In this issue of Blood, Gao et al describe the genomic landscape of pediatric acute lymphoblastic leukemia with intrachromosomal amplification of chromosome 21 (iAMP21-ALL).1
iAMP21-ALL is a relatively recently identified ALL subtype, first reported as a distinct entity in 20032,3 after occasional earlier case reports. It comprises ≈2% of childhood ALL, and was initially recognized because use of fluorescence in situ hybridization (FISH) probes to identify the ETV6::RUNX1 translocation detected cases with increased numbers of signals associated with the RUNX1 probe.4 Both the number of extra RUNX1 signals and the amplified region of chromosome 21 observed on karyotype are variable, posing challenges for both defining the diagnostic criteria and understanding the critical molecular driver of leukemogenesis. Uncertainty about the role of iAMP21 as a driver lesion in ALL also arises in a small subset of cases in which iAMP21 is identified as co-occurring with other ALL subtype-defining genetic lesions, including ETV6::RUNX1, high hyperdiploidy, BCR::ABL1, and P2RY8::CRLF2.4-6 Clinically, iAMP21-ALL is associated with an older age and lower white blood cell count at diagnosis, and relatively unfavorable outcomes, although survival is improved when patients are treated on higher-intensity treatment regimens.7,8 Thus, accurate identification of iAMP21-ALL cases is important because intensification of therapy partially compensates for the unfavorable prognosis associated with this subtype. In addition, an improved understanding of the key molecular drivers of iAMP21-ALL may inform development of targeted therapeutic strategies to improve outcomes.
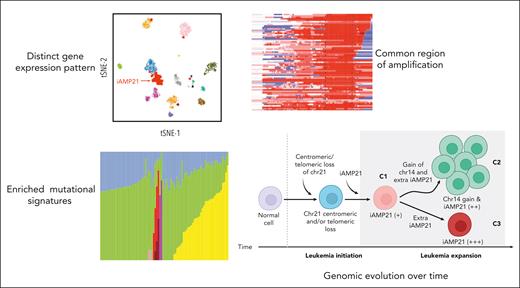
In the present study, Gao et al assembled an impressive cohort of 124 pediatric cases from St. Jude Children’s Research Hospital, the Children’s Oncology Group, and UK clinical trials, for comprehensive genomic characterization by whole-genome sequencing (WGS) and/or whole-transcriptome sequencing (WTS). Although 72 of these cases were previously reported,5 the expanded cohort reported here has enhanced power to provide a detailed characterization of the biology of iAMP21-ALL (see figure).
A key contribution of this study is to leverage the combined WGS and WTS analyses to define a common region of amplification in iAMP21-ALL, encompassing 71 genes, and to identify 43 genes in this region with differential expression compared with non–iAMP21-ALL cases. Many of these differentially expressed genes have been previously implicated in leukemia pathogenesis (eg, CHAF1B, DYRK1A, ERG, HMGN1, and RUNX1). More important, bioinformatic analyses did not identify any smaller subset of genes as key drivers, suggesting that coordinated deregulation of multiple genes is required for leukemogenesis in iAMP21-ALL.
In addition to defining the common region of amplification and key differentially expressed genes in iAMP21-ALL, the large size of this study permitted further characterization of the complexity of this ALL subtype. It has previously been recognized that 2 specific constitutional abnormalities of chromosome 21 (Robertsonian translocations between chromosomes 15 and 21 and ring chromosomes involving chromosome 21) are associated with an increased risk of iAMP21-ALL, likely because the abnormal chromosome 21 structure initiates formation of the iAMP21.9 Here, Gao et al use WGS to provide detailed characterization of the pattern of chromosome 21 copy number alterations in cases with these constitutional abnormalities, demonstrating that it differs from the mechanism of successive break-fusion-break cycles that generates iAMP21 in typical cases. They go on to identify an additional “Robertsonian-like” subgroup, with a copy number profile that resembles the Robertsonian variant, but without evidence of a constitutional Robertsonian translocation. Using WTS, they demonstrate that all 3 of these variant subgroups share the iAMP21 gene expression signature, supporting the appropriateness of classifying them as iAMP21-ALL. Conversely, they demonstrate that cases with other subtype-defining alterations (DUX4 rearrangement and ETV6::RUNX1) do not share the iAMP21 gene expression profile, and are therefore more appropriately classified as bearing iAMP21 as a secondary rather than primary alteration.
A key clinical implication of this study is that the original definition of iAMP21-ALL based solely on FISH (at least 5 RUNX1 signals per interphase cell, with ≥3 occurring on a single abnormal chromosome 21) is insufficient. Although this definition correctly classifies most cases, it does not identify variant cases with rearrangements of portions of chromosome 21 to other chromosomes, nor cases where RUNX1 is not contained in the highest region of amplification. The authors advocate for use of copy number profiling by single-nucleotide polymorphism array or WGS, as either a stand-alone diagnostic test or a back-up for evaluation of complex or atypical cases. This aligns with another recent report that found that 9% of 207 iAMP21-ALL cases were missed by the FISH-based definition, but identified by chromosomal microarray testing.6
Gao et al also use single-cell and mutational signature analyses to provide some intriguing insights into the timing of events in the pathogenesis of iAMP21-ALL. Their data suggest that the formation of the iAMP21 chromosome is an early event that evolves progressively over time, and intriguingly, that the iAMP21-ALL subtype is enriched for a UV-mutational signature. They propose a putative model that would account for this signature, positing that UV-mediated mutagenesis may occur during peripheral circulation of preleukemic iAMP21-chromosome–positive cells. Further research is needed to explain the functional basis for iAMP21-ALL mutational signatures.
Overall, this study elegantly integrates WGS and WTS data to provide a detailed characterization of iAMP21-ALL, defining the key region of amplification and the set of differentially expressed genes; highlighting the importance of copy number profiling to identify variant cases; and suggesting the timing of acquisition of somatic alterations. Because 91 of 124 (73%) of the cases in this cohort were of European ancestry, additional studies of racially and ethnically diverse populations are needed to investigate possible ancestry-associated differences in the genomics and/or clinical features of iAMP21-ALL. Further studies are also needed to investigate potential targeted therapeutic strategies based on the upregulated pathways identified in this study and others.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal