Key Points
iAMP21-ALL comprises multiple subgroups with distinct patterns of chromosome 21 structural variation but a common region of amplification.
Single-cell and molecular timing analyses demonstrate early acquisition of the iAMP21 chromosome followed by additional alterations.
Abstract
Intrachromosomal amplification of chromosome 21 defines a subtype of high-risk childhood acute lymphoblastic leukemia (iAMP21-ALL) characterized by copy number changes and complex rearrangements of chromosome 21. The genomic basis of iAMP21-ALL and the pathogenic role of the region of amplification of chromosome 21 to leukemogenesis remains incompletely understood. In this study, using integrated whole genome and transcriptome sequencing of 124 patients with iAMP21-ALL, including rare cases arising in the context of constitutional chromosomal aberrations, we identified subgroups of iAMP21-ALL based on the patterns of copy number alteration and structural variation. This large data set enabled formal delineation of a 7.8 Mb common region of amplification harboring 71 genes, 43 of which were differentially expressed compared with non-iAMP21–ALL ones, including multiple genes implicated in the pathogenesis of acute leukemia (CHAF1B, DYRK1A, ERG, HMGN1, and RUNX1). Using multimodal single-cell genomic profiling, including single-cell whole genome sequencing of 2 cases, we documented clonal heterogeneity and genomic evolution, demonstrating that the acquisition of the iAMP21 chromosome is an early event that may undergo progressive amplification during disease ontogeny. We show that UV-mutational signatures and high mutation load are characteristic secondary genetic features. Although the genomic alterations of chromosome 21 are variable, these integrated genomic analyses and demonstration of an extended common minimal region of amplification broaden the definition of iAMP21-ALL for more precise diagnosis using cytogenetic or genomic methods to inform clinical management.
Introduction
Recurrent cytogenetic abnormalities and cooperating genetic alterations define B-lineage acute lymphoblastic leukemia (B-ALL) subtypes and are used to diagnose and risk-stratify patients.1-7 B-ALL with intrachromosomal amplification of chromosome 21 (iAMP21-ALL)8,9 is defined by the presence of a grossly abnormal chromosome 21, harboring heterogeneous copy number changes and rearrangements.10-13 iAMP21-ALL is diagnosed via cytogenetics, fluorescence in situ hybridization (FISH), probes directed to the RUNX1 gene, or copy number profiling.14-17 Retrospective studies reported a high relapse rate of iAMP21-ALL with standard therapy,18,19 which was improved by more intensive treatments.20-22 In view of the potential therapy escalation for these patients, accurate diagnosis is critical.
Formation of the iAMP21 chromosome involves 1 or more breakage-fusion-bridge (BFB) cycles, followed by chromothripsis of chromosome 21.23 Rarely, iAMP21-ALL arises from the constitutional Robertsonian translocation between chromosomes 15 and 21, rob(15;21)c, or in carriers of a constitutional ring chromosome 21, r(21)c.24 In carriers of rob(15;21)c, the missegregation of the dicentric Robertsonian chromosome, followed by chromothripsis and stabilization of the derivative chromosome drives iAMP21 chromosome formation, without the BFB steps that initiate the typical iAMP21 chromosome.23 Recent data reporting the genomic landscape of more than 2700 childhood ALL cases noted enrichment of a UV-mutational signature in a subset of B-ALL cases with aneuploidy, including B-ALL with high hyperdiploidy and iAMP21-ALL, and inferred that UV mutagenesis followed whole chromosomal gains in high hyperdiploid ALL and preceded these gains in iAMP21-ALL.25 Although these studies provided an insight into the structural basis of genesis of the iAMP21 chromosome and implicated a role of UV mutagenesis in aneuploid ALL, these studies did not involve a comprehensive genome-wide analysis of genomic changes in this leukemia subtype, systematically examine cases with constitutional aberrations of chromosome 21 that predispose to iAMP21-ALL and compare these with cases lacking constitutional alterations, or examine the timing of acquisition of the iAMP21 chromosome. Also, these studies were not sufficiently powered or detailed to formally delineate the minimal region(s) of genomic alteration of chromosome 21 that may drive leukemogenesis. In this study, we report integrated whole genome sequencing (WGS) and whole transcriptome sequencing (WTS)analysis of a large cohort of patients with iAMP21-ALL, including mutational signature and single-cell analyses to define mutational ontogeny and evolution.
Methods
Patients and samples
We studied 124 pediatric patients with iAMP21-ALL enrolled in St. Jude Children’s Research Hospital (n = 21), Children’s Oncology Group (COG; n = 75), UK (n = 27), and ECOG (n = 1) clinical trials (supplemental Table 1, available on the Blood website). Patients were included in this study based on the availability of samples for genomic analysis. Diagnostic samples were classified as iAMP21-ALL based on the presence of a grossly abnormal chromosome 21 in the karyotype and/or 5 or more signals using probes directed to RUNX1 using FISH.26,27 All cases were verified as iAMP21-ALL based on copy number profiles. Informed consent was obtained from patients and/or guardians in accordance with the Declaration of Helsinki. Study approval was obtained from institutional review boards.
Genomic sequencing and analysis
Methods for WGS, single-cell WGS (scWGS), WTS, single-cell RNA sequencing (scRNA-seq), whole-exome sequencing (WES), single nucleotide polymorphism (SNP) arrays, and genomic data analyses are provided in supplemental Information.
Results
Clinical characteristics and outcomes of iAMP21-ALL
We studied diagnostic and remission bone marrow/blood samples obtained from 124 children with iAMP21-ALL via WGS (n = 102) and/or WTS (n = 92; supplemental Figure 1A; supplemental Table 1). Seventy-two samples were previously reported,25 and 52 were sequenced for this study. The median age at diagnosis was 10 years. There was no gender bias (female, n = 66 and male, n = 58; supplemental Table 1).22 Five-year event-free survival and overall survival were 76.0 ± 5.1% and 96.2 ± 2.3%, respectively. Ancestry analysis predicted patient origin to be African (n = 2), American (n = 20), Asian (n = 2), European (n = 91), and admixed (n = 9) (supplemental Table 2). These numbers were too small to comment on the association of iAMP21-ALL with ancestry.
Transcriptomic diversity of iAMP21-ALL
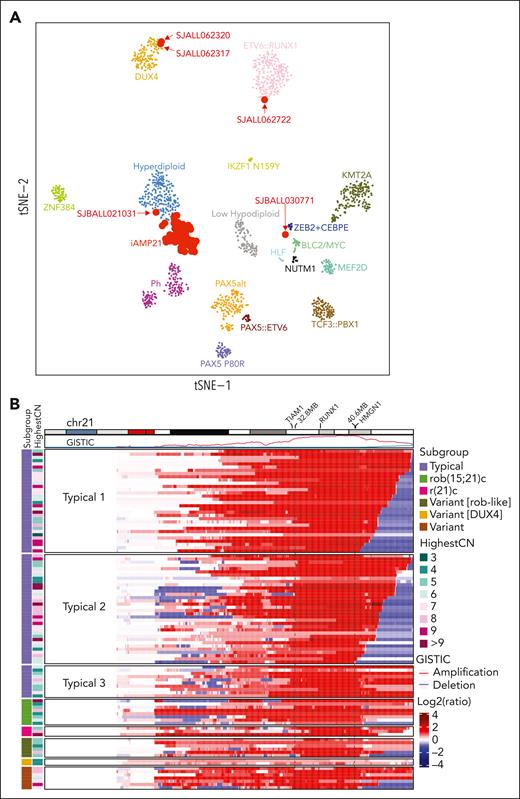
Analysis of the 92 iAMP21-ALL samples with WTS data and 1401 non-iAMP21–B-ALL samples6 showed that most (n = 87) samples, including 3 rob(15;21)c cases, clustered together, adjacent to high hyperdiploid B-ALL in two-dimensional gene expression space, suggesting that chromosome 21 gene gains is a central driver of the leukemic transcriptome in both subtypes (Figure 1A; supplemental Figure 1B). In 1 case (SJALL062722) with rob(15;21)c, a duplicated genome and biallelic ETV6 alterations clustered with ETV6::RUNX1, consistent with ETV6::RUNX1-like ALL.28,29 In 1 case (SJBALL021031) with additional gains of chromosomes 14 and 21, clustered close to high hyperdiploid ALL (supplemental Table 1), and in another case (SJBALL030771), the clusters were adjacent to those in cases with ZEB2 and/or CEBPE alterations, with no evidence of alterations in either of the genes. Two cases (SJALL062317 and SJALL062320) clustered with DUX4-rearranged ALL, and, in addition to the iAMP21 chromosome, harbored IGH::DUX4 rearrangements and ERG deletions. These patients were younger than other patients with iAMP21-ALL (3 and 5 years old at diagnosis), with long relapse-free survival on standard therapy.
Clustering pattern based on gene expression and chromosome 21 copy number variation of iAMP21-ALL. (A) t-distributed stochastic neighbor embedding plot showing gene expression profiling of 1493 B-ALL samples, including 92 iAMP21-ALL samples. Each dot represents a sample. The top 1000 most variable genes (on the basis of median absolute deviation) were selected and processed by the t-distributed stochastic neighbor embedding algorithm with a perplexity score of 30. Eighty-seven cases clustered together, forming an iAMP21-ALL cluster. One carrier of rob(15;21)c with a tetraploid clone clustered with the ETV6::RUNX1 cases (SJALL062722), 1 typical case clustered with the ZEB2 plus CEBPE group (SJBALL030771), and 1 typical case with gains of chromosomes 14 and 21 clustered with high hyperdiploidy (SJBALL021031), whereas 2 cases were located within the DUX4 cluster. (B) Heatmap showing DNA copy number profiles of chromosome 21 derived from WGS data of 102 cases. The log2 ratio of copy number was calculated based on the tumor and germ line samples for each case, with red and blue showing copy number amplification/gain and deletion, respectively. Each row represents a sample. The ideogram for chromosome 21 as well as GISTIC scores for copy number amplification and deletion is shown across the top. Subgroup information and the number of copies in the most highly amplified region (HighestCN) are shown on the left. The highlighted region between the 2 dashed lines represents the common region of gain derived from GISTIC analysis including all patients (32.8-40.6 Mb, between TIAM1 and HMGN1). RUNX1 is located within this common region. Typical 1 cases (n = 32) exhibit the deletion of the chromosome 21 sub-telomeric region without juxta-centromeric deletions. The size of the sub-telomeric deletions ranged from 0.1 Mb to 6.9 Mb. Typical 2 cases (n = 34) exhibit the deletion of both sub-telomeric and juxta-centromeric regions of chromosome 21. Typical 3 cases (n = 10) exhibit deletions of only juxta-centromeric regions.
Clustering pattern based on gene expression and chromosome 21 copy number variation of iAMP21-ALL. (A) t-distributed stochastic neighbor embedding plot showing gene expression profiling of 1493 B-ALL samples, including 92 iAMP21-ALL samples. Each dot represents a sample. The top 1000 most variable genes (on the basis of median absolute deviation) were selected and processed by the t-distributed stochastic neighbor embedding algorithm with a perplexity score of 30. Eighty-seven cases clustered together, forming an iAMP21-ALL cluster. One carrier of rob(15;21)c with a tetraploid clone clustered with the ETV6::RUNX1 cases (SJALL062722), 1 typical case clustered with the ZEB2 plus CEBPE group (SJBALL030771), and 1 typical case with gains of chromosomes 14 and 21 clustered with high hyperdiploidy (SJBALL021031), whereas 2 cases were located within the DUX4 cluster. (B) Heatmap showing DNA copy number profiles of chromosome 21 derived from WGS data of 102 cases. The log2 ratio of copy number was calculated based on the tumor and germ line samples for each case, with red and blue showing copy number amplification/gain and deletion, respectively. Each row represents a sample. The ideogram for chromosome 21 as well as GISTIC scores for copy number amplification and deletion is shown across the top. Subgroup information and the number of copies in the most highly amplified region (HighestCN) are shown on the left. The highlighted region between the 2 dashed lines represents the common region of gain derived from GISTIC analysis including all patients (32.8-40.6 Mb, between TIAM1 and HMGN1). RUNX1 is located within this common region. Typical 1 cases (n = 32) exhibit the deletion of the chromosome 21 sub-telomeric region without juxta-centromeric deletions. The size of the sub-telomeric deletions ranged from 0.1 Mb to 6.9 Mb. Typical 2 cases (n = 34) exhibit the deletion of both sub-telomeric and juxta-centromeric regions of chromosome 21. Typical 3 cases (n = 10) exhibit deletions of only juxta-centromeric regions.
Other fusion transcripts were detected in 38 patients, including P2RY8::CRLF2 (n = 14) and PAX5 rearrangements (n = 3), with individual fusions in the remaining cases (supplemental Table 3). Thirteen fusions were predicted to be in the frame, 5 of which were known, including ZCCHC7::PAX5, PAX5::MAPT, SCAPER::ABHD17C, SETD2::CCDC12, and DNAJC1::NEBL (supplemental Table 3). In total, 21 patients showed positive results for P2RY8::CRLF2, when deletions in pseudoautosomal region 1 (PAR1), which accompanies this rearrangement, were included (supplemental Table 4). Most PAR1 deletions had a variant allele frequency (VAF) ranging from 25% to 50%, suggesting that they were secondary to the iAMP21 chromosome (supplemental Figure 1C). WTS data from patients with iAMP21-ALL with P2RY8::CRLF2 or PAX5 fusions clustered with data of other iAMP21-ALL cases. Subtype prediction using a reference WTS data set of 19 canonical B-ALL subtypes helped classify these 92 patients with iAMP21-ALL as iAMP21-ALL (n = 84), high hyperdiploidy (n = 2), Ph-like ALL (n = 2), ETV6::RUNX1-like ALL (n = 1), DUX4-r (n = 2), and NUTM1-r (n = 1) (supplemental Table 5).
We identified 2827 (1894 upregulated and 933 downregulated; ≥twofold change, adjusted P < .05) differentially expressed genes between those cases in the iAMP21-ALL cluster (n = 87) compared with those in the non-iAMP21–B-ALL cluster (supplemental Table 6), with upregulation of 77 of 241 (32%) chromosome 21 protein-coding genes and negative enrichment of gene sets associated with lymphoid proliferation and maturation (supplemental Figures 1D-E; supplemental Tables 6-8).
DNA copy number profiles define subgroups of iAMP21-ALL
The hallmark of iAMP21-ALL is the variable genomic complexity of chromosome 21 alterations, comprising multiple regions of gain, inversion, and deletion.23 Analysis of WGS data helped detect a common region of chromosome 21 gain, with variability in the patterns of chromosome 21 copy number alterations (CNAs) (Figure 1B; supplemental Figure 2A). Most commonly (typical or sporadic23; n = 76), the CNA profile reflected at least 1 BFB cycle and a stepwise gain in copy number up to the highest region of gain on chromosome 21. These cases were subgrouped based on the copy number state of the centromeric and telomeric regions flanking the highest region of gain (typical, 1-3) (Figure 1B; supplemental Figure 2B). However, all except 2 (SJBALL021031 and SJBALL030771) of the 54 typical cases with WTS data clustered within the iAMP21-ALL gene expression two-dimensional t-distributed stochastic neighbor embedding space, validating their classification as iAMP21-ALL.
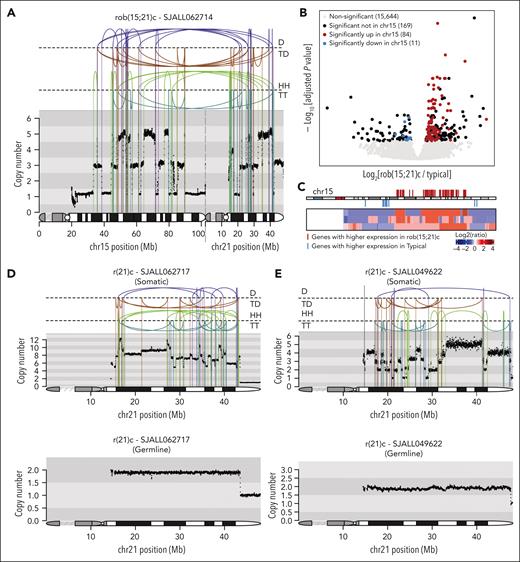
A distinct chromosome 21 profile was observed in 26 cases, which lacked evidence of the inversion foldback loops indicative of BFB cycles23 and/or the stepwise copy number changes of the aforementioned typical cases. We reported this profile, with rearrangements between chromosomes 15 and 21 in 4 carriers of rob(15;21)c who developed iAMP21-ALL23 and confirmed these alterations in 8 carriers of rob(15;21)c in this study (Figure 2A). Notably, 95 (36%) genes differentially expressed between rob(15;21)c and typical iAMP21-ALL were located on chromosome 15, with highly expressed genes located in amplification regions and underexpressed genes in deleted regions (Figure 2B-C; supplemental Table 9).
Copy number and gene expression profiles for rob(15;21)c and r(21)c iAMP21-ALL cases. (A) Example of the copy number and rearrangement profile of the der(15;21) chromosome for patient SJALL062714 with rob(15;21)c. Copy number is indicated on the y-axis, and the chromosome 15 and 21 ideograms are shown along the bottom of the plot to indicate breakpoint location. Rearrangements are separated based on their orientation: (B) Volcano plot comparing gene expression pattern between rob(15;21)c and typical subgroups. Only those cases in the main iAMP21-ALL cluster of Figure 1B with matched WGS and WTS were used in this analysis. Two hundred sixty-four genes showed significant differences (adjusted P value < .05), including 95 (36%) on chromosome 15 (84 and 11 upregulated and downregulated, respectively in rob(15;21)c compared with typical groups). (C) Heatmap showing the copy-number profile of chromosome 15 for 3 rob(15;21)c cases with matched WGS and WTS. Log2 ratios of copy number calculated based on the tumor and germ line samples for each case were used, with red and blue showing copy number amplification/gain and deletion, respectively. Each row represents a sample. The ideogram for chromosome 15 is shown on the top of the heatmap. Genes with significantly higher and lower expression in rob(15;21)c compared with those in typical cases are shown in red and blue, respectively, and listed in supplemental Table 8. (D-E) Rearrangement and copy number patterns of 2 cases with r(21)c: SJALL062717 (D) and SJALL049622 (E). Somatic and germ line profiles are shown at the top and bottom, respectively. Rearrangements are separated based on their orientation. D, deletion; HH, head-to-head inverted; TD, tandem duplication; TT, tail-to-tail inverted.
Copy number and gene expression profiles for rob(15;21)c and r(21)c iAMP21-ALL cases. (A) Example of the copy number and rearrangement profile of the der(15;21) chromosome for patient SJALL062714 with rob(15;21)c. Copy number is indicated on the y-axis, and the chromosome 15 and 21 ideograms are shown along the bottom of the plot to indicate breakpoint location. Rearrangements are separated based on their orientation: (B) Volcano plot comparing gene expression pattern between rob(15;21)c and typical subgroups. Only those cases in the main iAMP21-ALL cluster of Figure 1B with matched WGS and WTS were used in this analysis. Two hundred sixty-four genes showed significant differences (adjusted P value < .05), including 95 (36%) on chromosome 15 (84 and 11 upregulated and downregulated, respectively in rob(15;21)c compared with typical groups). (C) Heatmap showing the copy-number profile of chromosome 15 for 3 rob(15;21)c cases with matched WGS and WTS. Log2 ratios of copy number calculated based on the tumor and germ line samples for each case were used, with red and blue showing copy number amplification/gain and deletion, respectively. Each row represents a sample. The ideogram for chromosome 15 is shown on the top of the heatmap. Genes with significantly higher and lower expression in rob(15;21)c compared with those in typical cases are shown in red and blue, respectively, and listed in supplemental Table 8. (D-E) Rearrangement and copy number patterns of 2 cases with r(21)c: SJALL062717 (D) and SJALL049622 (E). Somatic and germ line profiles are shown at the top and bottom, respectively. Rearrangements are separated based on their orientation. D, deletion; HH, head-to-head inverted; TD, tandem duplication; TT, tail-to-tail inverted.
Three patients with iAMP21-ALL were carriers of rare constitutional ring chromosomes 21, r(21)c, with germ line copy number loss of the chromosome 21 telomeric region (Figure 2D-E; supplemental Figure 3A). These cases exhibited the same distinct chromosome 21 profile as that of carriers of rob(15;21)c. This profile was also noted in the 15 remaining cases, without evidence of associated constitutional chromosome 21 abnormalities, indicating that the lack of BFB/stepwise copy number changes are not restricted to carriers of chromosome 21–constitutional abnormalities. We refer to these cases lacking BFB/stepwise copy number changes as variant iAMP21-ALL.
Six additional variant cases had Robertsonian-like characteristics, because of to the presence of somatic genomic alterations involving other acrocentric chromosomes, consistent with rob(15;21)c (n = 4) or rob(21;22)c (n = 2) but without evidence of the constitutional Robertsonian rob(15;21)c or rob(21;22)c translocations (supplemental Figure 3B-D; supplemental Table 1). These cases commonly harbored dicentric chromosomes evident in their leukemic karyotypes but not observed upon analysis of WGS data (der[15;21]), der([21;22]), likely because of the lack of coverage of centromeric regions of acrocentric chromosomes. One additional case lacked WGS but had supportive evidence of Robertsonian-like rob(15;21) from WTS (supplemental Figure 3C). Two iAMP21-ALL cases with IGH::DUX4 were also classified as variants (supplemental Figure 3E). Examples of the remaining 7 variant cases are shown in supplemental Figure 4.
Of relevance to the clinical classification of iAMP21-ALL, the observation that the majority of non-IGH::DUX4 variant cases clustered with typical iAMP21-ALL cases upon the t-distributed stochastic neighbor embedding projection of gene expression profiling data, in addition to their similar median age and white blood cell counts, indicates that they should be classified as iAMP21-ALL regardless of variant type (supplemental Figure 1B).
Defining the common region of genomic alteration of chromosome 21
The integration of WGS and WTS data from this expanded cohort compared with those from prior studies defined common regions of genomic gain and the overexpression of chromosome 21 in iAMP21-ALL. Genomic identification of significant targets in cancer (GISTIC)30 analysis of 102 cases with WGS data showed the common region of genomic gain to extend from TIAM1 and HMGN1 (hg19 32.8 Mb to 40.6 Mb; gray, dashed lines in Figure 1B). Among the 71 genes in this region, 43 were differentially expressed compared with those of non-iAMP21–B-ALL (48, if the comparator group excluded high hyperdiploid cases; supplemental Figure 5A-B; supplemental Tables 5 and 7). These genes included those previously implicated in the pathogenesis of leukemia, with alterations of chromosome 21, including CHAF1B,31,DYRK1A,32,ERG,33,HMGN1,34 and RUNX1.15
We further performed integrative multiomic analysis of CNA and expression data from 70 iAMP21 ALL and 179 non-iAMP21/nonhigh hyperdiploid ALL cases,25 using multi-omics factor analysis (MOFA),35 an unsupervised approach to identify sources of heterogeneity in multiomic data sets (supplemental Figure 6A). Comparison of the latent factor (LF) scores, which measures sources of data variations between patients with iAMP21-ALL and other B-ALL, indicated that they were best separated by LF2 (P = 2.0 × 10-34, Wilcoxon rank-sum test; supplemental Figure 6B-C). Gene expression did not contribute to the variance explained by LF2 (supplemental Figure 6A), so we examined the distribution of the loading weight on LF2 associated with DNA copy number alterations (CNA) for chromosome 21 genes. This helped us identify a slightly wider region of chromosomal gain, confirming that the region from TIAM1 and HMGN1 was commonly gained, and the copy number pattern of this region distinguishes iAMP21-ALL from other B-ALL subtypes (supplemental Figure 6D). Thus, although multiple genes are overexpressed in the common region of amplification, the integration of expression and copy number data did not suggest that a single gene was the driver of iAMP21-ALL.
The genomic landscape of iAMP21-ALL
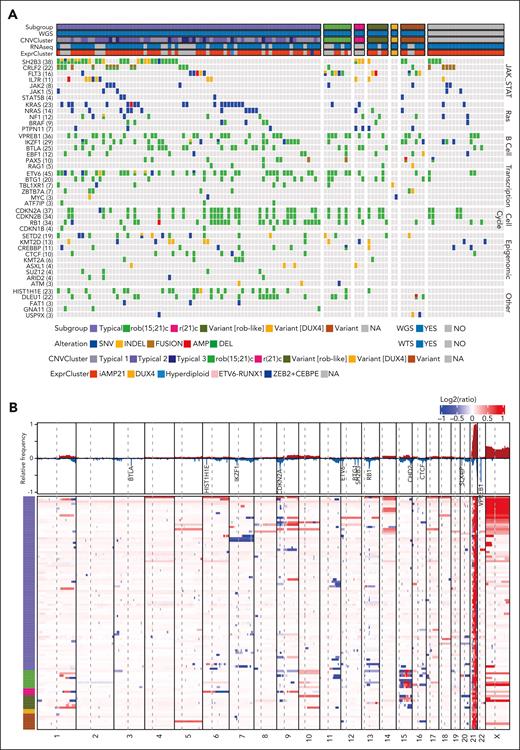
Forty-two genes were altered in at least 3 cases (Figure 3A; supplemental Table 10). The most commonly altered pathways were JAK-STAT signaling (n = 79 of 124 [64%]), Ras signaling (n = 51 [41%]), B-cell differentiation (n = 82 [66%]), other transcriptional regulators (n = 63 [51%]), cell cycle/apoptosis (n = 56 [45%]), and epigenomic/chromatin regulation (n = 56 [45%]). Multiple genes in these pathways, including KRAS (n = 22), SH2B3 (n = 15), NRAS (n = 13), FLT3 (n = 11), IL7R (n = 10), PTPN11 (n = 7), ASXL1 (n = 4), and STAT5B (n = 4), were recurrently mutated (supplemental Table 11). Genes with recurrent copy number variations, particularly focal deletions, included ETV6 (n = 41) (supplemental Figure 7A), VPREB1 (n = 36), CDKN2A (n = 36), CDKN2B (n = 34), RB1 (n = 33), SH2B3 (n = 29), BTLA (n = 25),IKZF1 (n = 24), DLEU1 (n = 22), HIST1H1E (n = 22), SLX4IP (n = 22), BTG1 (n = 20), CHD2 (n = 12), and CTCF (n = 9) (Figure 3B; supplemental Table 11). We did not observe associations between individual genetic alterations and different iAMP21-ALL subgroups.
Genomic profile of iAMP21-ALL. (A) Oncoprint of focal genomic changes in iAMP21-ALL. Sequence mutations were detected among WGS, WES, and WTS data. Copy number variations were detected using WGS and SNP array data. Internal tandem duplication and gene fusions were detected using WTS data. Variations in common pathways, including JAK-STAT signaling, Ras signaling, B-cell differentiation, transcriptional regulation, cell cycle/apoptosis, epigenomic, and PI3K-AKT signaling ubiquitination, are shown. Subgroup, data availability, and cluster location based on WTS data (ExprCluster) (Figure 1C) are shown across the top of the heatmap. (B) Heatmap showing genome-wide copy number profiles for 102 cases with high quality WGS. Log2 ratios of copy number calculated based on the tumor and germ line samples for each case were used, with red and blue indicating copy number amplification/gain and deletion, respectively. Each row represents a sample. The subgroup information is shown on the left. The relative frequencies for copy number amplification (red) and copy number deletion (blue) across all the cases are shown at the top. The genes with a high frequency of focal deletion are highlighted.
Genomic profile of iAMP21-ALL. (A) Oncoprint of focal genomic changes in iAMP21-ALL. Sequence mutations were detected among WGS, WES, and WTS data. Copy number variations were detected using WGS and SNP array data. Internal tandem duplication and gene fusions were detected using WTS data. Variations in common pathways, including JAK-STAT signaling, Ras signaling, B-cell differentiation, transcriptional regulation, cell cycle/apoptosis, epigenomic, and PI3K-AKT signaling ubiquitination, are shown. Subgroup, data availability, and cluster location based on WTS data (ExprCluster) (Figure 1C) are shown across the top of the heatmap. (B) Heatmap showing genome-wide copy number profiles for 102 cases with high quality WGS. Log2 ratios of copy number calculated based on the tumor and germ line samples for each case were used, with red and blue indicating copy number amplification/gain and deletion, respectively. Each row represents a sample. The subgroup information is shown on the left. The relative frequencies for copy number amplification (red) and copy number deletion (blue) across all the cases are shown at the top. The genes with a high frequency of focal deletion are highlighted.
Large regions (>30 Mb) of chromosome X gain were common (n = 36 [35%]); complex, large (>30 Mb) CNAs of chromosome 15 were present in rob(15;21)c and rob(15;21)-like cases (Figure 3B). Copy number–neutral loss-of-heterozygosity (CN-LOH) most commonly involved 9p (n = 12, resulting in the homozygous loss of CDKN2A/B), 12q (n = 14, resulting in the duplication of SH2B3 deletion/mutation), and 6p (n = 3, resulting in the homozygous loss of multiple histone genes) (supplemental Figure 7B). In total histone gene alternations involving 6p, including focal deletions, CN-LOH, single nucleotide variations, and small insertion-deletions, were observed in 27 (26%) cases (supplemental Figure 8).
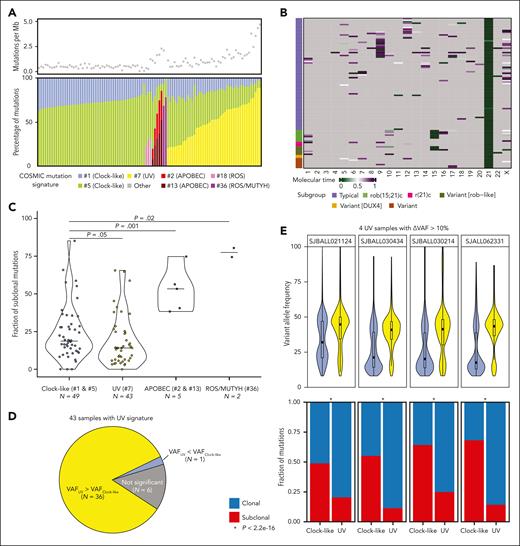
Enrichment of a UV-mutational signature in iAMP21-ALL
Mutational signature analysis of WGS data showed that, like other cancers,36 clock-like signatures (#1 and #5) were present in all samples (Figure 4A). The UV-mutational signature (#7) was observed in 43 cases, as recently reported,25,37 but was unrelated to specific structural variations, CNAs, or mutations (supplemental Figure 9; supplemental Tables 12-13). Although the numbers were small, the UV-mutational signature was not observed in the patients with DUX4-r (n = 2), rob(15;21)c (n = 8), and r(21)c (n = 3). Infrequently observed signatures were those of APOBEC (#2 and #13) (n = 5), ROS/MUTYH (#36) (n = 2), and ROS (#18) (n = 2) (supplemental Table 12). Samples with UV-mutational signatures had significantly higher mutation loads than those with only clock-like signatures (n = 49; P = 7.9 × 10-15, using Wilcoxon rank-sum test; Figure 4A). There was no age difference between patients with UV-mutational signature and those without it (P = .13; Wilcoxon rank-sum test), although there was no evidence of the UV-mutational signature in younger patients (≤6 years old; n = 10). Samples with APOBEC and ROS/MUTYH signature mutations also had higher mutation loads (P = 1.3 × 10-6 and P = 1.6 × 10-3, respectively), whereas the 2 samples with IGH::DUX4 fusions had ROS signatures and fewer mutations (P = .056; Figure 4A).
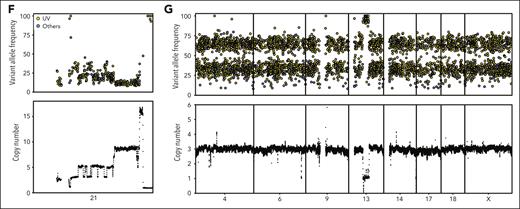
Mutation signature and molecular time of somatic mutations in iAMP21-ALL. (A) (Top) Scatterplot showing the number of mutations per Mb. (Bottom) Barplot showing faction of mutations in different COSMIC mutational signatures for each patient. Clock-like signatures (#1 and #5) are present in all the patients, 43 cases have UV signature mutations (#7), 5 cases have APOBEC signatures (#2 and #13), 2 cases have ROS/MUTYH signatures (#36), and 2 have ROS signatures (#18), both of which have DUX4-rearrangements. (B) Heatmap showing the molecular timing of copy number gains and CN-LOH. Chromothripsis of chromosomes 21 and 15 tend to occur early, whereas other variations have inconsistent patterns. (C) Violin plot showing the fraction of subclonal mutations across the samples with different mutation signatures. The fraction of subclonal mutations in the samples with UV-mutational signatures are significantly lower than that the samples with only a clock-like signature (P = .05), whereas the samples with APOBEC or ROS/MUTYH signatures are significantly higher (P = .001 and .02, respectively, two-sided Wilcoxon rank-sum test). (D) Pie chart showing 36 of 43 samples with UV-mutational signatures have significantly higher VAF for UV signature mutations than clock-like signature mutations. (E) (Top) Violin plot showing that 4 samples with UV-mutational signatures have VAF difference >10% between the UV signature and clock-like signature mutations. (Bottom) Barplot showing that for all 4 samples there are significantly higher fractions of clonal mutations in the UV signature than the clock-like signature. (F) Scatterplot showing VAF distribution for somatic mutations on chromosome 21 colored by mutation signatures (top) and the copy number profile (bottom) in patient SJALL062328. UV-induced mutations on chromosome 21 (n = 259) were not represented on the amplified allele (>9 copies), demonstrating that they occurred after iAMP21 chromosome formation. (G) Scatterplot showing VAF distribution for somatic mutations on chromosomes 4, 6, 9, 13, 14, 17, 18, and X colored based on mutation signatures (top) and their copy number profiles (bottom) in patient SJALL062328. The VAF was indicative of mutations arising on 1 of 3 and 2 of 3 alleles represented on the trisomic chromosomes, indicating that the mutations preceded chromosomal gain.
Mutation signature and molecular time of somatic mutations in iAMP21-ALL. (A) (Top) Scatterplot showing the number of mutations per Mb. (Bottom) Barplot showing faction of mutations in different COSMIC mutational signatures for each patient. Clock-like signatures (#1 and #5) are present in all the patients, 43 cases have UV signature mutations (#7), 5 cases have APOBEC signatures (#2 and #13), 2 cases have ROS/MUTYH signatures (#36), and 2 have ROS signatures (#18), both of which have DUX4-rearrangements. (B) Heatmap showing the molecular timing of copy number gains and CN-LOH. Chromothripsis of chromosomes 21 and 15 tend to occur early, whereas other variations have inconsistent patterns. (C) Violin plot showing the fraction of subclonal mutations across the samples with different mutation signatures. The fraction of subclonal mutations in the samples with UV-mutational signatures are significantly lower than that the samples with only a clock-like signature (P = .05), whereas the samples with APOBEC or ROS/MUTYH signatures are significantly higher (P = .001 and .02, respectively, two-sided Wilcoxon rank-sum test). (D) Pie chart showing 36 of 43 samples with UV-mutational signatures have significantly higher VAF for UV signature mutations than clock-like signature mutations. (E) (Top) Violin plot showing that 4 samples with UV-mutational signatures have VAF difference >10% between the UV signature and clock-like signature mutations. (Bottom) Barplot showing that for all 4 samples there are significantly higher fractions of clonal mutations in the UV signature than the clock-like signature. (F) Scatterplot showing VAF distribution for somatic mutations on chromosome 21 colored by mutation signatures (top) and the copy number profile (bottom) in patient SJALL062328. UV-induced mutations on chromosome 21 (n = 259) were not represented on the amplified allele (>9 copies), demonstrating that they occurred after iAMP21 chromosome formation. (G) Scatterplot showing VAF distribution for somatic mutations on chromosomes 4, 6, 9, 13, 14, 17, 18, and X colored based on mutation signatures (top) and their copy number profiles (bottom) in patient SJALL062328. The VAF was indicative of mutations arising on 1 of 3 and 2 of 3 alleles represented on the trisomic chromosomes, indicating that the mutations preceded chromosomal gain.
Molecular timing of somatic alterations in iAMP21-ALL
We inferred the molecular timing of genomic alterations (copy number gain, CN-LOH, chromothripsis, and somatic mutations), using MutationTimeR,38 for 102 patients with WGS data. Aggregated analysis showed that chromothripsis always occurred early, particularly that involving chromosome 21 in all cases and chromosome 15 in patients with rob(15;21)c (Figure 4B); in contrast, there was no clear trend for other copy number changes. Using FISH with combinations of RUNX1 and CRLF2 probes,11 in association with alternative allele frequency data, helped confirm that P2RY8::CRLF2 is secondary to iAMP21-ALL (supplemental Table 4; supplemental Figure 1C).
Samples with APOBEC and ROS/MUTYH signatures had higher fractions of subclonal mutations compared with those with only clock-like signatures (P = .001 and .02, respectively), whereas most somatic mutations in those cases with a UV-mutational signature were clonal (Figure 4C). In addition, the VAFs of UV mutations were significantly higher than clock-like mutations in 36 of the 43 samples with UV-mutational signatures (Figure 4D-E).
To examine the timing of iAMP21 chromosome formation and the acquisition of UV signature mutations, VAFs of UV-mutational signatures were compared with those of the iAMP21 chromosome copy number profile in 9 samples with at least 100 mutations on chromosome 21. The absence of UV signature mutations with high VAF on chromosome 21 indicated that UV signature mutations arise after the generation of the iAMP21 chromosome (Figure 4F; supplemental Figure 10). Similar analysis of the UV signature mutations located on triploid chromosomes in patients with iAMP21-ALL indicated that the UV signature mutations often occurred before whole chromosome gains (Figure 4G; supplemental Figure 10). These observations are consistent with the notion that iAMP21 chromosome formation is the earliest event, followed by UV signature mutations that precede an ongoing process of clock-like mutagenesis and other chromosomal abnormalities.
The IGH::DUX4 cases (SJALL062317 and SJALL062320) had the lowest level of copy number gain (n = 4) within the common region of chromosome 21 amplification (supplemental Table 1) among patients with iAMP21-ALL, and RUNX1 FISH showed the iAMP21 chromosome to be subclonal, present in 58% and 41% of the cell populations, respectively. Alongside these observations, the clonal deletion of ERG detected via WGS (supplemental Figure 3E) and validated via SNP arrays indicated that iAMP21 chromosome formation was likely a secondary event to IGH::DUX4 in these cases.
Clonal evolution of iAMP21-ALL
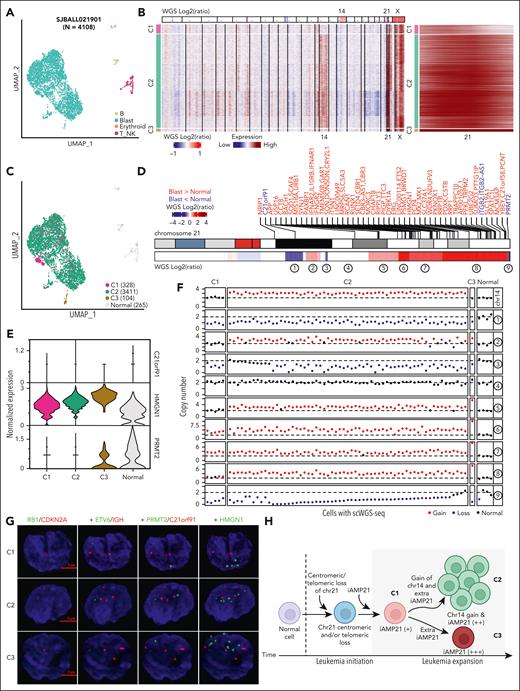
We performed scRNA-seq of samples from 4 patients with iAMP21-ALL and annotated cell types based on the expression of lineage-associated genes (Figure 5A; supplemental Figure 11A; supplemental Table 14). Inferred CNAs from these data were consistent with their copy- number profiles detected using WGS (Figure 5B; supplemental Figure 11B-E). Copy number gain of chromosome 21 was observed in all blast cells from the 4 samples, confirming that the generation of the iAMP21 chromosome is an early clonal event. However, copy number gain of chromosome 14 (patient SJBALL021901) and gain of 1q (patient SJBALL030871) were observed in only a subset of cells. These results are consistent with the inferred molecular timing analyses presented in the previous subsection.
Clonal evolution of iAMP21-ALL using scRNA-seq. (A) Uniform manifold approximation and projection (UMAP) representation of the scRNA-seq data set from patient SJBALL021901. Clusters of cells are colored based on the cell types. (B) scRNA-seq derived copy number profiles for blast cells in patient SJBALL021901. Copy number profiles from WGS are shown (top); chromosomes with copy number gains are labeled. Three clusters were observed via scRNA-seq: C1 has the lowest copy number of chromosome 21 and no copy number gain of chromosome 14. C2 has an intermediate copy number gain of chromosome 21 together with copy number gain of chromosome 14. C3 has the highest copy number of chromosome 21 with no copy number gain of chromosome 14. Chromosome 21 is shown (right). (C) UMAP representation of the scRNA-seq data for SJBALL021901 colored based on the 3 copy number clusters. Healthy cells are shown in gray. (D) Heatmap showing copy number profiles of chromosome 21 derived from WGS data of SJBALL021901. Differentially expressed genes between blasts and healthy cells based on scRNA-seq are labeled on the top in red and blue for genes with higher and lower expression in blasts, respectively. Nine segments with varying copy number patterns are labeled at the bottom. (E) Violin plot showing the expression profile of 3 selected genes across different copy number clusters and healthy cells in SJBALL021901. (F) Scatterplot showing copy numbers of chromosome 14 and the 9 segments on chromosome 21, as illustrated in panel D, for 70 cells with single-cell WGS in SJBALL021901. These cells were assigned to different clusters based on their copy number patterns. (G) DNA FISH image of 3 representative cells from different copy number clusters of SJBALL021901. Seven probes were used in total, paired in 4 sequential hybridizations with imaging after every hybridization. The first hybridization is RB1 (green) and CDKN2A (red), the second is ETV6 (green) and IGH (red), the third is PRMT2 (green) and C21orf91 (red), and the fourth is HMGN1 (green). The scales (5 μm) are shown in red. (H) Schematic representation of the clonal evolution model in sample SJBALL021901. C1 is an early clone with a centromeric and/or telomeric loss and lower copy number gain of the iAMP21 chromosome, followed by 2 evolutionary tracks: (1) gain of chromosome 14 and further amplification of the iAMP21 chromosome, resulting in the generation of the predominant clone C2 and (2) further amplification of the iAMP21 chromosome without the gain of chromosome 14 in C3.
Clonal evolution of iAMP21-ALL using scRNA-seq. (A) Uniform manifold approximation and projection (UMAP) representation of the scRNA-seq data set from patient SJBALL021901. Clusters of cells are colored based on the cell types. (B) scRNA-seq derived copy number profiles for blast cells in patient SJBALL021901. Copy number profiles from WGS are shown (top); chromosomes with copy number gains are labeled. Three clusters were observed via scRNA-seq: C1 has the lowest copy number of chromosome 21 and no copy number gain of chromosome 14. C2 has an intermediate copy number gain of chromosome 21 together with copy number gain of chromosome 14. C3 has the highest copy number of chromosome 21 with no copy number gain of chromosome 14. Chromosome 21 is shown (right). (C) UMAP representation of the scRNA-seq data for SJBALL021901 colored based on the 3 copy number clusters. Healthy cells are shown in gray. (D) Heatmap showing copy number profiles of chromosome 21 derived from WGS data of SJBALL021901. Differentially expressed genes between blasts and healthy cells based on scRNA-seq are labeled on the top in red and blue for genes with higher and lower expression in blasts, respectively. Nine segments with varying copy number patterns are labeled at the bottom. (E) Violin plot showing the expression profile of 3 selected genes across different copy number clusters and healthy cells in SJBALL021901. (F) Scatterplot showing copy numbers of chromosome 14 and the 9 segments on chromosome 21, as illustrated in panel D, for 70 cells with single-cell WGS in SJBALL021901. These cells were assigned to different clusters based on their copy number patterns. (G) DNA FISH image of 3 representative cells from different copy number clusters of SJBALL021901. Seven probes were used in total, paired in 4 sequential hybridizations with imaging after every hybridization. The first hybridization is RB1 (green) and CDKN2A (red), the second is ETV6 (green) and IGH (red), the third is PRMT2 (green) and C21orf91 (red), and the fourth is HMGN1 (green). The scales (5 μm) are shown in red. (H) Schematic representation of the clonal evolution model in sample SJBALL021901. C1 is an early clone with a centromeric and/or telomeric loss and lower copy number gain of the iAMP21 chromosome, followed by 2 evolutionary tracks: (1) gain of chromosome 14 and further amplification of the iAMP21 chromosome, resulting in the generation of the predominant clone C2 and (2) further amplification of the iAMP21 chromosome without the gain of chromosome 14 in C3.
Three clusters of cells (C1-C3) were identified in SJBALL021901, each with a distinct inferred copy number profile for the iAMP21 chromosome. In addition, chromosome 14 gain was present in the predominant clone C2, whereas no copy number gain of chromosome 14 was detectable in C1 and C3 (Figure 5B). Most of the cells in C1 and C3 formed independent clusters based on their genome-wide gene expression pattern (Figure 5C). The comparison between blasts with healthy cells revealed 70 differentially expressed genes on chromosome 21, with 4 genes showing lower expression in blasts, including C21orf91 and PRMT2 in the regions of centromere and telomere loss, respectively, in this case (Figure 5D; supplemental Table 15). A comparison of the 3 clusters showed a progressive change in the expression of chromosome 21 genes (Figure 5E; supplemental Figure 11F; supplemental Table 15).
To validate these inferred observations of clonal heterogeneity, we performed single-cell WGS of 71 flow-sorted blasts and healthy cells from SJBALL021901 to determine the copy number of chromosome 14 and each segment of chromosome 21 (Figure 5D; supplemental Tables 16-17). We observed 5 cells with the lowest level of chromosome 21 gain and 2 copies of chromosome 14 (representative of C1); 61 cells with an intermediate level of chromosome 21 gain and 3 copies of chromosome 14 (representative of C2); and 1 cell with the highest copy number gain of chromosome 21 and 2 copies of chromosome 14 (representative of C3), along with 4 healthy cells (Figure 5F; supplemental Table 18). Multiplex FISH of SJBALL021901 helped identify 42, 193, and 4 cells from C1, C2, and C3, respectively (Figure 5G; supplemental Tables 19-20). We performed single-cell WGS on another sample, SJBALL030072 and also found that the gain of the iAMP21 chromosome occurs before copy number changes on other chromosomes (supplemental Figure 12). These data and our model of clonal evolution indicate that the formation of the iAMP21 chromosome is an early event in leukemogenesis that may undergo progressive amplification (although the frequency of this phenomenon is not known), further increasing the expression of genes in the common region of amplification (Figure 5H).
Discussion
iAMP21-ALL is a high-risk subtype of childhood B-ALL.1 In this study, we confirmed the older age and lower white blood cell counts of children with iAMP21 B-ALL compared with children within other B-ALL subtypes.22,24 Patients in this study were treated in accordance with protocols in which risk stratification included iAMP21-ALL status, thus the high 5-year event-free survival and overall survival rates validate the success of treatment intensification, reinforcing the need for accurate diagnosis and risk stratification of patients with iAMP21-ALL.20-22
To date, large-scale genome-wide characterization of iAMP21-ALL has been lacking. By integrating WGS and WTS of more than 100 cases, we provide a comprehensive portrait of the genomic landscape, delineate driver events, and show their sequence of acquisition in the genesis of this disease. Several recurrently mutated genes and pathways are particularly frequent in this disease, including alterations of ETV6 and mutations activating JAK-STAT signaling, suggesting their roles in the acquisition of the full leukemic phenotype and potential for therapeutic targeting.
We identified several recurring patterns of rearrangements and CNAs of the iAMP21 chromosome. Most patients exhibited at least 1 BFB cycle and a stepwise gain in chromosome 21 copy number, characterizing a typical iAMP21 chromosome profile with variability in the presence of proximal and distal deletions. Here, this mechanism of iAMP21 chromosome formation was supported by single-cell analyses that document multiclonality with variegation in the magnitude of chromosome 21 amplification and associated gene expression.
In addition, we described patients with the variant iAMP21-ALL without the evidence of BFB and/or stepwise gain in copy number. These variants include patients with rob(15;21)c,23 but we also identified a subgroup of Robertsonian-like iAMP21-ALL cases, which exhibit somatic copy number and rearrangement profiles of chromosomes 15 and 21 as seen in carriers of rob(15;21)c, or less commonly, of chromosomes 21 and 22 as would be expected in carriers of rob(21;22)c but without the evidence of the constitutional Robertsonian chromosome. These cases highlight a role of acquired missegregation of acrocentric chromosomes similar to what we described about individuals with rob(15;21)c.23 In support of this notion, several acquired dicentric chromosomes were observed in the karyotypes of some cases, suggesting that the centromeres of other acrocentric chromosomes may also be involved with chromosome 21 in the formation of iAMP21 chromosomes at a higher incidence than that previously recognized.39 Our cohort included 3 carriers of r(21)c in whom the constitutional ring chromosome gave rise to the iAMP21 chromosome, an otherwise rare phenomenon.24 As ring chromosomes are inherently unstable in mitosis, telomere loss at the constitutional level may initiate the BFB cycle, giving rise to the iAMP21 chromosome in these cases.
Because most of the iAMP21-ALL samples exhibited a distinct gene expression signature,6,25 irrespective of the iAMP21 chromosome copy number profile or the concomitant constitutional or somatic genomic alterations, the classification of iAMP21-ALL can be expanded to include all variant types. Exceptions include uncommon cases with other subtype-defining alterations, such as DUX4-rearrangement or the previously described association with ETV6::RUNX1,22,40 in which the iAMP21 chromosome was a secondary event; such cases should be classified based on the primary genetic event. An analysis of recent genomic studies shows the co-occurrence of iAMP21 chromosome with these subtype-defining alterations to be rare6,25,41 and usually subclonal to founding fusions such as ETV6::RUNX1.22
The integration of WGS and WTS data enabled the delineation of 71 genes within the common region of amplification, with the overexpression of most genes compared with that of genes in other subtypes of B-ALL. The large size of this region and the inability to nominate 1 or few genes as drivers using in silico analyses suggests that the coordinated deregulation of multiple genes, including known oncogenes such as RUNX1, DYRK1A, CHAF1B, and ERG, is required for leukemogenesis in iAMP21-ALL.
We showed a striking enrichment of UV-mutational signatures in many (43%) patients with iAMP21-ALL, which were commonly clonal with high mutational burdens. Our findings confirm the enrichment of this signature in aneuploid subtypes of ALL characterized by the gain or preservation of chromosome 2125,37 and can be extended in several aspects. Systematic integrated genomic analysis of this large cohort enabled detailed analysis of mutational timing, demonstrating that the formation of the iAMP21 chromosome is an early, leukemia-initiating event, followed by the acquisition of UV signature mutations and sequence and structural alterations of ALL driver genes. Moreover, the heterogeneity of the iAMP21 chromosome raised the possibility that this alteration is not fixed but may evolve over time. This notion was supported by single-cell analysis using gene expression as a surrogate for CNAs, confirmed via single-cell WGS that demonstrated progressive evolution of the iAMP21 chromosome and its relationship to the acquisition of secondary genomic alterations.
Although alternative mutagenic mechanisms have been proposed for the enrichment of the UV signature,37 several observations suggest that UV exposure is responsible: the association of the UV-mutational signature with white, European ancestry, rather than African-American ancestry42; the lack of identification of alternative chemical inducers of the UV signature 443; the lack of correlation of the UV-mutational signature with age, consistent with intermittent environmental exposure; the ability of UV light to penetrate the skin and reach cutaneous blood vessels 444; the detection of aneuploid preleukemic cells in peripheral blood at birth, suggesting cells may be exposed to UV mutagenesis 445,46; and the identification of the UV-mutational signature in other lymphoid malignancies, such as T-cell lymphoma, suggesting that lymphoid cells are susceptible to UV mutagenesis.47 These observations imply a scenario in which the formation of the iAMP21 chromosome generates preleukemic cells which circulate and are exposed to UV-mediated mutagenesis and the acquisition of full leukemic potential.
Most cases in this study were originally diagnosed as iAMP21-ALL based on our earlier definition of 3 or more additional RUNX1 signals present on a single copy of an abnormal chromosome 21 using FISH, often associated with a grossly abnormal chromosome 21 in the karyotype.26 The finding of variant cases involving chromosome 21 rearrangements with other chromosomes and cases in which RUNX1 is located outside the highest region of chromosome 21 amplification,39 in addition to the 3 typical forms and other variants highlighted herein, indicates that the original definition needs to be broadened. Our observation that the full range of typical and variant cases share a common gene expression profile over an enlarged, common region of amplification confidently simplifies diagnosis. Although cytogenetics and RUNX1 FISH will help successfully identify most typical iAMP21-ALL cases, we recommend copy number profiling (SNP arrays or WGS, as shown in this study) to validate variant cases48 and as a standalone diagnostic test, because it successfully identifies the extensive range of chromosome 21 genomic profiles together with the secondary genetic changes of iAMP21-ALL.
Collectively, our data support the view that the presence of the iAMP21 chromosome defines a subtype of B-ALL, which rarely occurs as a secondary event. The broad region of chromosomal gain provides reassurance to a diagnostic approach that detects multiple additional copies of a region of chromosome 21 at or close to RUNX1 using cytogenetic and/or genomic approaches.
Acknowledgments
The authors thank the Blood Cancer UK Childhood Leukaemia Cell Bank, the Children's Oncology Group, and the Biorepository of St. Jude Children's Research Hospital for the provision of samples.
This study was supported by the National Institutes of Health, National Cancer Institute grants P30 CA021765 (St. Jude Children’s Research Hospital Comprehensive Cancer Center support grant) and R35 CA197695 (C.G.M.); a St. Baldrick’s Foundation Robert J. Arceci Innovation Award (C.G.M.); the Henry Schueler 41&9 Foundation (C.G.M.); the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital; Blood Cancer UK (15036) (C.J.H. and A.V.M.); European Research Council (249891) (C.J.H.); Newcastle University Annual Fund (C.J.H.); and Cancer Research UK (C60802/A27193) (S.L.R.).
Authorship
Contribution: Q.G., S.L.R., I.I., P.S.G., R.E.C., C.S., S.L., M.B., P.S., D.R.B., M.T.R., Z.K., T.J., K.G.R., Y.F., W.C., T.-C.C., G.W., A.C., N.H., and A.V.M. performed sequencing/analyzed genomic data; I.I. and P.B. performed single-cell RNA-seq experiments; A.H.E., L.S., S.P., D.P., C.C., and M.D. performed biostatistical analyses; V.V. and M.V. performed FISH experiments; W.Y. and J.J.Y. performed ancestry analyses; and C.J.H. and C.G.M. designed and oversaw the study and wrote the manuscript.
Conflict-of-interest disclosure: C.G.M. received research funding from Loxo Oncology, Pfizer, and AbbVie; honoraria from Amgen and Illumina; and holds stock in Amgen. M.T.R., D.R.B., Z.K., and T.J. are employees of Illumina Inc, a public company that develops and markets systems for genetic analysis. The remaining authors declare no competing financial interests.
Correspondence: Christine J. Harrison, Leukaemia Research Cytogenetics Group Translational and Clinical Research Institute, Newcastle University Centre for Cancer, Faculty of Medical Sciences, Newcastle University, Level 6, Herschel Building, Brewery Ln, Newcastle upon Tyne NE1 7RU, United Kingdom; e-mail: christine.harrison@newcastle.ac.uk; and Charles G. Mullighan, Department of Pathology, St. Jude Children’s Research Hospital, 262 Danny Thomas Pl, Mail Stop 342, Memphis, TN 38105; e-mail: charles.mullighan@stjude.org.
References
Author notes
∗Q.G. and S.L.R. contributed equally to this study.
†C.J.H. and C.G.M. are joint senior authors.
Genomic data generated for this study have been deposited in the European Genome-phenome Archive (accession numbers EGAS00001004998, EGAS00001006577, and EGAS00001006796).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal