Abstract
Iron-refractory, iron-deficiency anemia (IRIDA) is a familial disorder characterized by iron deficiency anemia unresponsive to oral iron treatment but partially responsive to intravenous iron therapy. Previously, we showed that IRIDA patients harbor loss-of-function mutations in TMPRSS6, a type II transmembrane serine protease primarily expressed by the liver. Both humans and mice with TMPRSS6 mutations show inappropriately elevated levels of the iron-regulatory hormone hepcidin, suggesting that TMPRSS6 acts to negatively regulate hepcidin expression. Here we investigate the relationship between Tmprss6 and the bone morphogenetic protein (BMP)–Smad signaling pathway, a key pathway promoting hepcidin transcription in hepatocytes. We show that livers from mice deficient for Tmprss6 have decreased iron stores and decreased Bmp6 mRNA, but markedly increased mRNA for Id1, a target gene of Bmp6 signaling. In contrast, mice deficient for both Tmprss6 and hemojuvelin (Hjv), a BMP coreceptor that augments hepcidin expression in hepatocytes, showed markedly decreased hepatic levels of hepcidin and Id1 mRNA, markedly increased hepatic Bmp6 mRNA levels, and systemic iron overload similar to mice deficient for Hjv alone. These findings suggest that down-regulation of Bmp/Smad signaling by Tmprss6 is required for regulation of hepcidin expression and maintenance of systemic iron homeostasis.
Introduction
Hepcidin, a small peptide hormone produced by the liver that is detectable in serum and urine, is a central regulator of iron homeostasis.1 Hepcidin binds to ferroportin, the only known mammalian cellular iron exporter, which is expressed on the basolateral membrane of enterocytes and the plasma membrane of macrophages. Hepcidin binding leads to the internalization and degradation of ferroportin in lysosomes; thus, hepcidin decreases both absorption of dietary iron and release of iron from macrophage stores.2-5 Hepcidin expression is induced by iron loading6 and repressed by anemia and hypoxia.7 Hepcidin is also induced by inflammatory cytokines,6,8 including interleukin-6,9 which stimulates the hepcidin promoter through signal transducer and activator of transcription 3 signaling.10,11 The central role of hepcidin in the regulation of systemic iron homeostasis is highlighted by the fact that loss-of-function mutations in the hepcidin gene (HAMP) cause juvenile hemochromatosis, an autosomal recessive disorder characterized by severe iron deposition in multiple organs.12
Signaling through hemojuvelin (HJV), a glycosylphosphatidylinositol–anchored protein of the repulsive guidance molecule family, has emerged as a key hepcidin-activating pathway. In both humans and mice, mutations in the gene encoding hemojuvelin (HFE2, referred to here as HJV) markedly decrease hepcidin levels; systemic iron overload similar to disease caused by HAMP mutations results, demonstrating the importance of HJV in hepcidin production.13-15 HJV functions as a coreceptor for bone morphogenetic proteins (BMPs), members of the transforming growth factor-β superfamily of ligands that signal through SMAD proteins for a variety of biologic purposes. Hepatic BMP signaling is augmented by HJV to induce hepcidin expression.16 Loss of BMP6 function in mice results in severe iron overload, suggesting that BMP6 is the endogenous ligand for Hjv.17,18 Liver-specific inactivation of Smad4, the common downstream mediator of transforming growth factor-β superfamily ligands, also results in systemic iron overload.19
TMPRSS6 (also known as matriptase-2), a transmembrane serine protease produced by the liver, has recently been identified as a key repressor of hepcidin expression. Loss of Tmprss6 function in both humans and mice leads to inappropriately elevated hepcidin, resulting in impaired intestinal iron absorption that leads to iron deficiency anemia.20-24 Like other members of the type II transmembrane serine protease family, TMPRSS6 is characterized structurally by a short amino-terminal cytoplasmic tail, a transmembrane region, a stem region containing several structural domains, and a carboxy-terminal trypsinlike serine protease domain.25,26 TMPRSS6 physically interacts with HJV when both proteins are overexpressed in HeLa cells, and TMPRSS6 is capable of cleaving HJV present on the plasma membrane.27 Furthermore, in transfected hepatoma cell lines, coexpression of TMPRSS6 inhibits the ability of HJV to activate the hepcidin promoter.20,27 Mutated forms of TMPRSS6 in which the protease domain has been altered by truncation or missense mutation are less effective at mediating this inhibition. Together, these in vitro findings suggest that the elevated hepcidin levels observed in humans and mice harboring TMPRSS6 mutations result from dysregulated BMP/SMAD signaling. In this report, we used targeted mouse models to investigate the relationship between Tmprss6 and the Bmp/Smad signaling pathway in the regulation of systemic iron homeostasis in vivo.
Methods
Animal strains
B6.129P2-Tmprss6tm1Dgen/Crl mice were generated by homologous recombination at Deltagen and had been backcrossed to C57BL/6N mice for 7 generations before study. This knockout strategy replaces the Tmprss6 genomic region spanning from the 3′ portion of exon 16 to the 5′ portion of exon 17. This genomic region encodes amino acids 654 to 727 of the Tmprss6 protein, which comprise a large portion of the protease domain (amino acids 577-811; Ensembl transcript ENSMUST00000017086).28 Sequencing of genomic DNA isolated from B6.129P2-Tmprss6tm1Dgen/Crl mice demonstrated that approximately 1.7 kb of endogenous Tmprss6 genomic sequence was replaced with approximately 2 kb of novel sequence derived from the targeting vector. Hfe2tm1Nca/tm1Nca (hereafter referred to as Hjv−/− mice), generated by our laboratory, were maintained on a 129S6/SvEvTac background. Animals were genotyped by polymerase chain reaction (PCR) performed on genomic DNA prepared from toe clips as previously described.29 The Tmprss6 mutant and wild-type alleles were genotyped by multiplex PCR using a common forward primer (5′-GGAAAGATGCGGCAGAACTCG-3′) and 2 reverse primers specific for the targeted (5′-CGAGACTAGTGAGACGTGCTACTTCC-3′) and wild-type (5′-CAGCCTGTGATCCAGCAGTG-3′) alleles, yielding 420- and 218-bp products, respectively. The Hjv mutant allele was genotyped by PCR using primers 5′-CGGATGGTTTTTGGCAGTTAG-3′ and 5′-GCCTTTACGATATCTCAGTCC-3′, yielding a 261-bp product. The Hjv wild-type allele was genotyped by PCR using primers 5′-GAATGGCTTCCTTCCATCAA-3′ and 5′-ATCTTCAAAGGCTGCAGGAA-3′, yielding a 433-bp product.
Animal care and analysis
All mice were born and housed in the barrier facility at Duke University according to protocols approved by the Duke University Institutional Animal Care & Use Committee. Animals were maintained on a 270 parts per million iron diet (5T40 Modified Laboratory Rodent Diet 5001, containing 150 parts per million fenbendazole for pinworm prophylaxis; PMI Nutrition International). Food and water were provided ad libitum. Because of sex-specific differences in iron homeostasis, only female mice were analyzed. Nonpregnant females were analyzed at 8 weeks of age. Pregnant females and their gestating litters were analyzed at gestational day 18.5. To date the onset of pregnancy, sexually mature females (7-11 weeks of age) were placed with a sexually mature male, and females were examined for the presence of a vaginal plug the following morning. Animals were photographed using an Olympus C-5060 Wide Zoom digital camera.
Iron analysis in blood and tissues
Under Avertin anesthesia (Sigma-Aldrich), whole blood was collected from the retro-orbital sinus through heparinized capillary tubes (Fisher Scientific) into ethylenediaminetetraacetic acid–coated microtainer tubes (Becton Dickinson). Complete blood counts were performed on a CellDyn 3700 (Abbott) by the Duke University Medical Center Veterinary Diagnostics Laboratory. Using blood collected through untreated capillary tubes (Fisher) into serum separator tubes (Becton Dickinson), serum was prepared, and serum iron levels were determined using a serum iron/UIBC kit (Thermo DMA) according to the manufacturer's instructions. Nonheme iron concentrations of liver, spleen, heart, pancreas, and skeletal muscle (quadriceps femoris) were determined as previously described.30,31
Histologic analysis
Tissue samples of liver and spleen were fixed in 10% buffered formalin for 24 hours, transferred to 70% ethanol, and embedded in paraffin. Tissue sections (8-micrometer each) were deparaffinized and stained with either Giemsa or Perls Prussian blue by the Duke University Medical Center Research Histology Laboratory. Brightfield images from Giemsa- and Perls-stained tissues were acquired at room temperature using the 4×/0.13 NA, 10×/0.30 NA, or 20×/0.50 NA objective lens of an Eclipse E600 microscope (Nikon) with attached Spot RT Digital Camera from Diagnostic Instruments. Images were captured using Spot 4.5 software from Diagnostic Instruments. The whole image was adjusted for brightness and contrast to match the original slide and labeled using Adobe Photoshop CS3 with no further modifications. Three animals per genotype were examined, yielding similar results.
RNA extraction, reverse-transcription PCR, and quantitative reverse-transcription PCR
Total liver RNA was isolated from flash-frozen tissue using TRIzol (Invitrogen) and reverse transcribed as previously described.32 Expression of a partial Tmprss6 transcript was sought by PCR using primer sets in which the forward and reverse primers were located in different exons of the Tmprss6 gene. Quantitative PCR to assess Tmprss6 transcript abundance was performed using forward primer 5′-GAGGCAGAAGTACAACCGAC-3′, located within exon 10, and reverse primer 5′-CTGGGAGGTGAAGTTGATGG-3′, located within exon 11. Quantitative PCR to assess hepcidin,32 Id1,33 and Bmp633 transcript abundance was performed as previously described.
Statistical analysis
Unpaired, 2-tailed Student t tests were performed in Microsoft Excel:mac 2008 software. P values less than .05 were considered statistically significant.
Results
Phenotype of Tmprss6tm1Dgen mice
We obtained mice from Deltagen that harbor a targeted disruption of the Tmprss6 locus (Tmprss6tm1Dgen) on a C57BL/6N genetic background. The targeting construct disrupts the genomic region encoding 74 amino acids within the Tmprss6 protease domain, including Asp668, 1 of the 3 amino acids (serine, aspartate, histidine) located within the serine protease active site that is directly involved in catalysis. By disrupting the catalytic triad, this targeting construct is thus predicted to result in a loss of protease activity.
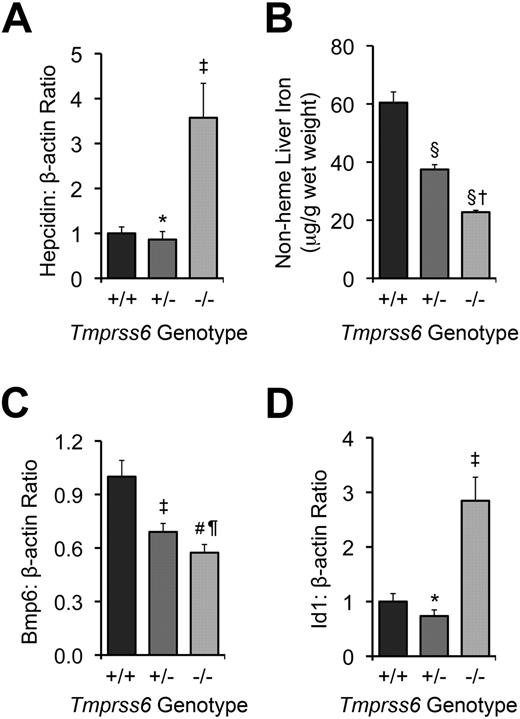
We intercrossed mice heterozygous for the Tmprss6tm1Dgen allele (hereafter referred to as Tmprss6+/− mice) to generate Tmprss6+/+, Tmprss6+/−, and Tmprss6−/− offspring for phenotypic characterization. Tmprss6−/− pups were smaller than littermate controls and underrepresented compared with expected Mendelian genotype frequencies (∼ 63% of expected number; χ2 = 4.079, 2 degrees of freedom, P = .13), consistent with decreased viability during the gestational and/or neonatal periods. Tmprss6−/− mice developed truncal alopecia before weaning (data not shown), and at 8 weeks of age, showed microcytic anemia, decreased serum iron and transferrin saturation, and increased hepatic hepcidin mRNA (Table 1; Figure 1A). These results demonstrate that mice homozygous for the Tmprss6tm1Dgen allele are phenotypically similar to previously reported Tmprss6 mutants.20,22
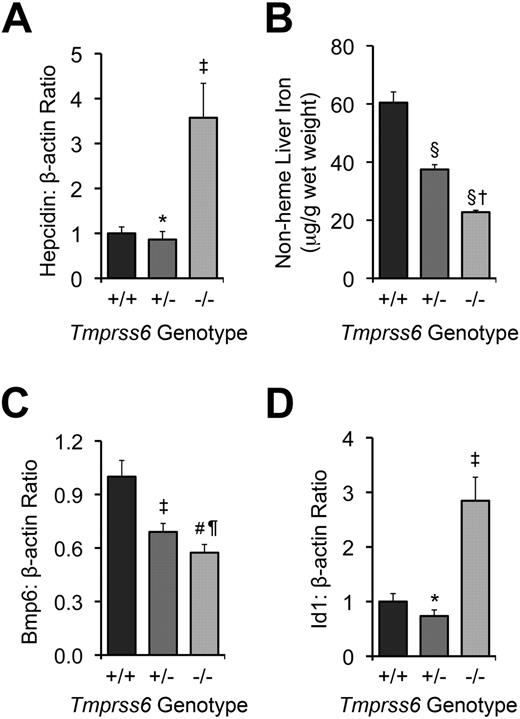
Tmprss6−/− and >Tmprss6+/− mice show abnormal systemic iron homeostasis. (A) Hepatic hepcidin (Hamp) mRNA expression relative to β-actin (Actb) mRNA expression in Tmprss6+/+, Tmprss6+/−, and Tmprss6−/− mice. (B) Nonheme liver iron concentrations (μg/g wet weight) of Tmprss6+/+, Tmprss6+/−, and Tmprss6−/− mice. (C) Hepatic Bmp6 mRNA expression relative to β-actin mRNA expression in Tmprss6+/+, Tmprss6+/−, and Tmprss6−/− mice. (D) Hepatic Id1 mRNA expression relative to β-actin mRNA expression in Tmprss6+/+, Tmprss6+/−, and Tmprss6−/− mice. Mean values from analysis of 8-week-old female mice (6-8 mice per genotype) are graphed. In panels A, C, and D, mRNA expression ratios are normalized to a Tmprss6+/+ mean value of 1. Error bars represent SEM. ‡P < .05, #P < .005, §P < .001, and *P value not significant, compared with Tmprss6+/+ mice. †P < .001, ¶P value not significant, compared with Tmprss6+/− mice.
Tmprss6−/− and >Tmprss6+/− mice show abnormal systemic iron homeostasis. (A) Hepatic hepcidin (Hamp) mRNA expression relative to β-actin (Actb) mRNA expression in Tmprss6+/+, Tmprss6+/−, and Tmprss6−/− mice. (B) Nonheme liver iron concentrations (μg/g wet weight) of Tmprss6+/+, Tmprss6+/−, and Tmprss6−/− mice. (C) Hepatic Bmp6 mRNA expression relative to β-actin mRNA expression in Tmprss6+/+, Tmprss6+/−, and Tmprss6−/− mice. (D) Hepatic Id1 mRNA expression relative to β-actin mRNA expression in Tmprss6+/+, Tmprss6+/−, and Tmprss6−/− mice. Mean values from analysis of 8-week-old female mice (6-8 mice per genotype) are graphed. In panels A, C, and D, mRNA expression ratios are normalized to a Tmprss6+/+ mean value of 1. Error bars represent SEM. ‡P < .05, #P < .005, §P < .001, and *P value not significant, compared with Tmprss6+/+ mice. †P < .001, ¶P value not significant, compared with Tmprss6+/− mice.
Because the Tmprss6tm1Dgen allele disrupts only a portion of the Tmprss6 genomic locus, we asked whether an abnormal form of the Tmprss6 mRNA might still be expressed in Tmprss6−/− mice. Using complementary DNA prepared from Tmprss6−/− liver as template, we detected transcription of Tmprss6 exons located 5′ to the targeted region. However, we could not detect transcription of Tmprss6 exons located 3′ to the targeted region, confirming targeting of the Tmprss6 genomic locus. Quantitative PCR analysis demonstrated that the abnormal Tmprss6 transcript present in Tmprss6−/− mice was expressed at approximately 20% of wild-type levels.
Effect of Tmprss6 deficiency on tissue iron stores
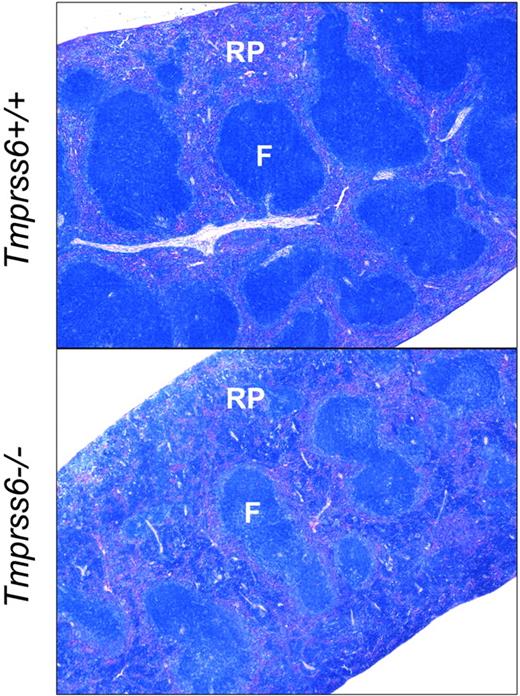
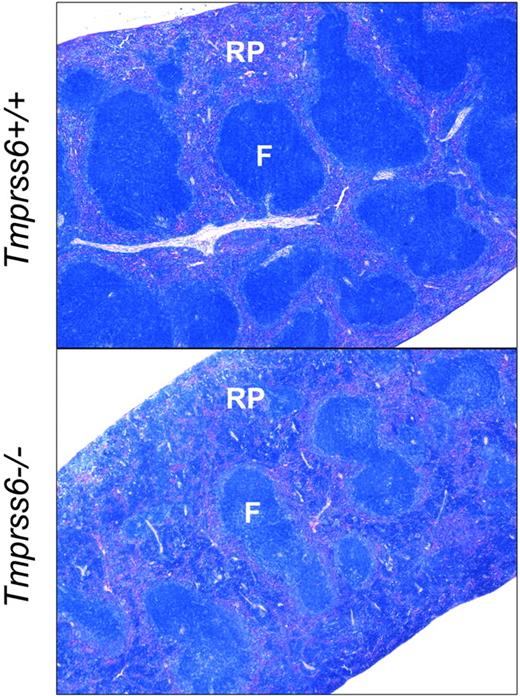
We next evaluated the effect of loss of Tmprss6 function on tissue iron stores of 8-week-old female mice. Hepatic nonheme iron concentration, reflecting the total body iron endowment, was significantly lower in Tmprss6−/− mice compared with littermate controls (Figure 1B), whereas splenic nonheme iron concentration, which primarily reflects macrophage iron stores generated by the recycling of hemoglobin from senescent erythrocytes, was similar between genotypes (Table 2). However, Tmprss6−/− mice showed a trend toward increased spleen weight (P = .20 vs Tmprss6+/+), due to increased splenic hematopoiesis (Figure 2), yielding a trend toward increased total splenic nonheme iron content (P = .14 vs Tmprss6+/+). These findings suggest that inappropriately elevated hepcidin expression in Tmprss6−/− mice results in chronically impaired uptake of dietary iron, reflected in decreased hepatic iron stores, as well as chronically impaired mobilization of iron from splenic macrophage stores in the setting of iron deficiency.
Tmprss6−/− mice show increased extramedullary hematopoiesis. Giemsa-stained histologic sections of spleens from Tmprss6+/+ (top) and Tmprss6−/− (bottom) mice. RP indicates splenic red pulp; and F, lymphoid follicle. Original magnification ×40.
Tmprss6−/− mice show increased extramedullary hematopoiesis. Giemsa-stained histologic sections of spleens from Tmprss6+/+ (top) and Tmprss6−/− (bottom) mice. RP indicates splenic red pulp; and F, lymphoid follicle. Original magnification ×40.
Effect of Tmprss6 deficiency on Bmp/Smad signaling
We next sought to determine whether the elevated hepcidin mRNA levels present in Tmprss6−/− mice correlate with dysregulated Bmp/Smad signaling. Hepatic mRNA levels of Bmp6, a Bmp ligand recently identified as a key regulator of hepcidin expression in vivo,17,18 were significantly lower in Tmprss6−/− mice than Tmprss6+/+ mice (Figure 1C). These findings are compatible with a previous report demonstrating that hepatic Bmp6 mRNA expression correlates with dietary iron content and tissue iron burden in wild-type mice of C57BL/6 and DBA/2 genetic backgrounds.33 In contrast, hepatic levels of Id1, a transcriptional target of Bmp6 signaling,17,18 were significantly increased in Tmprss6−/− mice compared with littermate controls (Figure 1D). Although we did not measure Bmp6 protein levels directly, the finding that Tmprss6−/− mice showed decreased Bmp6 mRNA levels but increased Id1 mRNA levels compared with Tmprss6+/+ mice suggests that the elevated Bmp/Smad signaling in Tmprss6−/− mice does not result from increased input of the Bmp6 ligand but rather a failure to dampen Bmp/Smad signaling at a point genetically downstream of the Bmp6 ligand. In addition, the decreased Bmp6 hepatic mRNA levels detected in Tmprss6−/− mice, which have decreased hepatic stores, suggest that Tmprss6 is not required for the transcriptional regulation of Bmp6 by iron.
Heterozygous loss of Tmprss6 causes iron deficiency
We next sought to determine whether heterozygous loss of Tmprss6 function alters systemic iron homeostasis. When analyzed at 8 weeks of age, Tmprss6+/− mice showed trends toward decreased serum iron and transferrin saturation compared with Tmprss6+/+ controls (P = .13 and P = .08, respectively; Table 1). Although Tmprss6+/− mice maintained a normal hematocrit, they nevertheless showed small but significant reductions in erythrocyte mean corpuscular volume and mean corpuscular hemoglobin, suggesting that the availability of iron for erythropoiesis was slightly restricted (Table 1). Tmprss6+/− splenic nonheme iron concentration was similar to that of Tmprss6+/+ mice (Table 2). However, Tmprss6+/− hepatic nonheme iron concentration, although significantly greater than that of Tmprss6−/− mice, was also significantly lower than that of Tmprss6+/+ mice (Figure 1B). Together, these findings suggest that mice cannot fully compensate for heterozygous loss of Tmprss6 to achieve normal systemic iron homeostasis.
As expected in the setting of decreased hepatic iron stores, hepatic Bmp6 mRNA levels were significantly lower in Tmprss6+/− mice compared with Tmprss6+/+ mice (Figure 1C). However, Tmprss6+/− mice showed hepatic hepcidin mRNA levels that were similar to those of Tmprss6+/+ controls (Figure 1A). Thus, Tmprss6+/− hepcidin mRNA levels, although similar to Tmprss6+/+ levels, were nevertheless inappropriately elevated relative to their lower hepatic iron stores. Hepatic levels of Id1 mRNA were also similar in Tmprss6+/− and Tmprss6+/+ mice (Figure 1D). Because Tmprss6+/− mice express lower levels of Bmp6 mRNA, we propose that decreased Bmp6 signaling input, combined with a decreased ability to down-regulate Bmp/Smad signaling due to Tmprss6 haploinsufficiency, might result in net hepcidin and Id1 levels that are similar to wild-type levels. At younger ages, when iron demands for body growth and the accompanying expansion of the red cell mass are increased, the hepcidin regulatory defect present in Tmprss6+/− mice might be more pronounced, leading to the development of the decreased hepatic iron stores we observed in the 8-week-old mice. Interestingly, in humans with iron-refractory, iron deficiency anemia (IRIDA) due to homozygous TMPRSS6 mutations, the severity of the anemia has been shown to improve with age, consistent with the hypothesis that the phenotypic effects of Tmprss6 deficiency are more severe at younger ages.24
Effect of heterozygous loss of Tmprss6 on response to increased iron demands
The finding that mice with heterozygous loss of Tmprss6 showed decreased iron stores compared with wild-type controls raised the possibility that Tmprss6+/− mice might display impaired regulation of systemic iron homeostasis in settings where iron demands are increased. To investigate this possibility, we compared parameters of iron homeostasis in pregnant Tmprss6+/+ and Tmprss6+/− females at gestational day 18.5, a time when maternal iron absorption is increased in response to increased iron needs.34 To control for possible effects of fetal genotypes on maternal iron status, Tmprss6+/− females were mated with Tmprss6+/+ males, and Tmprss6+/+ females were mated with Tmprss6+/− males, so that each gestating litter would contain fetuses of both Tmprss6+/+ and Tmprss6+/− genotypes in approximately equal proportions.
At gestational day 18.5, both Tmprss6+/+ and Tmprss6+/− dams showed significant reductions in liver nonheme iron concentration compared with nonpregnant females of the corresponding genotypes (Table 3), consistent with mobilization of hepatic iron stores to support the increased iron demands of pregnancy. Compared with Tmprss6+/+ dams, Tmprss6+/− dams showed spleen nonheme iron concentrations that were slightly lower and hepcidin mRNA levels that were slightly higher; however, these differences did not reach statistical significance. In both Tmprss6+/+ and Tmprss6+/− pregnant females, hepatic hepcidin mRNA levels were, however, markedly reduced compared with nonpregnant levels, demonstrating that mice with a single wild-type Tmprss6 allele retain the ability to reduce hepcidin expression. Pregnant Tmprss6+/+ and Tmprss6+/− dams displayed blood hemoglobin levels, serum iron, and serum transferrin saturation levels that were similar to nonpregnant genotype-matched controls (Table 4).
Interestingly, although maternal parameters of iron homeostasis were not significantly different between pregnant Tmprss6+/+ and Tmprss6+/− females, the Tmprss6 genotype of a pregnant female did have a significant effect on the liver iron stores of her gestating fetuses. The mean liver nonheme iron concentration of fetuses from Tmprss6+/− dams was significantly lower than fetuses of Tmprss6+/+ dams (mean ± SEM of 75 ± 2 μg/g wet weight for fetuses [n = 42] of Tmprss6+/− dams vs 85 ± 2 μg/g wet weight for fetuses [n = 36] of Tmprss6+/+ dams; P = .007). Because nonpregnant Tmprss6+/− females showed significantly lower hepatic iron stores than nonpregnant Tmprss6+/+ females when measured at 8 weeks of age, the decreased hepatic iron stores in fetuses of Tmprss6+/− females may reflect the influence of lower maternal hepatic iron stores at the onset of pregnancy and/or subtle differences in iron homeostasis during pregnancy.
