Insight into the clonal dynamics of leukemia is vital for our understanding of its pathogenesis and progression. In this issue of Blood, Belderbos et al1 use cellular barcoding to map the clonal fate of xenografted primary human leukemic cells in both time and space.
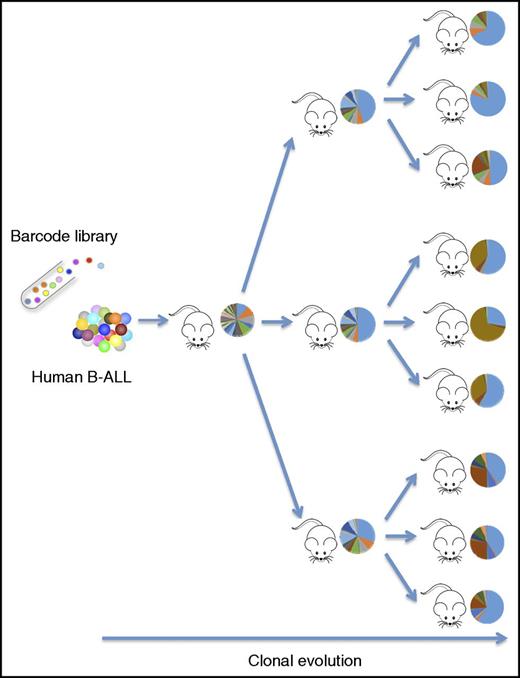
Cellular barcoding provides a clonal fate map of primary human ALL cells passaged through serial transplantation in immunodeficient mice. The clonal distribution patterns (circles) are adapted from 1 experiment in the study by Belderbos et al that begins on page 3210. B-ALL, B-progenitor cell ALL.
Cellular barcoding provides a clonal fate map of primary human ALL cells passaged through serial transplantation in immunodeficient mice. The clonal distribution patterns (circles) are adapted from 1 experiment in the study by Belderbos et al that begins on page 3210. B-ALL, B-progenitor cell ALL.
Tumors emerge through an evolutionary process where specific traits triggered by accumulating mutations are selected for and against, ultimately leading to full-blown cancer. The importance of such clonal evolutionary patterns in understanding and treating cancer is becoming increasingly appreciated, especially in the context of treatment resistance and relapsing disease.2 While assessing clonal hierarchies poses a great challenge in most solid tissues, the hematopoietic organ is more accessible due to its liquid nature and provides unique opportunities to track and monitor specific cell populations. As an example, large-scale sequencing studies of somatic mutations in acute myeloid leukemia have revealed clear hierarchies and detailed sequential clonal evolutionary patterns with relatively high precision.3 However, in other leukemias, such as acute lymphoblastic leukemia (ALL), the situation appears more complex, and analysis of mutations has not been able to provide a similar detailed view of clonal dynamics during disease progression.
Here, Belderbos et al reasoned that cellular barcoding could provide a more direct and dynamic assessment of clonal distribution and competition in leukemia pathogenesis that experimentally could complement and validate current models of clonal evolution derived from mutational landscapes. Cellular barcoding is based on the genomic integration of specific DNA sequences or “barcodes” in target cells that later can be tracked by next-generation sequencing (NGS) of amplified DNA segments from the cellular offspring.4,5 In this manner, NGS platforms can function as highly efficient “barcode scanners,” allowing precise detection and quantification of thousands of genetically tagged cell clones from a single pool of cells at relatively low cost.
Building on their previous expertise with barcoding experiments, Belderbos et al first set out to generate a new and improved lentiviral barcode library. Rather than employing a pool of random barcode vectors, which was previously the standard approach, the authors generated several hundred individual barcode preps that they then assembled at equimolar ratios into predefined libraries with a known composition and complexity. This strategy should ensure optimal and even distribution of barcodes and also enhance the reproducibility and the general robustness of the approach, as identical libraries can be multiply regenerated. A predefined library composed of known barcodes further minimizes the risk of detecting false barcodes due to sequencing errors. The authors used experiments in a leukemic cell line to validate the library composition and establish methods for detection and tracking of the barcodes both in vitro and in vivo. Overall, their findings confirmed an even distribution and high complexity of the library and a capacity for high-resolution tracking of individual cell clones.
The key experiment of this study (see figure) was performed in primary diagnostic samples of pediatric B-cell ALL. The authors transduced leukemic cells with barcode libraries and serially passaged them in vivo through tertiary transplantations in NSG mice. They then analyzed the barcode distribution following each round of transplantation in multiple distinct hematopoietic locations, including both skeletal (long bones, spine, and sternum) and extramedullary (blood, spleen, and liver) sites. Two general patterns were observed from these experiments. Firstly, from a temporal perspective, the engrafted cells showed a highly polyclonal and random pattern in the primary recipients, while the clonal complexity decreased gradually and acquired a more stable pattern following secondary and tertiary transplantation (see figure). Secondly, in terms of location, Belderbos et al found that the clones were unevenly distributed not only among the various skeletal sites analyzed but also when comparing all skeletal sites to the extramedullary sites. They were thus able to get a precise clonal fate map of the leukemic cells in both time and space during the course of the serial transplants. However, more specific conclusions regarding the observed clonal patterns were restricted by the fact that only 2 individual ALL cases were analyzed. Several barcoded samples failed to engraft in NSG mice, and the approach would likely benefit from employing some of the more sensitive xenograft models that have been introduced in the field recently.6,7
So what are the implications of this study? The main advancement lies in the technical and analytical aspects, which have been carefully and elegantly worked out. The paper represents the first demonstration of a detailed spatial and temporal fate mapping of primary human leukemia in vivo using barcoding, and it provides a paradigm for future studies in this area. As more cases are analyzed in the future, the barcoding approach in combination with detailed mutational analysis promises to reveal important distinctions in the clonal evolutionary patterns of different subtypes of ALL and other leukemias. Additional layers of information can further be retrieved by tracking the clonal dynamics in the presence or absence of therapeutic agents. Ultimately, this may guide the prediction of disease development in the clinic and influence the choice of therapeutic approach.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal