In this issue of Blood, Rahman et al1 and Hu et al2 identify somatic alterations targeting noncoding elements in the vicinity of LMO1 and LMO2 as novel mechanisms of oncogene activation in human T-cell acute lymphoblastic leukemia (T-ALL).
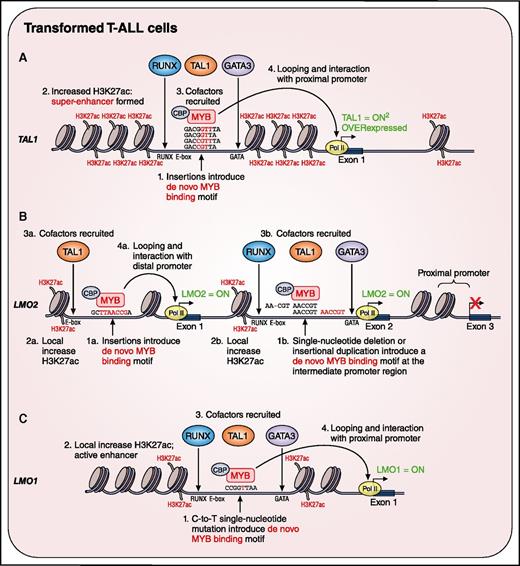
Somatic noncoding mutations create de novo transcription factor binding sites near the TAL1, LMO1, and LMO2 transcription oncogenes in T-ALL. (A) Insertions in the upstream noncoding elements of TAL1 create a de novo MYB binding site that leads to recruitment of coactivators and the formation of an H3K27ac-positive super-enhancer that drives oncogenic TAL1 expression. (B) Similarly, indels create de novo MYB binding sites at either (1) an upstream distal enhancer leading to distal promoter activation or (2) an intermediate promoter in intron 1, which both can drive LMO2 oncogene activation. (C) A single nucleotide mutation creates a de novo MYB binding motif in a distal enhancer element of the LMO1 proto-oncogene. CBP, CREB-binding protein. Professional illustration by Patrick Lane, ScEYEnce Studios.
Somatic noncoding mutations create de novo transcription factor binding sites near the TAL1, LMO1, and LMO2 transcription oncogenes in T-ALL. (A) Insertions in the upstream noncoding elements of TAL1 create a de novo MYB binding site that leads to recruitment of coactivators and the formation of an H3K27ac-positive super-enhancer that drives oncogenic TAL1 expression. (B) Similarly, indels create de novo MYB binding sites at either (1) an upstream distal enhancer leading to distal promoter activation or (2) an intermediate promoter in intron 1, which both can drive LMO2 oncogene activation. (C) A single nucleotide mutation creates a de novo MYB binding motif in a distal enhancer element of the LMO1 proto-oncogene. CBP, CREB-binding protein. Professional illustration by Patrick Lane, ScEYEnce Studios.
Over the last few years, genetic sequencing studies of human cancer have largely been focused on the coding part of the human genome. However, more recently, emerging evidence suggests that somatically acquired noncoding alterations could also be critically involved in tumor formation, as exemplified by the identification of CTCF/cohesin-binding site mutations in colorectal cancer3 or TERT promoter mutations in melanoma.4
In the context of T-ALL, an aggressive hematologic cancer characterized by aberrant activation of transcription factor oncogenes,5 2 landmark studies recently identified somatic insertions in a regulatory element upstream of the transcriptional start site (TSS) of the TAL1 proto-oncogene.6,7 Notably, these small noncoding alterations introduced a de novo recognition site for MYB,7 resulting in the generation of a somatically acquired super-enhancer that triggered aberrant monoallelic expression of TAL17 (see figure panel A). However, up until now, it remained largely unclear if this concept of somatically acquired enhancer activity would also be applicable for other transcription factor oncogenes involved in T-ALL disease biology.
LMO1 and LMO2 are critical regulators of hematopoietic differentiation and lineage commitment, and are part of a macromolecular transcriptional complex that consists of LIM domain binding proteins, GATA, ETS, and basic helix-loop-helix transcription factors.8 Under normal physiological conditions, LMO1 and LMO2 expression is strongly decreased during T-cell development shortly after lineage commitment.8 However, sustained or forced expression of these proteins in mice results in a gain of preleukemic thymocyte self renewal, allowing for clonal expansion and accumulation of secondary mutations, eventually leading to overt T-cell leukemia.9 Also, in human T-ALL, aberrant expression of LMO1 and/or LMO2 has been observed in a subset of patients as a result of chromosomal translocations or cryptic deletions targeting a negative regulatory element in the LMO2 locus,5 or creating loss of insulated neighborhood boundaries.10 Nevertheless, a number of clonal T-cell tumors still display aberrant LMO1 or LMO2 levels, even in the absence of such translocations or deletions, suggesting alternative mechanisms of oncogene activation in human T-ALL.
Indeed, in this issue, Rahman et al showed that T-ALL cell lines and patient samples that display aberrant and monoallelic LMO2 expression in the absence of a translocation or deletion at this locus, are characterized by an active intermediate promoter in the noncoding intron 1 of LMO2 (see figure panel B). Interestingly, sequencing analysis of this particular intron revealed the presence of recurrent somatic insertions and/or deletions (indels), creating de novo transcription factor binding sites for MYB, ETS1, or RUNX1 (see figure panel B). Given that these factors are all part of an oncogenic protein complex11 that also includes TAL1 and LMO2 itself, it is tempting to speculate that these newly identified noncoding mutations will ultimately trigger the formation of an autoregulatory loop that will further boost oncogenic transcription during malignant T-cell transformation.11 Only the intronic LMO2 mutations that created a de novo MYB binding motif were functionally validated. Nevertheless, the fact that specific mutations also created ETS1 or RUNX1 recognition sites in close proximity of a preexisting MYB motif suggests that cooperative but interchangeable activity of different transcription factors could contribute to oncogenic LMO2 activation. This seems to be different as compared with the MYB centered mechanism-of-action that was suggested for the previously identified TAL1 enhancer mutations.7 Interestingly, another study recently reported a very similar mechanism of LMO2 oncogene activation in T-ALL.12 However, in that case, the somatically acquired noncoding mutations, which also created a de novo MYB binding motif, were located further upstream of the LMO2 TSS and created a somatically acquired enhancer that triggered aberrant monoallelic expression of LMO212 (see figure panel B).
In a complementary study, also presented in this issue, Hu et al performed whole genome sequencing analysis on a cohort of 31 pediatric T-ALLs and confirmed the presence of the previously described TAL1 noncoding mutations,6,7 as well as the intronic LMO2 alterations mentioned earlier. However and most notably, their unbiased approach also enabled the identification of a recurrent point mutation in a noncoding area upstream of the TSS of the LMO1 proto-oncogene (see figure panel C). Similar to what has been described for TAL16,7 and LMO2, this somatically acquired single-nucleotide variant created a de novo MYB recognition site, triggering the formation of an aberrant transcriptional enhancer complex that drives high levels of monoallelic LMO1 expression (see figure panel C). Interestingly, this oncogenic enhancer mutation at the LMO1 locus might occur as part of an APOBEC-induced mutational signature, as recently described by Li et al.13
In conclusion, Rahman et al and Hu et al confirm that somatic alterations that drive aberrant activation of neomorphic promoters or enhancers14 can be critically involved in malignant T-cell transformation. These studies further complement our molecular genetic understanding of human T-ALL and reinforce the critical importance of the oncogenic TAL-LMO complex in T-ALL disease biology. In addition, it is tempting to speculate that other oncogenes involved in the biology of this disease, such as NOTCH1, MYC, TLX1, TLX3, HOXA, NKX2.1, or NKX2.2,5 could also be activated through similar mechanisms. In contrast, one could also imagine a scenario in which noncoding alterations disrupt certain transcription factor binding motifs, thereby causing inactivation of an enhancer in the proximity of a certain tumor suppressor gene. Altogether, these studies provide a strong rationale to further explore the landscape of somatic noncoding alterations in human cancer.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal