Key Points
Recurrent intronic mutations that create probable MYB, ETS1, and RUNX1 binding sites occur at the LMO2 promoter in some T-ALL patients.
CRISPR/Cas9-mediated disruption of the mutant MYB site in PF-382 cells markedly downregulates LMO2 expression.
Abstract
Somatic mutations within noncoding genomic regions that aberrantly activate oncogenes have remained poorly characterized. Here we describe recurrent activating intronic mutations of LMO2, a prominent oncogene in T-cell acute lymphoblastic leukemia (T-ALL). Heterozygous mutations were identified in PF-382 and DU.528 T-ALL cell lines in addition to 3.7% of pediatric (6 of 160) and 5.5% of adult (9 of 163) T-ALL patient samples. The majority of indels harbor putative de novo MYB, ETS1, or RUNX1 consensus binding sites. Analysis of 5′-capped RNA transcripts in mutant cell lines identified the usage of an intermediate promoter site, with consequential monoallelic LMO2 overexpression. CRISPR/Cas9-mediated disruption of the mutant allele in PF-382 cells markedly downregulated LMO2 expression, establishing clear causality between the mutation and oncogene dysregulation. Furthermore, the spectrum of CRISPR/Cas9-derived mutations provides important insights into the interconnected contributions of functional transcription factor binding. Finally, these mutations occur in the same intron as retroviral integration sites in gene therapy–induced T-ALL, suggesting that such events occur at preferential sites in the noncoding genome.
Introduction
LIM-domain-only protein 2 (LMO2) plays a crucial bridging role in the formation of a large multimeric transcriptional complex that includes TAL1, LDB1, GATA, RUNX1, ETS1, and MYB.1 In mice, Lmo2 is progressively silenced after the early T-cell progenitor (ETP) stage of thymic development and leads to T-cell acute lymphoblastic leukemia (T-ALL) when overexpressed in transgenic models.2-4 In human thymi, LMO2 is similarly downregulated after commitment to the T-cell lineage as indicated by DNA microarray analyses.5 Overexpression of LMO2 in human hematopoietic stem cells also leads exclusively to preleukemic alterations in thymocytes and T cells but not in other lineages.6 Reported mechanisms of aberrant LMO2 expression in human T-ALL include recurrent chromosomal translocations, such as t(11;14)(p13;q11) and t(7;11)(q35;p13); cryptic deletions of an upstream negative regulatory region, as in del(11)(p12p13); and retroviral insertional mutagenesis at the LMO2 locus during gene therapy.7-11 Although ∼50% of T-ALL patients overexpress LMO2, only about 10% of patients have a detectable cytogenetic lesion.12 Notably, many of these patients will overexpress LMO2 from a single allele, a feature reminiscent of TAL1 overexpressing T-ALL cases driven by small somatic indel mutations that create binding sites for MYB, thus generating a neomorphic enhancer.13,14 We hypothesized that cis-acting mechanisms may account for T-ALL cases with monoallelic LMO2 expression that lack abnormalities of the LMO2 locus.15,16
Study design
Detailed methods are described in the supplemental Data available on the Blood Web site. Chromatin immunoprecipitation sequencing (ChIP-seq) was performed on T-ALL cell lines after immunoprecipitation with antibodies against MYB and acetylated H3K27 (H3K27ac). Analysis of motif enrichment was used to confirm enrichment of MYB motifs in the MYB ChIP-seq data (supplemental Tables 1 and 2). LMO2 messenger RNA levels were quantified by quantitative reverse transcription polymerase chain reaction (qRT-PCR). Mutation screening of primary T-ALL samples was achieved by denaturing high-performance liquid chromatography of LMO2 intron 1 PCR products. Luciferase reporter constructs consisting of 469 bp PCR products inserted upstream of an SV40 promoter and firefly luciferase gene were electroporated into Jurkat cells. CRISPR/Cas9 genome editing was used to target the LMO2 intron 1 mutations in the PF-382 T-ALL cell line.
Results and discussion
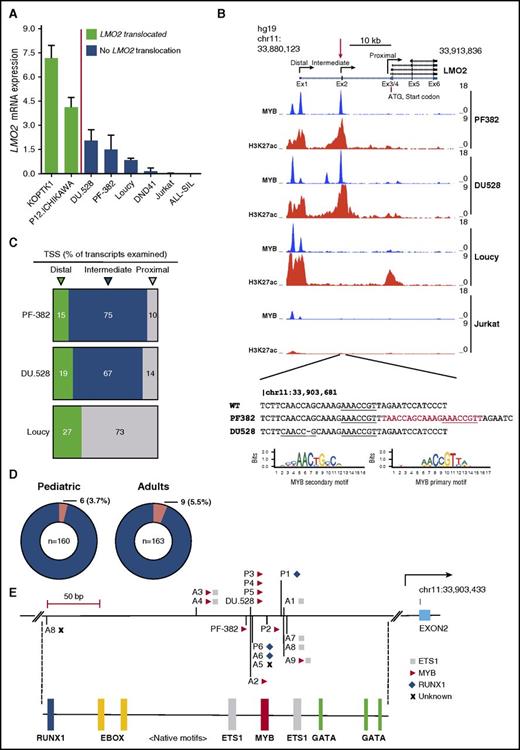
To test this hypothesis, we first assessed LMO2 expression by qRT-PCR in several T-ALL cell lines arrested at different stages of thymic differentiation. The ETP-like T-ALL cell line Loucy expressed LMO2 at levels significantly higher than the more mature T-ALL cell lines (DND-41, ALL-SIL, Jurkat), reflecting physiological expression of LMO2 at the ETP stage of thymic development (Figure 1A). The TAL1-positive cell lines DU.528 and PF-382 both exhibited upregulated LMO2 expression, yet crucially have no reported chromosomal lesions affecting this locus (Figure 1A).17,18 In contrast to Loucy cells, aberrant H3K27ac marks (indicative of active chromatin) were identified before and encompassing the noncoding exon 2 of the LMO2 gene by ChIP-seq in PF-382 and DU.528 T-ALL cell lines (Figure 1B; supplemental Figure 1). Sequencing across these peaks revealed a heterozygous 20 bp duplication in PF-382 cells and a heterozygous 1 bp deletion in DU.528 cells located close to a region recently described as an intermediate promoter for reasons that were not then apparent (Figure 1B).19 Notably, the mutations were not described as normal germ line variants in the Single Nucleotide Polymorphism Database. In silico analysis of the reference sequence identified a high-confidence primary MYB binding motif (AACCGTT) that was duplicated in the PF-382 cell line, whereas the single bp deletion in DU.528 cells creates a CAACCGC sequence that closely resembles a secondary MYB binding motif (Figure 1B; supplemental Tables 3 and 4).
LMO2 intron 1 mutations in pediatric and adult human T-ALL. (A) LMO2 expression as determined by qRT-PCR in LMO2 translocated T-ALL cell lines (KOPT-K1 and P12-Ichikawa) and nontranslocated T-ALL cell lines (DU.528, PF-382, Loucy, DND41, Jurkat, and ALL-SIL). (B) ChIP-Seq tracks at the LMO2 locus for MYB and H3K27ac in PF-382, DU.528, Loucy, and Jurkat T-ALL cell lines. Y-axis values are reads per bin per million mapped reads. Mutations are shown below as identified by Sanger sequencing of PF-382 and DU.528 DNA, with inserted sequences shown in red and MYB motifs underlined. The position weight matrices for the primary and secondary MYB binding sites are from UniPROBE.27 (C) Pie chart summarizing the percentage of LMO2 transcripts identified by rapid amplification of 5′ complementary DNA ends that start from the distal, intermediate, and proximal promoters for the PF-382, DU.528, and Loucy T-ALL cell lines. A total of 20, 21, and 22 LMO2 transcripts was examined for PF-382, DU.528, and Loucy T-ALL cell lines, respectively. (D) Pie chart summarizing mutation recurrence within pediatric and adult human T-ALL cohorts. (E) Indels mapped to the LMO2 intron 1 mutation hotspot labeled with the associated de novo consensus site as aligned to the UniPROBE or HOCOMOCO position weight matrices, in which MYB, ETS1, and RUNX1 sites are marked as a triangle, square, and diamond, respectively. Below, motif analysis of the region shows the native binding sites for members of the TAL1 complex, including RUNX1, E-box (for TAL1 binding), ETS1, MYB, and GATA. TSS, transcription start site.
LMO2 intron 1 mutations in pediatric and adult human T-ALL. (A) LMO2 expression as determined by qRT-PCR in LMO2 translocated T-ALL cell lines (KOPT-K1 and P12-Ichikawa) and nontranslocated T-ALL cell lines (DU.528, PF-382, Loucy, DND41, Jurkat, and ALL-SIL). (B) ChIP-Seq tracks at the LMO2 locus for MYB and H3K27ac in PF-382, DU.528, Loucy, and Jurkat T-ALL cell lines. Y-axis values are reads per bin per million mapped reads. Mutations are shown below as identified by Sanger sequencing of PF-382 and DU.528 DNA, with inserted sequences shown in red and MYB motifs underlined. The position weight matrices for the primary and secondary MYB binding sites are from UniPROBE.27 (C) Pie chart summarizing the percentage of LMO2 transcripts identified by rapid amplification of 5′ complementary DNA ends that start from the distal, intermediate, and proximal promoters for the PF-382, DU.528, and Loucy T-ALL cell lines. A total of 20, 21, and 22 LMO2 transcripts was examined for PF-382, DU.528, and Loucy T-ALL cell lines, respectively. (D) Pie chart summarizing mutation recurrence within pediatric and adult human T-ALL cohorts. (E) Indels mapped to the LMO2 intron 1 mutation hotspot labeled with the associated de novo consensus site as aligned to the UniPROBE or HOCOMOCO position weight matrices, in which MYB, ETS1, and RUNX1 sites are marked as a triangle, square, and diamond, respectively. Below, motif analysis of the region shows the native binding sites for members of the TAL1 complex, including RUNX1, E-box (for TAL1 binding), ETS1, MYB, and GATA. TSS, transcription start site.
To assess whether the mutations form aberrant sites of MYB binding, we performed ChIP-seq for MYB and analyzed peaks of MYB enrichment at the LMO2 locus. There was a complete absence of MYB binding at the intermediate promoter in cells that were wild-type at this locus, suggesting that the presence of the single native MYB motif in itself is insufficient to recruit MYB. In contrast, both PF-382 and DU.528 cells that harbor dual MYB motifs displayed precisely aligned MYB binding at the mutation site (Figure 1B). To determine whether the mutations affected promoter usage, we performed rapid amplification of 5′ complementary DNA ends in LMO2 mutant and wild-type cell lines by using a common primer in exon 6 capable of capturing the transcription start site of all LMO2 isoforms. Whereas the majority (73%) of 5′ capped transcripts in Loucy cells originated from the proximal promoter, both PF-382 and DU.528 cells demonstrated preferential usage of the recently described intermediate promoter (75% and 67% of transcripts, respectively; Figure 1C).
Our observations were not limited to T-ALL cell lines because heterozygous mutations at LMO2 intron 1 were detected in diagnostic samples from 3.7% of pediatric (6 of 160) and 5.5% of adult (9 of 163) T-ALL patients (Figure 1D). Absence of the mutations in 7 available patient-matched remission samples confirmed that they were somatic (supplemental Figure 2). Notably, the mutations were densely distributed around highly conserved native ETS1, MYB, and GATA motifs (supplemental Figure 3). Including the cell lines, 7 mutations introduced an additional MYB site, resulting in 2 MYB motifs spaced 10 or 20 bp apart, equivalent to 1 or 2 helical coils of DNA, respectively (Figure 1E). Three mutations created potential binding sites for both MYB and ETS1, 3 formed potential ETS1 sites, and 3 produced potential new RUNX1 binding sites (Figure 1E; supplemental Tables 3 and 4). Given that NOTCH and TAL1 have been shown to collaborate with LMO2 to promote leukemogenesis in murine models of T-ALL, it is noteworthy that of the 15 patients with LMO2 promoter mutations, 7 had NOTCH-1 mutations and 8 had TAL1 activating lesions, including 2 with TAL1-enhancer mutations (both creating new MYB motifs; supplemental Table 5).20,21 Such collaboration between TAL1, LMO2, and NOTCH-1 has also been described in gene therapy–induced T-ALL, including 1 patient who harbored both a retroviral integration upstream of LMO2 and an episomal reintegration at the TAL1 locus.9,13,22
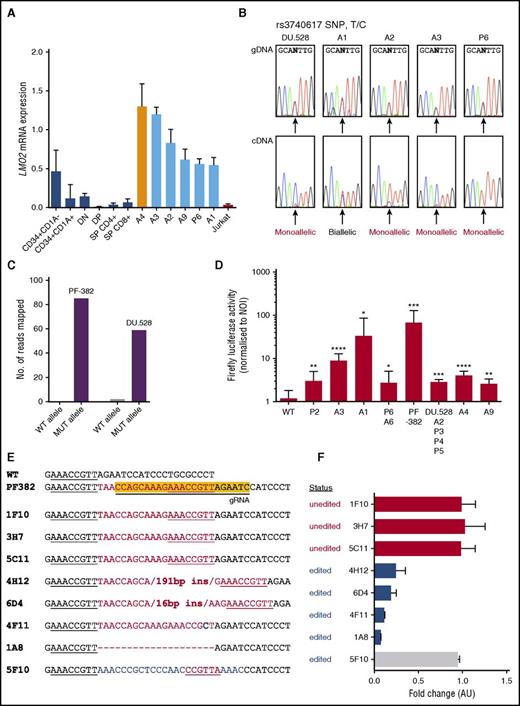
To ascertain whether LMO2 promoter mutations in T-ALL led to aberrant expression compared with its matched thymic counterpart, we assessed LMO2 expression by qRT-PCR in thymic subsets sorted for different levels of thymic differentiation.5 Validating earlier reports that used microarrays, LMO2 expression was highest in the most immature, precommitment stages of T-cell development and was expressed at low levels from the double-negative stage onward, when thymocytes had undergone biallelic TCR-γ rearrangement (Figure 2A).5 To determine the level of differentiation arrest of the 15 mutant patient samples, we analyzed the TCR-γ locus by qPCR (supplemental Figure 4); 12 of the 15 samples (including 5 of the 6 patients with available RNA) had biallelic TCR-γ deletion (position weight matrices; supplemental Table 5), indicating that maturation arrest occurred after the pro–T-cell stage of differentiation, and that the majority of patients did not have the ETP ALL phenotype. Thus, compared with their physiological counterparts, those patients with RNA available for LMO2 qRT-PCR exhibited aberrant LMO2 overexpression (P < .002 vs double-negative and double-positive subsets; Figure 2A). Although we were unable to confirm LMO2 overexpression in all mutant samples because of the unavailability of RNA, all classes of mutation (additional MYB, ETS1, RUNX1, or MYB+ETS1 sites) were represented in the 6 patients with LMO2 overexpression. Exploiting a heterozygous germ line single nucleotide polymorphism (rs3740617), DU.528 cells and 3 of 4 informative patient samples displayed skewed allelic expression of LMO2 (Figure 2B). The observation of biallelic expression in sample A1 suggests a potential lesion on the second allele that remains undefined. Consistent with their cis-activating potential, ≥96% of reads from MYB ChIP-seq performed in DU.528 and PF-382 cells aligned to the mutant rather than the wild-type allele (Figure 2C; supplemental Figure 5). Furthermore, the gain-of-function nature of the mutations was confirmed by luciferase reporter assays conducted in Jurkat cells where all mutations markedly activated luciferase activity compared with the wild-type sequence (Figure 2D; supplemental Figure 6A).
LMO2 intron 1 indels are predominantly monoallelically activating, and CRISPR/Cas9-mediated knockout of the PF-382 mutant allele downregulates LMO2 expression. (A) LMO2 expression as determined by qRT-PCR in human sorted thymic subsets, primary patient samples with LMO2 intron 1 indels, and the wild-type Jurkat cell line. P < .002 for samples A1, A2, A3, A9, and P6 vs double negative and double positive by two-tailed t test. Primary patient samples were assessed for the absence of biallelic TCR-γ deletion (ABD), of which patient sample A4 (orange bar) exhibited ABD, whereas all other patients were non-ABD. (B) The informative single nucleotide polymorphism (SNP) (rs3740617) was amplified in 4 patient samples and the DU.528 cell line from both genomic DNA (gDNA) and complementary DNA (cDNA) templates to infer monoallelic expression. To do this, if one chromatogram peak is detected at a heterozygous SNP within the cDNA, the expression can be interpreted as coming from one allele. (C) Quantification of the number of reads mapped to the wild-type (WT) or mutant (MUT) allele where 54 of 56 reads and 85 of 85 reads mapped to the mutant alleles for DU.528 and PF-382, respectively. (D) Firefly luciferase activity after renilla and no-insert vector normalization for patient-derived indels. Data shown are from ≥3 independent experiments performed in triplicate. Values are mean ± standard deviation and P values were calculated by a two-tailed Student t test. (E) The yellow highlighted sequence is the target region for the CRISPR/Cas9 guide RNA. Aligned sequences are from CRISPR/Cas9-edited PF-382 single-cell clones showing the associated genomic edits generated. Red sequences are inserted sequences, blue are altered, and dashes represent deleted bases. Underlined region shows the presence of the native and mutant MYB binding sites. (F) Gene expression of LMO2 for each PF-382 clone, as determined by qRT-PCR. Data are expressed as fold change relative to the mean expression of the unedited clones in arbitrary units (AU). Clones are labeled as “unedited” when CRISPR/Cas9 did not edit the region targeted by the guide RNA and “edited” when successful targeting led to the formation of an indel. *P ≤ .05, **P ≤ .01, ***P ≤ .001, and ****P ≤ .0001. NOI, no-insert control; T/C, T to C single nucleotide polymorphism.
LMO2 intron 1 indels are predominantly monoallelically activating, and CRISPR/Cas9-mediated knockout of the PF-382 mutant allele downregulates LMO2 expression. (A) LMO2 expression as determined by qRT-PCR in human sorted thymic subsets, primary patient samples with LMO2 intron 1 indels, and the wild-type Jurkat cell line. P < .002 for samples A1, A2, A3, A9, and P6 vs double negative and double positive by two-tailed t test. Primary patient samples were assessed for the absence of biallelic TCR-γ deletion (ABD), of which patient sample A4 (orange bar) exhibited ABD, whereas all other patients were non-ABD. (B) The informative single nucleotide polymorphism (SNP) (rs3740617) was amplified in 4 patient samples and the DU.528 cell line from both genomic DNA (gDNA) and complementary DNA (cDNA) templates to infer monoallelic expression. To do this, if one chromatogram peak is detected at a heterozygous SNP within the cDNA, the expression can be interpreted as coming from one allele. (C) Quantification of the number of reads mapped to the wild-type (WT) or mutant (MUT) allele where 54 of 56 reads and 85 of 85 reads mapped to the mutant alleles for DU.528 and PF-382, respectively. (D) Firefly luciferase activity after renilla and no-insert vector normalization for patient-derived indels. Data shown are from ≥3 independent experiments performed in triplicate. Values are mean ± standard deviation and P values were calculated by a two-tailed Student t test. (E) The yellow highlighted sequence is the target region for the CRISPR/Cas9 guide RNA. Aligned sequences are from CRISPR/Cas9-edited PF-382 single-cell clones showing the associated genomic edits generated. Red sequences are inserted sequences, blue are altered, and dashes represent deleted bases. Underlined region shows the presence of the native and mutant MYB binding sites. (F) Gene expression of LMO2 for each PF-382 clone, as determined by qRT-PCR. Data are expressed as fold change relative to the mean expression of the unedited clones in arbitrary units (AU). Clones are labeled as “unedited” when CRISPR/Cas9 did not edit the region targeted by the guide RNA and “edited” when successful targeting led to the formation of an indel. *P ≤ .05, **P ≤ .01, ***P ≤ .001, and ****P ≤ .0001. NOI, no-insert control; T/C, T to C single nucleotide polymorphism.
To assess causality between the mutations and LMO2 dysregulation, we used CRISPR/Cas9 genome editing with a guide RNA designed to target the duplicated MYB site in PF-382 cells (supplemental Figure 6B). Crucially, clone 4F11, which had a single T>C substitution disrupting the MYB binding site, and clone 1A8, in which the mutant allele had been reverted to wild-type, resulted in the most dramatic downregulation of LMO2 (Figure 2E-F; supplemental Figure 7). Interestingly, 2 clones (4H12 and 6D4) that increased the distance between the native and the mutant MYB sites resulted in a marked reduction in LMO2 expression, supporting the hypothesis that MYB binding is augmented when additional motifs are orientated on the same side of the DNA helix.23 This was further validated by the lack of reduction in LMO2 expression in a clone (5F10) in which the sequence between the 2 MYB sites was altered but the spacing distance was unchanged.
In conclusion, we identified and functionally validated a novel recurrent mutation hotspot occurring in a noncoding site that drives LMO2 overexpression from a neomorphic promoter in a substantial proportion of both adult and pediatric T-ALL patients. Remarkably, the mutations create potential binding sites for MYB, ETS1, or RUNX1, all of which are members of a highly oncogenic TAL1-LMO2 complex in T-ALL; this indicates that LMO2 is a component of an autoregulatory self-sustaining positive feedback loop in these cells, analogous to the autoregulation of TAL1 we recently described in Jurkat cells.14,24 To prove that the newly formed ETS1 and RUNX1 sites are sufficient to drive LMO2 expression, we attempted to but ultimately were unable to knock in these mutations in vitro. Thus, the oncogenic potential of these particular mutations remain an area of ongoing study. There is still a question regarding exactly how various members of the TAL1 complex position themselves on DNA with regard to spacing, orientation, and order of motifs, so-called syntax.25 Thus, identifying gain-of-function noncoding mutations that have been selected for during tumorigenesis in vivo offers important insights into the optimal DNA syntax required for nucleation of such multiprotein transcription factor complexes. For instance, it may become apparent why a single MYB binding site is sufficient to drive expression from certain loci, such as at the TAL1 enhancer, whereas others require dual MYB motifs. Finally, we note that these mutations occur within the same intron as retroviral integration sites described in 2 cases of gene therapy–induced T-ALL (supplemental Figure 8).22,26 This raises the possibility that formation of aberrant promoters and enhancers, either by mutation or retroviral insertion, occur at preferred rather than random sites in the noncoding genome.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients, families, and clinical teams who have been involved in both trials. Primary childhood leukemia samples used in this study were provided by the Bloodwise Childhood Leukemia Cell Bank, working with the laboratory teams in the Bristol Genetics Laboratory, Southmead Hospital, Bristol, United Kingdom; Molecular Biology Laboratory, Royal Hospital for Sick Children, Glasgow, United Kingdom; Molecular Haematology Laboratory, Royal London Hospital, London, United Kingdom; and Molecular Genetics Service and Sheffield Children’s Hospital, Sheffield, United Kingdom.
This work was supported by National Institutes of Health, National Cancer Institute grants 1R01CA176746-01, 5P01CA109901-08, and 5P01CA68484 (A.T.L.), by Bloodwise and Gabrielle’s Angels Foundation (M.R.M. and S.R.), by the Freemason’s Grand Charity (M.M.), by Alex’s Lemonade Stand Foundation for Childhood Cancer (Z.L.), by grants from Bloodwise (formerly known as Leukaemia and Lymphoma Research, UK) and the Medical Research Council (UK) for the UKALL2003 trial (ISRCTN07355119), and by Cancer Research UK for the UKALL14 trial (ISRCTN66541317). This work was undertaken at University College London, which receives funding from the Department of Health’s National Institute of Health Research Biomedical Research Centre. B.J.A. is a Hope Funds for Cancer Research Grillo-Marxuach Family Fellow.
Authorship
Contribution: S.R., M.M., T.E.L., N.F., A.P., Z.L., S.B., C.A., T.P., K.P.-O., L.G.-P., and B.J.A. performed experimental work; K.Z.A., R.J.M., T.N., A.K.F., R.E.G., K.P.-O., L.G.-P., and F.J.T.S. provided primary samples; S.R., M.M., T.E.L., N.F., D.C.L., R.A.Y., F.J.T.S., and A.T.L. analyzed data; S.R., R.E.G., F.J.T.S., D.C.L., and M.R.M. wrote the manuscript; M.R.M. designed the study; and all authors approved the final manuscript.
Conflict-of-interest disclosure: R.A.Y. is a founder and member of the Board of Directors of Syros Pharmaceuticals, which develops therapies that target gene regulatory elements. The remaining authors declare no competing financial interests.
Correspondence: Marc R. Mansour, University College London Cancer Institute, Department of Haematology, 72 Huntley St, London WC1E 6BT, United Kingdom; e-mail: m.mansour@ucl.ac.uk.