To the editor:
l-Asparaginase (ASNase) is a key component of protocols used to treat acute lymphoblastic leukemia (ALL). Poorly understood interpatient differences in ASNase pharmacokinetics demand therapeutic drug monitoring to prevent patients from receiving an inadequate dose.1 Although it is unclear whether there is a causal relation between elevated ASNase levels and toxicities, underexposure compromises therapeutic benefits. A recent report demonstrated that lysosomal proteases degrade ASNase in vitro.2 However, to which extent these proteases contribute to ASNase clearance in patients remains unclear. Here we link a strongly prolonged ASNase turnover to a germ line mutation in the gene encoding cathepsin B.
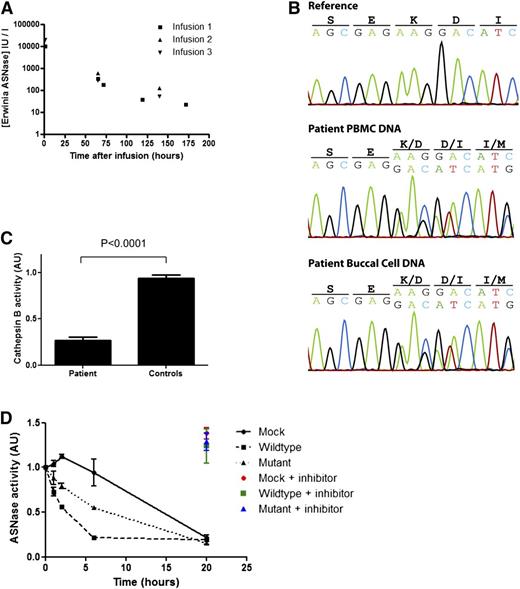
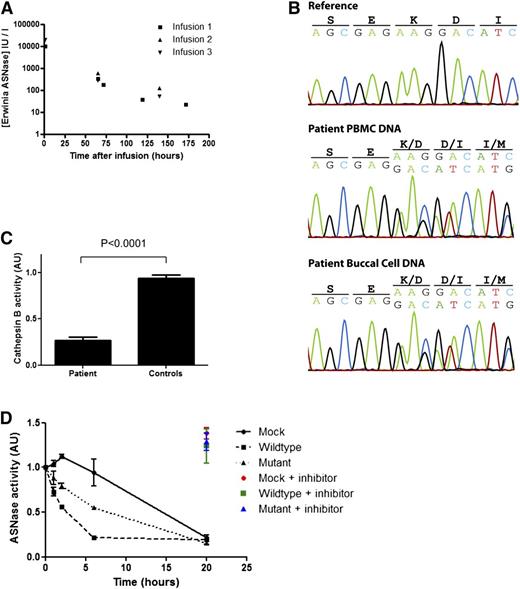
A pediatric patient, treated for common B-cell progenitor ALL, developed encephalopathy associated with hyperammonemia (354 µmol/L) after the sixth dose of Erwinase. She required plasmapheresis to reduce serum ammonia levels. Serum analysis revealed abnormally high concentrations of Erwinase at the time of plasmapheresis (2167 IU/mL, 2 days after the last dose). After recovery, Erwinase treatment was restarted under therapeutic drug monitoring. This revealed a strongly increased half-life of Erwinase (Figure 1A), which could explain the high Erwinase serum concentrations (see supplemental Case Report for detailed information; available on the Blood Web site).
A mutation in the gene encoding cathepsin B in a patient with strongly reduced metabolism asparaginase. (A) Pharmacokinetics of Erwinase serum levels indicate a strongly delayed metabolism of ASNase in this patient. The half-life (ln2/Kel) of 28.5 hours (normal 7-15 hours5 ) was calculated using best-fit nonparametric modeling of the data points. (B) DNA sequencing of the cathepsin B gene reveals a heterozygous deletion of a single codon (c.709_711delAAG) in DNA isolated from PBMCs and buccal cells of the patient. (C) Cathepsin B activity in lysates of EBV-transformed B cells of the patient (carrying the mutation) and age-matched controls (N = 5) was determined by measuring cleavage of the fluorescent substrate Ac-RR-AFC. The plot shows an average of 2 experiments with standard deviation. One of the control samples was set to 1, and all samples were correlated to this sample. Unpaired 2-tailed t test was used to determine significance. (D) ASNase was incubated in lysate of HEK293 cells expressing wild-type or mutant cathepsin B. After incubation, residual ASNase activity was assayed as described in the supplemental Methods section. Cathepsin inhibitor CA-074 was included in selected samples to confirm the contribution of cathepsin B in this degradation. The plot shows an average of 3 independent experiments with standard error of the mean. Analysis of variance statistical analysis was applied to test for significance.
A mutation in the gene encoding cathepsin B in a patient with strongly reduced metabolism asparaginase. (A) Pharmacokinetics of Erwinase serum levels indicate a strongly delayed metabolism of ASNase in this patient. The half-life (ln2/Kel) of 28.5 hours (normal 7-15 hours5 ) was calculated using best-fit nonparametric modeling of the data points. (B) DNA sequencing of the cathepsin B gene reveals a heterozygous deletion of a single codon (c.709_711delAAG) in DNA isolated from PBMCs and buccal cells of the patient. (C) Cathepsin B activity in lysates of EBV-transformed B cells of the patient (carrying the mutation) and age-matched controls (N = 5) was determined by measuring cleavage of the fluorescent substrate Ac-RR-AFC. The plot shows an average of 2 experiments with standard deviation. One of the control samples was set to 1, and all samples were correlated to this sample. Unpaired 2-tailed t test was used to determine significance. (D) ASNase was incubated in lysate of HEK293 cells expressing wild-type or mutant cathepsin B. After incubation, residual ASNase activity was assayed as described in the supplemental Methods section. Cathepsin inhibitor CA-074 was included in selected samples to confirm the contribution of cathepsin B in this degradation. The plot shows an average of 3 independent experiments with standard error of the mean. Analysis of variance statistical analysis was applied to test for significance.
We hypothesized that a defect in one of the lysosomal proteases, previously reported to be capable of degrading asparaginase in vitro,2 might be responsible for the prolonged half-life observed in this patient. Sequencing of DNA isolated from peripheral blood mononuclear cells (PBMCs) and from buccal cells revealed a heterozygous single codon deletion (c.709_711delAAG) in the gene encoding cathepsin B in the germ line of the patient, which is not listed in the Database of Single Nucleotide Polymorphisms (dbSNP; National Center for Biotechnology Information, Bethesda, MD) or in our in-house database containing the exome sequence data of 1154 individuals (Figure 1B). This mutation results in a deletion of a highly conserved lysine residue in the C terminus of the protein (p.K237del), which is predicted3 to lead to a loss of structural integrity of the protein (supplemental Figure 1A-B).
Cathepsin B is synthesized as a 44-kDa pre-proenzyme that is processed in late endosomes to a 33-kDa active single chain and matured into an active 2-chain form consisting of a 24-kDa heavy chain (and a 27-kDa glycosylated form) and a 5-kDa light chain.4 We expressed both wild-type and mutant cathepsin B, cloned from RNA extracted from patient PBMCs, in HEK293 cells to follow the maturation process. Biochemical analysis revealed defective maturation of the mutant cathepsin B, which was confirmed by an aberrant subcellular localization (supplemental Figure 1C-F).
To test whether this mutation affects the protease activity, we assessed cathepsin B activity in Epstein-Barr virus (EBV)–immortalized B cells obtained from the patient. Indeed, cathepsin B activity in the B cells obtained from the patient was strongly reduced as compared with B cells from age-matched donors (Figure 1C), indicating that deletion of residue K237 results in a loss of function.
