcscidII2rgtm1Wjl/SzJ recipient mice. Mice bearing genome-edited Jurkat cells were split into 2 groups: with vs without doxycycline (Dox) treatment (ie, with vs without LMO1 enhancer deletion). (F) Leukemia progress was monitored weekly by complete blood count and flow cytometry quantification of human CD45 cells in peripheral blood. Median survival for mice bearing parental cells or isgLMO1 cells without doxycycline treatment was 29.5 and 32.5 days, respectively, whereas doxycycline-treated mice bearing isgLMO1 cells remained alive at the end of this experiment. Statistical significance of the differences was estimated by using 2-sided Mann-Whitney-Wilcoxon test (A-C, E), 2-way analysis of variance (D), or log-rank test (F); *P < .05, **P < .01, and ***P < .001. NS, not significant.
LMO1 enhancer mutation analyses for transcription activity in vitro and leukemia progression in vivo. (A) Jurkat cells were transfected with a reporter gene construct with WT sequence or LMO1 enhancer mutation, and transcription activation was quantified by luciferase activity relative to Renilla. LMO1 mutant enhancer activity was significantly diminished upon downregulation of MYB by short interfering RNA (siRNA). (B) In HEK293T cells, LMO1 enhancer mutation drove luciferase transcription only when MYB was ectopically expressed. (C) LMO1 overexpression was abrogated upon conversion of the LMO1 mutant allele to the WT allele in Jurkat cells, using CRISPR/Cas9 genomic engineering. (D) Engineered Jurkat cells with WT genotype at the LMO1 enhancer locus also showed a significantly lower rate of proliferation compared with parental Jurkat cells. (E) Jurkat cells were transduced with lentiviral vectors encoding constitutive Cas9 and isgLMO1. Parental Jurkat or CRISPR/Cas9-engineered cells were injected into sublethally irradiated NOD.Cg-PrkdcscidII2rgtm1Wjl/SzJ recipient mice. Mice bearing genome-edited Jurkat cells were split into 2 groups: with vs without doxycycline (Dox) treatment (ie, with vs without LMO1 enhancer deletion). (F) Leukemia progress was monitored weekly by complete blood count and flow cytometry quantification of human CD45 cells in peripheral blood. Median survival for mice bearing parental cells or isgLMO1 cells without doxycycline treatment was 29.5 and 32.5 days, respectively, whereas doxycycline-treated mice bearing isgLMO1 cells remained alive at the end of this experiment. Statistical significance of the differences was estimated by using 2-sided Mann-Whitney-Wilcoxon test (A-C, E), 2-way analysis of variance (D), or log-rank test (F); *P < .05, **P < .01, and ***P < .001. NS, not significant.
TAL1, LMO1, and LMO2 encode transcription regulators that are tightly regulated during thymocyte differentiation,4,5,16-19 upregulation of which has been reported in patients with T-ALL and can result in leukemogenesis with varying latency in vivo.20,21 Synergistic interactions among these 3 oncogenes have been suggested to affect progenitor cell self-renewal,20 but we did not observe a clear pattern of cosegregation of TAL1, LMO1, and LMO2 noncoding mutations or overexpression. It is intriguing that genomic lesions at the TAL1 and LMO1 loci exclusively occur in the noncoding region and result in transcription deregulation, whereas somatic alterations in other T-ALL oncogenes largely involve coding mutations only (eg, NOTCH1). Such gene-dependent preference in a mutation profile is also observed in several other cancers,22 but the underlying biological mechanisms remain unclear. Compared with TAL1 enhancer mutations,6 noncoding mutation at the LMO1 locus in T-ALL displayed remarkable positional conservation. This highly restrictive pattern suggested a strong selection pressure during leukemogenesis, plausibly in favor of mutations causing the most significant upregulation of LMO1. The site of recurrent LMO1 enhancer mutation encompasses binding motifs for several T-ALL transcription regulators (MYB, CREBBP, and RUNX1) and may be uniquely poised for the formation of an enhancer complex, although this needs to be tested in primary samples in the future. In fact, this is also true with the TAL1 enhancer mutation, even though the exact composition of enhancer protein complex differs.6 Compared with TAL1 and LMO1 loci, the functional consequences of LMO2 intronic mutations are unclear. Compared with LMO2 enhancer mutations that have been reported recently,23,24 noncoding LMO2 mutations in our cohort were also restricted to the same hotspot (chr11:33903600-70), but none created an MYB-binding site and were not significantly associated with LMO2 expression. Given the oncogenic effects of LMO2 overexpression,4,16 we postulate that MYB-specific transcriptional activation is required for the pathogenic effects of noncoding variants at this locus.
The online version of this article contains a data supplement.
Authorship
Acknowledgments: The authors thank the patients and parents who participated in the clinical trials included in this study.
This work was supported by grants from the National Institutes of Health, National Cancer Institute (CA021765 and CA176063), the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital, the National Science Foundation of China (81370627 and 81170513), the Jiangsu Province Key Project (BL2013014), the National Medical Research Council in Singapore (NMRC/CSA/0053/2013) (A.E.-J.Y.), and the National Science Foundation of China (81300401) (H.Z.). H.Z. is a St. Baldrick’s International Scholar (grant 318318).
Contribution: S.H. and J.J.Y. jointly supervised research; C.-H.P., A.J.S., S.H., and J.J.Y. conceived and designed the experiments; H.Z., J.Y., X.Z., G.D., T.-N.L., and Y.C. performed the experiments; M.Q., Y.G., and H.Z. performed statistical analysis; M.Q., Y.G., and M.C. analyzed the data; H.H., J.L., J.P., C.L., S.K.-Y.K., T.C.Q., H.A., A.-M.T., A.E.-J.Y., A.J.S., and S.H. contributed to reagents/materials/analysis tools; and M.Q., H.Z., A.J.S., and J.J.Y. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jun J. Yang, 262 Danny Thomas Pl, MS313, Memphis, TN 38105; e-mail: jun.yang@stjude.org; and Shaoyan Hu, 303 Jingde Rd, Pingjiang Qu, Suzhou, Jiangsu, China 215003; e-mail: hsy139@126.com.
References
Author notes
S.H., M.Q., H.Z., and Y.G. contributed equally to this study.

![Figure 1. Genome-wide analysis and comprehensively characterization of somatic non-coding regulatory mutations in T-ALL. (A) Hotspot analysis identified loci with significant enrichment of recurrent noncoding mutations (single nucleotide variants and/or short insertion/deletions). Statistical significance (−log10 [P value], y-axis) is plotted against the respective chromosomal position of each mutation hotspot (x-axis). Dashed horizontal line, genome-wide significance threshold (P = 5 × 10−8); open circles, mutations significantly associated with expression of adjacent genes (TAL1, LMO1); dots below y = 1, sites with only 1 mutation. (B) Association of LMO1 enhancer mutation (11:8289481G>A) with LMO1 overexpression in the discovery cohort. FPKM, fragments per kilobase of transcript per million mapped reads. (C) Patients with TAL1 enhancer mutations (Mut) had the highest TAL1 expression compared with those with an intrachromosomal rearrangement (STIL-TAL1) or those with a wild-type (WT) genotype. (D) Among a panel of T-ALL cell lines, Jurkat cells were identified as having the LMO1 enhancer mutation and LMO1 overexpression. Gray and blue rectangles, T-ALL cases or cell lines with or without the LMO1 enhancer mutation, respectively. (E) All LMO1 enhancer mutation events were completely conserved across patients and created an MYB binding site. (F) In Jurkat cells, binding of MYB, CREBBP, and RUNX1 was detected by ChIP-seq, with the peak precisely superimposing the LMO1 enhancer mutation (red arrow). RNA polymerase II (RNAPII) binding, DNase I hypersensitivity, and H3K27 acetylation confirmed active transcription at this locus. ChIP-seq peak calling was performed using the MACS2 algorithm (enrichment P < 10−7). (G) Further analysis of the ChIP-seq reads of each allele indicates that transcription factor binding and transcription activation were specific to the mutant allele. ChIP-seq and DNase I hypersensitivity signals (y-axis) were normalized to reads per million reads sequenced in each sample. Statistical significance of the differences was estimated by using 2-sided Mann-Whitney-Wilcoxon test (B-C).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/24/10.1182_blood-2017-03-771162/4/m_blood771162f1.jpeg?Expires=1766316886&Signature=y7NMVo8OdhhSfyBTLFfMcHPu6zX4fq4MbI5SnujbQf0WyXuf1IgZGp4nM8lJFCcCziNN4UXd23uCFleu0ZTO6q3m~5K0XSQopmeS5TbxT190mXBDH9tpglK8Yjw8d0RlYyVv6Ja-arKGMkn07SZqBEh0JugI2lbMyAR1B1HlP3Vt3lElRDS4HvqsDbibjvXtZ6aZdnVjvMoF9y3GGTPA54vL8zx28NDTsZWpHnLnrwAjP3~c-WrUkpoRqS4el02FlyBZUaPv~hRpsnk6IWb4v5npjiWGY9joGlxEyZWBS7wPwqakdWHABuKsFOSzO2fd4gCSha7K2d2pWyFGqlTFMw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)