Tmprss6 regulates systemic iron homeostasis in a hemojuvelin-dependent manner
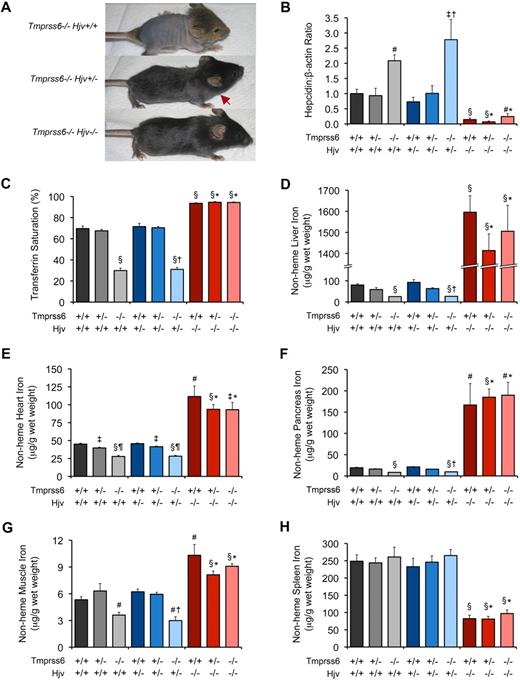
To further characterize the relationship between Tmprss6 and the Bmp/Smad signaling pathway, we asked whether iron deficiency in Tmprss6−/− mice is dependent on the presence of functional Hjv. By breeding Tmprss6+/− mice to Hjv−/− mice, a model of juvenile hemochromatosis present on a 129S6/SvEvTac genetic background,13 we generated F1 Tmprss6+/−Hjv+/− mice, which we then intercrossed to yield F2 mice of all Tmprss6-Hjv genotype combinations for phenotypic characterization. Tmprss6−/−Hjv+/+ pups of mixed genetic background developed overt truncal alopecia, similar to Tmprss6−/− mice of C57BL/6N background, however Tmprss6−/−Hjv+/− pups showed less dense hair growth on the trunk, and Tmprss6−/−Hjv−/− mice retained full body hair (Figure 3A). As hair loss in Tmprss6-deficient mice results from iron deficiency,20,22 these results suggest that Hjv activity is required for the development of iron deficiency in Tmprss6−/− mice. Of note, in contrast to Tmprss6−/− pups of C57BL/6N genetic background, F2 Tmprss6−/−Hjv+/+ pups of C57BL/6N;129S6/SvEvTac mixed genetic background were indeed represented in the expected Mendelian genotype frequencies. This improvement in survival can likely be attributed to the difference in genetic background, as the 129S6/SvEvTac inbred strain has been shown to exhibit higher hepatic and splenic iron concentrations than inbred C57BL substrains.35
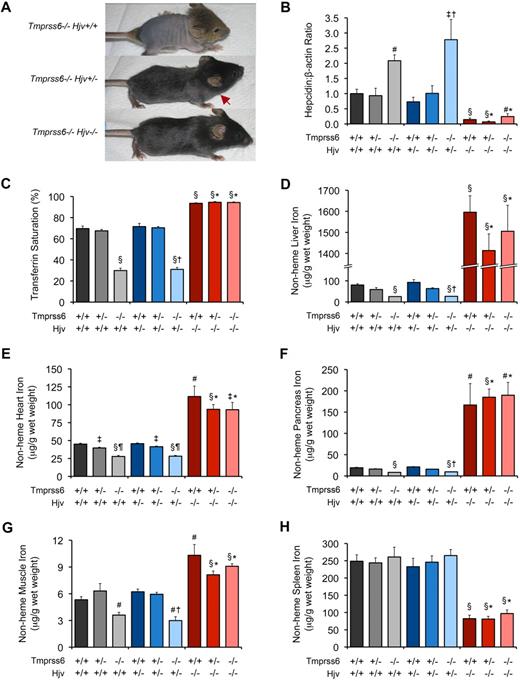
Tmprss6−/−Hjv−/− and Tmprss6+/+Hjv−/− mice show similar hepcidin mRNA levels and tissue iron concentrations. (A) Effect of Hjv genotype on iron deficiency–related hair loss in 19-day-old Tmprss6-deficient littermates. Tmprss6−/−Hjv+/+ pups (top) develop overt alopecia before weaning age. Tmprss6−/−Hjv+/− mice (middle) exhibit normal hair growth on the head but less dense hair on the body; red arrow indicates hair growth transition zone. Tmprss6−/−Hjv−/− mice (bottom) show normal body hair. (B-F) Phenotypic analysis of 8-week-old female mice of all Tmprss6-Hjv genotype combinations. Graphed are mean values obtained from analyses of hepcidin (Hamp) mRNA expression relative to β-actin (Actb) mRNA expression (B), serum transferrin saturation (C), and nonheme iron concentrations of liver (D), heart (E), pancreas (F), quadriceps muscle (G), and spleen (H). For each parameter, 6 to 8 female mice per genotype were analyzed. For panel B, mRNA expression ratios are normalized to a Tmprss6+/+Hjv+/+ mean value of 1. Error bars represent SEM. §P < .001 compared with Tmprss6+/+Hjv+/+; #P < .005 compared with Tmprss6+/+Hjv+/+; ‡P < .05 compared with Tmprss6+/+Hjv+/+; †P value not significant compared with Tmprss6−/−Hjv+/+; *P value not significant compared with Tmprss6+/+Hjv−/−; and ¶P < .001 compared with Tmprss6+/−Hjv+/+ mice.
Tmprss6−/−Hjv−/− and Tmprss6+/+Hjv−/− mice show similar hepcidin mRNA levels and tissue iron concentrations. (A) Effect of Hjv genotype on iron deficiency–related hair loss in 19-day-old Tmprss6-deficient littermates. Tmprss6−/−Hjv+/+ pups (top) develop overt alopecia before weaning age. Tmprss6−/−Hjv+/− mice (middle) exhibit normal hair growth on the head but less dense hair on the body; red arrow indicates hair growth transition zone. Tmprss6−/−Hjv−/− mice (bottom) show normal body hair. (B-F) Phenotypic analysis of 8-week-old female mice of all Tmprss6-Hjv genotype combinations. Graphed are mean values obtained from analyses of hepcidin (Hamp) mRNA expression relative to β-actin (Actb) mRNA expression (B), serum transferrin saturation (C), and nonheme iron concentrations of liver (D), heart (E), pancreas (F), quadriceps muscle (G), and spleen (H). For each parameter, 6 to 8 female mice per genotype were analyzed. For panel B, mRNA expression ratios are normalized to a Tmprss6+/+Hjv+/+ mean value of 1. Error bars represent SEM. §P < .001 compared with Tmprss6+/+Hjv+/+; #P < .005 compared with Tmprss6+/+Hjv+/+; ‡P < .05 compared with Tmprss6+/+Hjv+/+; †P value not significant compared with Tmprss6−/−Hjv+/+; *P value not significant compared with Tmprss6+/+Hjv−/−; and ¶P < .001 compared with Tmprss6+/−Hjv+/+ mice.
To directly examine the effect of Hjv deficiency on systemic iron homeostasis in Tmprss6−/− mice, we compared red blood cell parameters, hepatic hepcidin mRNA levels, serum transferrin saturation, and tissue nonheme iron concentrations in mice of all Tmprss6-Hjv genotype combinations at 8 weeks of age. Tmprss6−/−Hjv+/+ mice showed microcytic anemia, and compared with Tmprss6+/+Hjv+/+ controls, showed elevated hepatic hepcidin mRNA, decreased transferrin saturation, and decreased nonheme iron concentrations of liver, heart, pancreas, and skeletal muscle (Table 5; Figure 3B-G). Although Tmprss6−/−Hjv+/+ mice showed greater hair loss than Tmprss6−/−Hjv+/− as pups (Figure 3A), these genotypes showed no significant differences in parameters of iron homeostasis measured at 8 weeks of age. Splenic nonheme iron concentrations in Tmprss6−/−Hjv+/+ and Tmprss6−/−Hjv+/− mice were similar to Tmprss6+/+Hjv+/+ levels (Figure 3H).
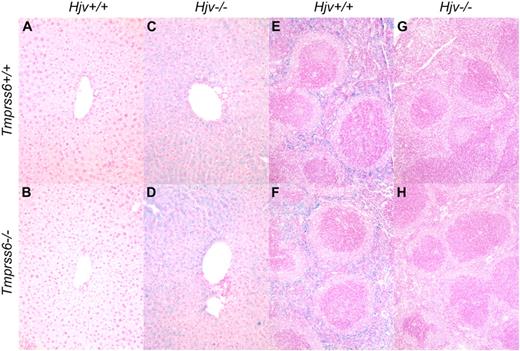
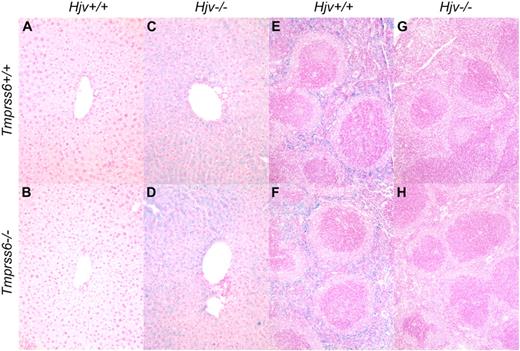
Consistent with a prior study of Hjv−/− mice,13 Tmprss6+/+Hjv−/− mice showed normal red blood cell parameters and hepcidin mRNA levels that were significantly decreased compared with Tmprss6+/+Hjv+/+ controls (Table 5; Figure 3B). Interestingly, Tmprss6+/−Hjv−/− and Tmprss6−/−Hjv−/− mice also showed normal red blood cell parameters, and their hepcidin mRNA levels were decreased to Tmprss6+/+Hjv−/− levels. As expected in the setting of low hepcidin, Tmprss6+/+Hjv−/−, Tmprss6+/−Hjv−/−, and Tmprss6−/−Hjv−/− mice all showed transferrin saturations and nonheme iron concentrations of liver, heart, pancreas, and muscle that were significantly increased compared with Tmprss6+/+Hjv+/+ controls (Figure 3C-G). In contrast to Tmprss6+/+Hjv+/+ and Tmprss6−/−Hjv+/+ livers (Figure 4A-B), livers of Tmprss6+/+Hjv−/− and Tmprss6−/−Hjv−/− mice showed marked iron accumulation in hepatocytes (Figure 4C-D). As predicted in the setting of low hepcidin, Tmprss6+/+Hjv−/−, Tmprss6+/−Hjv−/−, and Tmprss6−/−Hjv−/− mice showed spleen nonheme iron concentrations that were significantly decreased compared with Tmprss6+/+Hjv+/+ controls (Figure 3H). Whereas plentiful iron was detected in splenic macrophages of Tmprss6+/+Hjv+/+ and Tmprss6−/−Hjv+/+ mice (Figure 4E-F), minimal iron was observed in splenic macrophages of both Tmprss6+/+Hjv−/− and Tmprss6−/−Hjv−/− mice (Figure 4G-H).
Tmprss6−/−Hjv−/− and Tmprss6+/+Hjv−/− mice show similar iron distribution in liver and spleen. Perls Prussian blue staining of histologic sections of liver (A-D) and spleen (E-H). Blue staining represents iron accumulation in cells. Tmprss6+/+Hjv+/+ (A) and Tmprss6−/−Hjv+/+ (B) mice lack histologic evidence of excess iron deposition, whereas Tmprss6+/+Hjv−/− (C) and Tmprss6−/−Hjv−/− (D) mice show marked iron accumulation in hepatocytes. Splenic macrophages of Tmprss6+/+Hjv+/+ (E) and Tmprss6−/−Hjv+/+ (F) mice show abundant iron, whereas minimal iron is detected in splenic macrophages of Tmprss6+/+Hjv−/− (G) and Tmprss6−/−Hjv−/− (H) mice. Original magnification ×200 (A-D) and ×100 (E-H).
Tmprss6−/−Hjv−/− and Tmprss6+/+Hjv−/− mice show similar iron distribution in liver and spleen. Perls Prussian blue staining of histologic sections of liver (A-D) and spleen (E-H). Blue staining represents iron accumulation in cells. Tmprss6+/+Hjv+/+ (A) and Tmprss6−/−Hjv+/+ (B) mice lack histologic evidence of excess iron deposition, whereas Tmprss6+/+Hjv−/− (C) and Tmprss6−/−Hjv−/− (D) mice show marked iron accumulation in hepatocytes. Splenic macrophages of Tmprss6+/+Hjv+/+ (E) and Tmprss6−/−Hjv+/+ (F) mice show abundant iron, whereas minimal iron is detected in splenic macrophages of Tmprss6+/+Hjv−/− (G) and Tmprss6−/−Hjv−/− (H) mice. Original magnification ×200 (A-D) and ×100 (E-H).
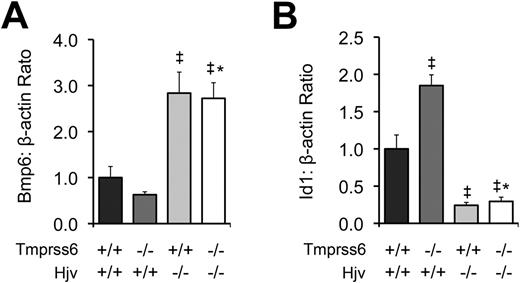
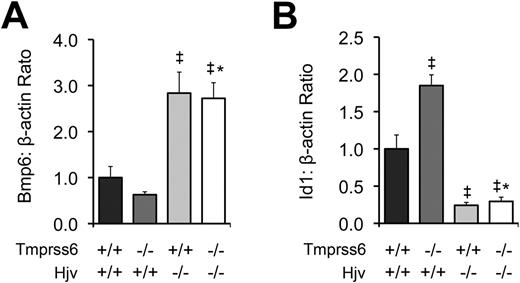
To further characterize the relationship between Tmprss6 and the Bmp/Smad pathway, we measured hepatic mRNA levels of Bmp6 and Id1 in F2 mice of selected Tmprss6-Hjv genotype combinations at 8 weeks of age. As predicted from our prior comparison of Tmprss6−/− and Tmprss6+/+ mice of C57BL/6N background (Figure 1C), Tmprss6−/−Hjv+/+ mice of mixed genetic background showed lower mean Bmp6 mRNA levels than Tmprss6+/+Hjv+/+ controls (Figure 5A). In contrast, both Tmprss6+/+Hjv−/− and Tmprss6−/−Hjv−/− mice showed Bmp6 levels that were significantly higher than Tmprss6+/+Hjv+/+ controls. As again predicted from our prior comparison of Tmprss6−/− and Tmprss6+/+ mice (Figure 1D), Tmprss6−/−Hjv+/+ mice showed significantly higher Id1 mRNA levels than Tmprss6+/+Hjv+/+ controls (Figure 5B). However, both Tmprss6+/+Hjv−/− and Tmprss6−/−Hjv−/− mice showed Id1 levels that were significantly lower than Tmprss6+/+Hjv+/+ controls. The finding that mice lacking Hjv show markedly decreased hepcidin and Id1 mRNA accompanied by systemic iron overload, regardless of their Tmprss6 genotype, together with the finding that Hjv is required for the development of elevated hepcidin mRNA, elevated Id1 mRNA, and systemic iron deficiency in mice lacking Tmprss6, suggests that Tmprss6 negatively regulates hepcidin expression, and thus regulates systemic iron homeostasis, by acting genetically upstream of Hjv in the Bmp/Smad signaling pathway. In addition, the elevated hepatic Bmp6 mRNA detected in Tmprss6+/+Hjv−/− and Tmprss6−/−Hjv−/− mice, both of which show increased hepatic iron stores, suggests that Hjv is not required for the transcriptional regulation of Bmp6 by iron.
Tmprss6−/−Hjv−/− and Tmprss6+/+Hjv−/− mice show similar Bmp6 and Id1 levels. (A) Hepatic Bmp6 mRNA expression relative to β-actin mRNA expression in Tmprss6+/+Hjv+/+, Tmprss6−/−Hjv+/+, Tmprss6+/+Hjv−/−, and Tmprss6−/−Hjv−/− mice. (B) Hepatic Id1 mRNA expression relative to β-actin mRNA expression in Tmprss6+/+Hjv+/+, Tmprss6−/−Hjv+/+, Tmprss6+/+Hjv−/−, and Tmprss6−/−Hjv−/− mice. Mean values from analysis of 8-week-old female mice are graphed. For each parameter, 4 female mice per genotype were analyzed. Error bars represent SEM. ‡P < .05 compared with Tmprss6+/+Hjv+/+; *P value not significant compared with Tmprss6+/+Hjv−/−.
Tmprss6−/−Hjv−/− and Tmprss6+/+Hjv−/− mice show similar Bmp6 and Id1 levels. (A) Hepatic Bmp6 mRNA expression relative to β-actin mRNA expression in Tmprss6+/+Hjv+/+, Tmprss6−/−Hjv+/+, Tmprss6+/+Hjv−/−, and Tmprss6−/−Hjv−/− mice. (B) Hepatic Id1 mRNA expression relative to β-actin mRNA expression in Tmprss6+/+Hjv+/+, Tmprss6−/−Hjv+/+, Tmprss6+/+Hjv−/−, and Tmprss6−/−Hjv−/− mice. Mean values from analysis of 8-week-old female mice are graphed. For each parameter, 4 female mice per genotype were analyzed. Error bars represent SEM. ‡P < .05 compared with Tmprss6+/+Hjv+/+; *P value not significant compared with Tmprss6+/+Hjv−/−.
Discussion
Recent studies in mice and humans have demonstrated that the transmembrane serine protease TMPRSS6 plays a key role in the maintenance of systemic iron homeostasis.20-24 In both species, TMPRSS6 mutation results in iron deficiency anemia accompanied by inappropriately elevated hepcidin levels, suggesting that the normal function of TMPRSS6 is to repress hepcidin expression in response to iron deficiency. We demonstrate here that although mice homozygous for a targeted disruption of Tmprss6 (Tmprss6tm1Dgen) display decreased hepatic mRNA levels of Bmp6, a key endogenous regulator of hepcidin expression, they display elevated hepatic mRNA levels of Id1, a transcriptional target of Bmp6 signaling. We also show that the elevated hepatic hepcidin and Id1 mRNA levels present in Tmprss6−/− mice are dependent on the presence of functional Hjv. Together, these findings suggest that Tmprss6 acts genetically upstream of Hjv to down-regulate Bmp/Smad signaling in the liver. We thus propose that loss of Tmprss6 function results in excess Bmp signaling through hemojuvelin, leading to elevated Id1 and hepcidin levels, the latter of which promotes systemic iron deficiency by reducing dietary iron absorption. By placing Tmprss6 genetically upstream of Hjv in hepcidin signaling, our results are compatible with the recent finding that Tmprss6 inhibits hepcidin activation by cleaving the plasma membrane–bound form of Hjv.27 Together, these findings raise the possibility that selective mutations in HJV, which alter its ability to interact with TMPRSS6 yet preserve its ability to facilitate BMP signaling, might also lead to the IRIDA phenotype.
In addition to Tmprss6, other negative regulators of hepcidin expression have been described. Hypoxia-inducible transcription factor-1α (HIF-1α), a regulatory subunit of hypoxia-inducible factor, has been shown to bind the hepcidin promoter in liver, and mice harboring a liver-specific deletion of HIF-1α failed to fully down-regulate hepcidin expression when fed an iron-deficient diet.36 Although these results suggested that HIF-1α may contribute to hepcidin repression in the setting of iron deficiency, another study failed to replicate these findings,37 and thus the role of HIF-1α in hepcidin regulation remains to be clarified. A soluble form of HJV (sHjv), generated by furin-mediated cleavage of the glycosylphosphatidylinositol-linked form of HJV, appears to down-regulate hepcidin expression by sequestering BMPs.38-40 Accordingly, administration of recombinant soluble Hjv suppresses hepcidin expression in mice.41 Our results raise the possibility that exogenous administration of agents that down-regulate BMP signaling, such as recombinant sHJV, might be effective at reducing hepcidin expression in IRIDA.
The phenotype of mice homozygous for the Tmprss6tm1Dgen allele reported here appears to differ slightly from that of a Tmprss6 knockout mouse model previously reported by Folgueras et al.22 When intercrossing mice heterozygous for the Tmprss6tm1Dgen allele, we found that Tmprss6−/− pups were underrepresented among live offspring compared with expected Mendelian genotype frequencies. In contrast, decreased viability was not observed in the Tmprss6 knockout mice studied by Folgueras et al, which were of mixed C57BL/6;129/Ola genetic background. We propose that this phenotypic difference likely reflects differences in the genetic background harboring the mutated Tmprss6 allele. In support of this hypothesis, we found that the viability of Tmprss6tm1Dgen homozygotes was fully restored after transfer to the C57BL/6N;129S6/SvEvTac mixed genetic background. As the iron content of the rodent chow used in the studies of Folgueras et al was not specified, we cannot exclude the additional possibility that dietary iron content may have also contributed to this phenotypic difference. Viability was not discussed in the report by Du et al20 describing the Tmprss6msk/msk mouse, which harbors a chemically induced splice site mutation that disrupts the Tmprss6 catalytic domain.
Although previous reports of Tmprss6 mouse models described phenotypic differences between wild-type mice and mice homozygous for a Tmprss6 mutation,20,22 we also observed several phenotypic differences between Tmprss6+/− mice and Tmprss6+/+ controls. Specifically, we found that mice harboring 1 defective Tmprss6 allele had moderately decreased liver iron concentrations as well as laboratory signs of iron-restricted erythropoiesis, demonstrating that normal murine systemic iron homeostasis could not be achieved through activity of a single wild-type Tmprss6 allele. These results raise the possibility that human heterozygous carriers of TMPRSS6 mutations may harbor subtle abnormalities of iron homeostasis that increase their risk for developing iron deficiency in response to certain physiologic stresses, such as pregnancy, decreased dietary intake, or inflammation. Although the frequency of such pathogenic TMPRSS6 mutations is likely to be extremely low, common sequence variants in the TMPRSS6 gene, including missense polymorphisms, have also been described in multiple ethnic populations. Indeed, several recent genome-wide association studies conducted in human populations have demonstrated associations between single nucleotide polymorphisms in the TMPRSS6 gene and parameters of systemic iron homeostasis.42-46 Although the results of these genome-wide association studies do not prove that TMPRSS6 polymorphisms predispose persons to overt iron deficiency, these findings nevertheless demonstrate that common genetic variants in TMPRSS6 impact iron homeostasis. Interestingly, in our studies reported here, we found that although mice with heterozygous loss of Tmprss6 were indeed able to markedly down-regulate their hepcidin expression during pregnancy, their fetuses displayed significantly lower hepatic iron stores than fetuses of wild-type females.
During the revision of this paper, Truksa et al47 reported a comparison of parameters of systemic iron homeostasis between the Tmprss6msk/msk mice (C57BL/6J genetic background), the Hjv−/−(Hfe2tm1Nca/tm1Nca) mice generated by our laboratory (129S6/SvEvTac genetic background), and double mutants of mixed genetic background generated by crossing these 2 strains.47 Using data pooled from male and female mice, these authors found that Tmprss6msk/msk mice lacking Hjv showed lower cardiac nonheme iron concentrations than the parental Hjv−/− strain; based on this result, the authors postulated that genetic loss of Tmprss6 was cardioprotective in regard to iron loading. In contrast, in our comparison of female F2 Tmprss6+/+Hjv−/− and Tmprss6−/−Hjv−/− mice of mixed C57BL/6N;129S6/SvEvTac genetic background, Tmprss6 genotype had no effect on cardiac iron concentration in Hjv−/− mice. We also observed no effect of Tmprss6 genotype on cardiac iron concentration in a separate analysis of male F2 mice (data not shown). Interestingly, however, the cardiac iron concentrations in the F2 Tmprss6+/+Hjv−/− mice of mixed C57BL/6N;129S6/SvEvTac background appeared lower than concentrations previously reported in Hjv−/− mice of 129S6/SvEvTac genetic background.13 For this reason, we conclude that genetic loss of Tmprss6 does not afford a cardioprotective effect in regard to iron loading.
In the study by Truksa et al,47 spleen iron levels were found to be significantly lower in Tmprss6msk/msk mutant mice of C57BL/6J background compared with C57BL/6J controls. In contrast, using sex- and age-matched littermate controls, we found that Tmprss6tm1Dgen/tm1Dgen female mice of the closely related C57BL/6N genetic background showed a trend toward higher splenic iron concentrations compared with Tmprss6+/+ littermates (Table 2). In a separate analysis of 8-week-old Tmprss6tm1Dgen/tm1Dgen male mice, we found that the difference in spleen iron concentrations between Tmprss6tm1Dgen/tm1Dgen and Tmprss6+/+ littermate controls reached statistical significance (mean ± SEM 120 ± 15 μg/g wet weight in Tmprss6tm1Dgen/tm1Dgen [n = 7 mice] vs 85 ± 9 μg/g wet weight in Tmprss6+/+ mice [n = 8 mice]; P < .05). We suggest that this phenotypic difference may reflect that fact that our study used littermate controls, which control for effects of maternal genotype on the resulting tissue iron content of the offspring.
Like HJV, other gene products mutated in hereditary forms of hemochromatosis also participate in hepcidin signaling. Hfe, which competes with transferrin for binding to transferrin receptor 1, induces hepcidin expression when not in complex with transferrin receptor 1.32 Interaction of HFE with transferrin receptor 2 is required for the induction of hepcidin by holotransferrin.48 Whether TMPRSS6 modulates the activity of HFE and/or transferrin receptor 2 remains to be determined.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Kristina Roberts, Tom Bartnikas, and Paul Schmidt for helpful discussions, and Dean Campagna for technical assistance.
This work was supported by a Cooley's Anemia Foundation Research Fellowship (K.E.F.) and by National Institutes of Health grants K08 DK084204 (K.E.F.) and R01 DK066373 (N.C.A.).
National Institutes of Health
Authorship
Contribution: K.E.F. designed and performed research, analyzed results, and wrote the paper; R.L.W. performed research, analyzed results, and assisted with preparation of the paper; and M.D.F. and N.C.A. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Karin E. Finberg, Duke University Medical Center, Department of Pathology, Box 3712, Durham, NC 27710; e-mail: karin.finberg@duke.edu.