Next, we determined whether the reduced protease activity of the mutant cathepsin B would result in a diminished degradation of ASNase. Therefore, we incubated Erwinase in lysates of cells expressing either wild-type or mutant cathepsin B and analyzed samples taken at the indicated time points for residual ASNase activity (Figure 1D). Mock transfected cells showed limited protease activity and cleared the Erwinase after 20 hours of incubation. Of note, this degradation was inhibited by the addition of a specific cathepsin B inhibitor, CA-074 (color marked data points, Figure 1D), indicating that endogenous cathepsin B is responsible for ASNase degradation in these lysates. Overexpression of the wild-type cathepsin B resulted in a rapid clearance of the Erwinase from the lysate, which again was fully inhibited by the addition of the cathepsin B inhibitor CA-074. Expression of the K237del mutant cathepsin B protein was still capable of degrading ASNase, but the rate of clearance was significantly reduced (P < .05) in comparison with the wild-type protein, consistent with the delayed ASNase clearance observed in this patient. Western blot analysis of asparaginase incubated in these cell lysates confirmed that degradation of asparaginase protein rather than the inhibition of enzymatic activity causes the decrease in asparaginase activity that we measured in the previous assay (supplemental Figure 1G-H).
It is unknown where degradation of ASNase occurs. Low levels of cathepsin B activity are detected in human serum (data not shown), but these amounts are insufficient to degrade Erwinase or Escherichia coli ASNase in vitro (supplemental Figure 1). Instead, it appears that cathepsin B–mediated degradation of ASNase occurs intracellularly, after being removed from the blood by phagocytic cells, which is consistent with the fact that pegylated forms of this protein show an increased serum half-life.5
Current knowledge of factors influencing ASNase pharmacokinetics is limited.5,6 Only the presence of inhibitory antibodies that bind asparaginase is known to significantly shorten the half-life of ASNase.7 In our patient, we found no evidence of an immune response targeting the ASNase. Our observations support the hypothesis that the lysosomal protease cathepsin B is an essential component in the regulation of ASNase turnover. Mutations in the cathepsin B gene, as observed in our patient, are rare events. However, because cathepsin B activity is tightly regulated, not only at the level of gene expression, but also by protein maturation and enzyme activity,8,9 many additional factors may contribute to variations in cathepsin B activity and can therefore contribute to the interpatient differences in ASNase kinetics.
The online version of this article contains a data supplement.
Authorship
Acknowledgments: The authors thank Dr Jeroen Middelbeek for help with microscopy imaging and Simon van Reijmersdal for technical assistance. This work was supported in part by research funding from KiKa (grant #134) (L.T.v.d.M.).
Contribution: L.T.v.d.M. performed experiments, analyzed and interpreted data, and wrote the manuscript; E.W. designed and performed the genomic analysis; M.L. performed research; H.V. performed in silico structure analysis; D.R. performed research; J.B. and C.L. performed drug monitoring and designed research; R.J.B. performed the pharmacokinetic studies; R.P.K. analyzed and interpreted the genomic analysis; P.H. designed the research and interpreted the data; F.N.v.L. and D.M.t.L. designed and supervised the research; and all authors edited and approved the manuscript.
Conflict-of-interest disclosure: J.B. served personally as a consultant and participated in advisory boards and safety boards for different asparaginase-selling companies including EUSA Pharma and former Erwinase license holders, Bayer and Medac GmbH. The laboratory at the University of Münster is also involved in scientific collaborations with EUSA, Medac, and Sigma-Tau, which to some extent contributed financially to the analytical procedures. The remaining authors declare no competing financial interests.
Current affiliation for D.R. is Department of Experimental Rheumatology, Radboud University Medical Center, Nijmegen, The Netherlands.
Correspondence: Frank N. van Leeuwen, 412 Laboratory of Pediatric Oncology, Department of Pediatrics, Radboud University Medical Centre, PO Box 9101, 6500 HB Nijmegen, The Netherlands; e-mail: frankn.vanleeuwen@radboudumc.nl.
References
Author notes
F.N.v.L. and D.M.t.L. contributed equally to this study.