Key Points
High-throughput drug screening identified gilteritinib and venetoclax as a highly synergistic drug combination in FLT3 wild-type AML.
Gilteritinib-venetoclax suppressed MCL-1 and decreased venetoclax-azacitidine–resistant FLT3 wild-type AML viability in vitro and in vivo.
Abstract
BCL-2 inhibition has been shown to be effective in acute myeloid leukemia (AML) in combination with hypomethylating agents or low-dose cytarabine. However, resistance and relapse represent major clinical challenges. Therefore, there is an unmet need to overcome resistance to current venetoclax-based strategies. We performed high-throughput drug screening to identify effective combination partners for venetoclax in AML. Overall, 64 antileukemic drugs were screened in 31 primary high-risk AML samples with or without venetoclax. Gilteritinib exhibited the highest synergy with venetoclax in FLT3 wild-type AML. The combination of gilteritinib and venetoclax increased apoptosis, reduced viability, and was active in venetoclax-azacitidine–resistant cell lines and primary patient samples. Proteomics revealed increased FLT3 wild-type signaling in specimens with low in vitro response to the currently used venetoclax-azacitidine combination. Mechanistically, venetoclax with gilteritinib decreased phosphorylation of ERK and GSK3B via combined AXL and FLT3 inhibition with subsequent suppression of the antiapoptotic protein MCL-1. MCL-1 downregulation was associated with increased MCL-1 phosphorylation of serine 159, decreased phosphorylation of threonine 161, and proteasomal degradation. Gilteritinib and venetoclax were active in an FLT3 wild-type AML patient-derived xenograft model with TP53 mutation and reduced leukemic burden in 4 patients with FLT3 wild-type AML receiving venetoclax-gilteritinib off label after developing refractory disease under venetoclax-azacitidine. In summary, our results suggest that combined inhibition of FLT3/AXL potentiates venetoclax response in FLT3 wild-type AML by inducing MCL-1 degradation. Therefore, the venetoclax-gilteritinib combination merits testing as a potentially active regimen in patients with high-risk FLT3 wild-type AML.
Introduction
With a median age at onset of 65 years, acute myeloid leukemia (AML) is predominantly a disease of the elderly, with limited intensive chemotherapy options.1 Unfit patients are offered low-dose therapy concepts, such as hypomethylating agents (HMAs) or low-dose cytarabine (LDAC), associated with low remission rates and poor median survival.2,3 Recently, the addition of the BCL-2-inhibitor venetoclax to either LDAC4 or HMAs5 vastly improved response rates. These findings led to US Food and Drug Administration (FDA) and European Medicines Agency (EMA) approval for the combination of HMAs and venetoclax in newly diagnosed patients ineligible for intensive chemotherapy. Nonetheless, a considerable fraction of patients with AML do not respond, and most patients relapse after initially achieving remission with venetoclax-azacitidine. Moreover, relapsed patients are resistant to all currently used therapies and usually die within a short timespan.5,6 The combinations used today were established based on FDA-approved preexisting low-intensity therapies in AML. However, a systematic evaluation of the most effective and synergistic venetoclax combination partners is still lacking.
Venetoclax acts as a small-molecule BCL-2 homology domain 3–mimetic drug. Interestingly, a recent study highlighted that lymphoid cells can escape venetoclax by reprogramming energy metabolism and overexpressing MCL-1 during complex clonal shifts.7 In line with this, Jones et al8 described shifts in metabolism in venetoclax-resistant AML. Increased levels of the antiapoptotic BCL-2 family members MCL-1 or BCL-XL were also observed in venetoclax-resistant AML cells.9,10 Moreover, it has been reported that BCL-2 is differentially expressed in subpopulations of AML cells, with the highest expression in malignant stem and progenitor cells and lowest expression in AML with a monocytic phenotype, which expresses MCL-1 instead and has evolved to be refractory to venetoclax.11 However, MCL-1 inhibition carries the risk of profound toxicity to normal tissues, especially cardiac toxicity.12 Therefore, indirect targeting of MCL-1 in combination with BCL-2 inhibition might be a promising therapeutic approach.
With the aim of identifying a combinatorial treatment more effective than venetoclax-azacitidine, many therapy options have been suggested in AML. These options include inhibition of PI3K, CDKs, SMAC, or complex I.13,14 So far, response mechanisms are not well understood, and the most effective treatment combinations are not known.
In this study, we developed and used a synergism-focused drug-targeting pipeline to identify the most potent venetoclax combination partners in high-risk AML.
Materials and methods
Drug screening
Drug response assays were performed with primary AML blasts cultured in RPMI-1640 (Invitrogen, Carlsbad, CA) supplemented with penicillin/streptomycin (Invitrogen), l-glutamine (Invitrogen), and 10% pooled and heat-inactivated AB-type human serum albumin (Sigma-Aldrich, St Louis, MO). Cells in medium plus human serum albumin were subjected to rolling for 3 hours in the dark. Only samples with viability >90% after 3-hour prestimulation were included in the screen and added to drug-precoated 384-well plates (781904; Greiner Bio One, Frickenhausen, Germany). Plates were coated with 64 drugs in 5 concentrations with and without venetoclax in 2 concentrations (1 and 20 nM). Cell viability was assessed after 48 hours with a PerkinElmer EnSight using CellTiter-Glo (G7572; Promega, Fitchburg, MA) and normalized to dimethyl sulfoxide controls, as previously described.15
Patient specimens
Primary AML samples were obtained from the German Study Alliance Leukemias AML Register Dresden and the BioMaterialBank Heidelberg. All patients provided informed consent in accordance with the Declaration of Helsinki, and biobanking procedures were approved by the ethics committee of the University of Heidelberg. Mononuclear cells of patients with AML were density gradient isolated from bone marrow aspirations taken at diagnosis. Only patients with AML with high-risk disease were included in the drug screen (supplemental Table 1; supplemental Figure 1A). High-risk disease was defined either according to the European LeukemiaNet classification16 risk group (n = 19) or if patients were refractory after induction chemotherapy (n = 12). Three primary samples were included in in vitro validation studies, 2 of which (01 and 70) had already been included in the drug screen (supplemental Table 1).
Treatment of patients 02, 03, 05, and 70 with venetoclax-gilteritinib was performed after written informed consent to the off-label use following the principles of Helsinki.
Cell culture
HL60, MOLM13, OCI-AML2, OCI-AML3, and MV4-11 AML cell lines were purchased from the German Collection of Microorganisms and Cell Cultures and cultured in RPMI 1640 medium (21875091; Thermo Fisher Scientific) supplemented with 10% (HL60) or 20% (other cell lines) fetal bovine serum (FBS.S 0615; Bio&SELL GmbH, Feucht, Germany). Venetoclax-azacitidine–resistant HL60 cells were generated by treating cells twice weekly with increasing doses of venetoclax (S8048; Selleckchem, Houston, TX) and azacitidine (S1782; Selleckchem) for several months.
Primary human bone marrow or peripheral blood samples were obtained from patients with AML who provided informed consent. Biobank procedures were approved by the Ethikkomission Heidelberg. Mononuclear cells were density gradient isolated and cultured as described by Pabst et al.17
Colony-formation assays
A total of 250 cells (cell line) or 4000 cells (primary sample) were seeded into 12-well plates with 550 μL of methylcellulose (04230 or 04034; STEMCELL Technologies) supplemented with penicillin streptomycin (A2213; Biochrom GmbH) and the indicated drugs. After 10 days, colonies were counted. All experiments were performed as technical triplicates, and each experiment was performed at least 3 times.
Apoptosis assays
Apoptosis was assessed by staining 1 to 5 × 105 cells per sample with annexin V antibody and propidium iodide (FITC Annexin V Apoptosis Detection Kit with PI; 556547; BD Biosciences) according to the manufacturer’s protocol. Analysis was carried out by flow cytometry. Experiments were performed with 2 technical replicates, and each experiment was performed at least 2 times.
Viability assays
Cell viability was assessed in 96-well plates with a density of 1 to 5 × 104 cells per well. After 48 hours of treatment, cells were stained with trypan blue (T8154; Sigma-Aldrich Chemie GmbH), and viable cells were counted. Alternatively, cells were stained with MTS reagent (G3582; Promega) and analyzed on an Infinite M1000 PRO plate reader (Tecan, Männedorf, Switzerland).
Immunoblotting
Cells were pelleted and lysed using RIPA buffer at 4°C (89900; Thermo Fisher Scientific) supplemented with protease (11836170001; Roche, Basel, Switzerland) and phosphatase inhibitor (04906845001; Roche). Primary AML samples were lysed using SDS lysis buffer (0.1% SDS and 50 mM of Tris; pH, 8) supplemented with protease and phosphatase inhibitor. After centrifugation, protein concentration was determined with a Pierce BCA Protein Assay Kit (23227; Thermo Fisher Scientific). Equal amounts of whole-cell lysate were mixed with 4X LDS sample buffer (NP0008; Thermo Fisher Scientific) and 10X sample-reducing agent (NP0009; Thermo Fisher Scientific), heated for 10 minutes at 70°C and loaded on a 4% to 12% tris-glycine gradient gel (XP04122BOX; Thermo Fisher Scientific) for sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis. Proteins were transferred onto a nitrocellulose membrane (GE10600001; Sigma-Aldrich), which was blocked with 5% bovine serum albumin (T844.2; Carl Roth GmbH & Co KG) in TBST buffer. Membranes were incubated at 4°C overnight with anti–BCL-2 (1:4000; ab692; Abcam, Cambridge, United Kingdom), anti–B-actin (1:5000; A5441; Sigma-Aldrich), anti-ERK (1:1000; 4695; Cell Signaling Technology), anti-GSK3A/B (1:200; sc-7291; Santa Cruz Biotechnology, Dallas, TX), anti–MCL-1 (1:4000; ab32087; Abcam), anti–phosphorylated ERK (pERK) Thr202/Tyr204 (1:2000; 4370; Cell Signaling Technology), anti-pGSK3A/B Ser21/9 (1:1000; 8566; Cell Signaling Technology), anti–pMCL-1 serine 159 (S159; 1:500; ab111574; Abcam), anti–pMCL-1 threonine (T163; 1:1000; 14765S; Cell Signaling Technology), and secondary antibodies against mouse or rabbit immunoglobulin (1:4000; P044701-2 and P044801-2; Dako/Agilent, Santa Clara, CA). Proteins were visualized using ECL reagent (RPN2232; GE Healthcare, Chicago, IL) with an Amersham imager 600 (Cytiva, Chalfont St Giles, United Kingdom).
Overexpression and site-directed mutagenesis
Lentiviral vectors encoding for MCL-1 (140746) and corresponding empty vector (17452) were purchased from Addgene. Gene fragments for overexpression of ERK were obtained from Integrated DNA Technologies (Coralville, IA) and corresponding empty vector from System Biosciences (CD811A-1; Palo Alto, CA). Lentivirus was produced by transfecting lentiviral vectors together with psPAX2 (12260; Addgene, Watertown, MA) and pMD2.G (12259; Addgene) into HEK293T cells. Virus supernatant was used to infect HL60 cells, which then underwent puromycin selection (0.7 μg/mL; P8833-10MG; Sigma-Aldrich).
Site-directed mutagenesis was used to introduce the phosphorylation site mutation S159A in MCL-1. The mutation was introduced by a polymerase chain reaction (Platinum SuperFi DNA Polymerase; 12351010; Thermo Fisher Scientific) with back-to-back primers (GTAGTGCCCCGTCCGTAC TGGTG and CCTCGACGCCGCCGCCAGCAG) followed by KLD treatment. Mutagenesis results were confirmed by Sanger sequencing.
PDX model
Patient-derived xenograft (PDX) cells of a highly aggressive AML with complex karyotype and TP53 mutation (AML-372)18 were injected IV into immunocompromised NSG mice at minimal age 8 weeks. Mice were randomly assigned to 1 of 4 treatment groups: gilteritinib at 85 mg/kg, venetoclax at 40 mg/kg, gilteritinib and venetoclax, or vehicle (sterile water with 1% Tween and 3% ethanol). Treatment was started when engraftment levels of 0.2% of human CD45+ cells were reached and was conducted for 4 weeks. Bone marrow aspirations were performed 4 weeks from start of treatment. Two weeks after the end of treatment, mice were euthanized, and bone marrow was isolated and analyzed by fluorescence-activated cell sorting for CD45 expression (clone HI30; BD Biosciences). Animal experiments were approved and performed in accordance with all regulatory guidelines of the official committee (Regierungspräsidium Karlsruhe).
Drug sensitivity and statistical analysis
All tests were performed using R (version 4.0.4) and RStudio Server (version 1.4.1106-5) or GraphPad Prism (version 9.2.0). Data were analyzed for normal distribution before statistical analyses. Values are presented as mean ± standard deviation of replicates. Two-tailed Student t test was used to determine statistical significance unless stated otherwise. For ex vivo drug screens, synergy scores were computed according to the Bliss independence model19 and the zero interaction potency model20 with the synergyfinder R package (version 2.4.15).21 Fifty percent inhibitory concentration scores were computed automatically using this R package, normalized for <70% variance and outliers. Relative inhibition (RI) scores were computed according to the area under the curve of the viability curves. The RI scores indicate the proportion of the maximum possible inhibition of each drug. The shiny app for data sharing utilizes r-base:4.1.3 installed from docker. Previously analyzed results are presented using ggplot2 3.3.5, ComplexHeatmap 2.10.0, and corrplot 0.92. Synergy for individual samples was freshly calculated with synergyfinder 3.2.6. The graphical abstract was created with BioRender.com.
Results
High-throughput drug screening identifies venetoclax combination partners for high-risk AML
We conducted a high-throughput ex vivo drug perturbation experiment with venetoclax and drugs targeting relevant pathways in myeloid malignancies (Figure 1A). Thirty-one samples from patients with high-risk AML (supplemental Tables 1A and 2-4; supplemental Figure 1A) were incubated with venetoclax (0, 1, and 20 nM) and 64 drugs in 5 concentrations. High–risk was defined as either refractory to conventional chemotherapy or high-risk status according to European LeukemiaNet 2017 guidelines.16
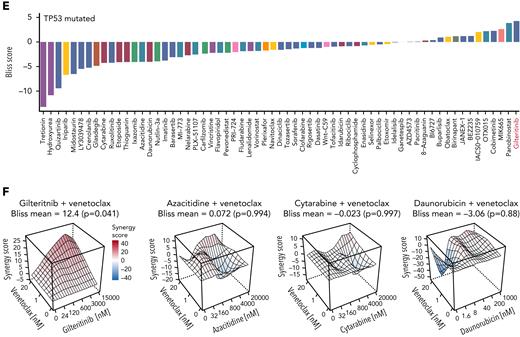
High-throughput drug screening approach identifies gilteritinib as a synergistic combination partner for venetoclax. (A) Experimental setup of the drug screening approach. A total of 31 high-risk AML patient samples were treated with venetoclax (0, 1, and 20 nM) with 64 different drugs in 5 different concentrations for 48 hours. Maximum concentrations used in the drug screen were 50% inhibitory concentrations found in literature; all other concentrations were deduced from division steps by 5. Viability was determined as a readout using CellTiter-Glo, and synergism scores (Bliss and zero interaction potency) were calculated using the synergyfinder R package (version 2.4.13).21 (B) Waterfall plot of mean Bliss scores of all drugs combined with venetoclax (calculated as the mean over all Bliss scores reached with each drug in 5 concentrations combined with venetoclax in 2 concentrations). Bliss synergy score was calculated as described by Bliss et al.19 Colors indicate targets of the respective drugs. Waterfall plot is shown for all primary AML samples (n = 31). (C) RI reached by gilteritinib monotherapy (left) and maximum Bliss synergy scores for venetoclax-gilteritinib reached in all tested concentrations (right) in FLT3 ITD mutated (n = 10) or FLT3 wild-type (WT; n = 17) samples. RI scores were computed according to the area under the curve of the viability curves. The RI scores indicate the proportion of the maximum possible inhibition of each drug independent of a single concentration. Mean RI and mean maximum Bliss scores, respectively, of individual patient samples are shown. Colors indicate the FLT3 mutational status. Statistical significance was assessed using a 2-tailed Student unpaired t test. (D) Heat map depicting Bliss scores for venetoclax combined with different FLT3 inhibitors in FLT3 WT and FLT3-mutated patient samples. (E) Waterfall plot of mean Bliss scores of all drugs combined with venetoclax (calculated as the mean over all Bliss scores reached with each drug in 5 concentrations combined with venetoclax in 2 concentrations). Bliss synergy score was calculated as described by Bliss et al.19 Colors indicate targets of the respective drugs. Waterfall plot is shown for the subgroup of patients with TP53 mutations obtained at first diagnosis (n = 6). (F) Bliss synergy scores of venetoclax in combination with gilteritinib, azacitidine, cytarabine, and daunorubicin, respectively, in a patient sample with TP53 mutation and FLT3 WT. Colors indicate synergism calculated as described by Bliss et al.19 Synergy scores of ≥0 are regarded as synergistic. ∗P ≤ .05. ITD, internal tandem duplication.
High-throughput drug screening approach identifies gilteritinib as a synergistic combination partner for venetoclax. (A) Experimental setup of the drug screening approach. A total of 31 high-risk AML patient samples were treated with venetoclax (0, 1, and 20 nM) with 64 different drugs in 5 different concentrations for 48 hours. Maximum concentrations used in the drug screen were 50% inhibitory concentrations found in literature; all other concentrations were deduced from division steps by 5. Viability was determined as a readout using CellTiter-Glo, and synergism scores (Bliss and zero interaction potency) were calculated using the synergyfinder R package (version 2.4.13).21 (B) Waterfall plot of mean Bliss scores of all drugs combined with venetoclax (calculated as the mean over all Bliss scores reached with each drug in 5 concentrations combined with venetoclax in 2 concentrations). Bliss synergy score was calculated as described by Bliss et al.19 Colors indicate targets of the respective drugs. Waterfall plot is shown for all primary AML samples (n = 31). (C) RI reached by gilteritinib monotherapy (left) and maximum Bliss synergy scores for venetoclax-gilteritinib reached in all tested concentrations (right) in FLT3 ITD mutated (n = 10) or FLT3 wild-type (WT; n = 17) samples. RI scores were computed according to the area under the curve of the viability curves. The RI scores indicate the proportion of the maximum possible inhibition of each drug independent of a single concentration. Mean RI and mean maximum Bliss scores, respectively, of individual patient samples are shown. Colors indicate the FLT3 mutational status. Statistical significance was assessed using a 2-tailed Student unpaired t test. (D) Heat map depicting Bliss scores for venetoclax combined with different FLT3 inhibitors in FLT3 WT and FLT3-mutated patient samples. (E) Waterfall plot of mean Bliss scores of all drugs combined with venetoclax (calculated as the mean over all Bliss scores reached with each drug in 5 concentrations combined with venetoclax in 2 concentrations). Bliss synergy score was calculated as described by Bliss et al.19 Colors indicate targets of the respective drugs. Waterfall plot is shown for the subgroup of patients with TP53 mutations obtained at first diagnosis (n = 6). (F) Bliss synergy scores of venetoclax in combination with gilteritinib, azacitidine, cytarabine, and daunorubicin, respectively, in a patient sample with TP53 mutation and FLT3 WT. Colors indicate synergism calculated as described by Bliss et al.19 Synergy scores of ≥0 are regarded as synergistic. ∗P ≤ .05. ITD, internal tandem duplication.
To assess the reliability of our ex vivo drug screening platform, we clustered the drugs targeting various pathways (supplemental Figure 1B) based on the similarity of their response profiles across all AML samples (supplemental Figure 1C). The clustering reflected drug target identity and relatedness of drugs (supplemental Figure 1D). Patients’ in vivo responses to cytarabine induction therapy could be reproduced in the ex vivo drug screen (supplemental Figure 2A).
Drug synergy effects were assessed with the Bliss independence model.19 For validation, we applied the zero interaction potency model.20 The top 3 venetoclax combination partners identified in our screen were MIK665 (MCL-1 inhibitor), OTX015 (BET inhibitor), and gilteritinib (FLT3 inhibitor; Figure 1B). The MCL-1 inhibitor MIK665 showed the strongest effects in combination with venetoclax, supporting growing evidence that the antiapoptotic protein MCL-1 confers resistance to BCL-2 inhibition in AML.22 The bromodomain inhibitor OTX015 has been described in preclinical studies to act synergistically with venetoclax by reducing MCL-1 levels.23 Our screening approach further identified the clinically approved drug gilteritinib as a highly active combination partner for venetoclax (Figure 1B; supplemental Figure 2B).
We further analyzed combination effects in subgroups of patients with different mutations (supplemental Figures 2C-E and 3A-D). As expected, there was a trend toward a higher monotherapeutic effect of gilteritinib in FLT3-mutated AML compared with FLT3 wild-type specimens (P = .075; Figure 1C). However, the synergistic effect of venetoclax-gilteritinib was stronger in FLT3 wild-type than in FLT3-mutated AML (P = .0427; Figure 1C; supplemental Figures 2C-D and 3A-B). In line, RI and Bliss scores were higher in samples with FLT3 wild–type than in samples with FLT3 ITD (supplemental Figure 4A-B). The highest synergy was observed for gilteritinib or MIK665 combined with venetoclax when compared with standard AML treatments (supplemental Figure 4C). The synergistic effect with venetoclax was restricted to gilteritinib and was not observed for other FLT3 inhibitors, such as quizartinib, midostaurin, or sorafenib (Figure 1D; supplemental Figure 4D). Interestingly, gilteritinib was the top synergistic combination partner for venetoclax in the subgroup of patients with TP53 mutation (Figure 1E; supplemental Figure 4E; supplemental Tables 3 and 4), for whom reduced responses to venetoclax with HMAs or LDAC have been reported.24 Results of patients included at first diagnosis without FLT3 ITD, but with TP53 mutation, showed that gilteritinib exerted high synergism with venetoclax, whereas other standard therapeutic approaches did not synergize or weakly synergized with venetoclax (Figure 1F; supplemental Figure 5A-E).
Taken together, data from our ex vivo drug perturbation approach identifies gilteritinib and venetoclax as effective combination partners with high synergy in high-risk AML with FLT3 wild type and TP53 mutation.
Proteomic profiling reveals upregulation of FLT3 signaling in venetoclax-resistant AML
Next, we aimed to understand what drives resistance to the FDA/EMA-approved venetoclax-azacitidine combination in FLT3 wild-type samples. We chose 6 primary FLT3 wild-type samples based on their drug screen ex vivo responses to venetoclax-azacitidine and the availability of sufficient material for fluorescence-activated cell sorting and mass spectrometry (Figure 2A). Two primary samples were classified as venetoclax-azacitidine sensitive and 4 primary samples as venetoclax-azacitidine insensitive according to calculated synergy scores (Figure 2B). Venetoclax-azacitidine–insensitive AML samples were enriched for proteins involved in FLT3 pathway activity (eg, RAF/MAP, FLT3, and MAPK1/MAPK3; normalized enrichment score for FLT3 signaling, 1.83; adjusted P = .0001; Figure 2C-D; supplemental Tables 5 and 6). These findings suggest that high FLT3 signaling is associated with venetoclax-azacitidine resistance in FLT3 wild-type AML.
Proteomics of primary AML patient samples reveal upregulation of FLT3 and MAPK signaling in venetoclax-azacitidine–resistant samples. (A) Experimental setup of the proteomic experiments conducted with primary patient samples. FLT3 wild-type samples (n = 6) were divided into 2 groups according to Bliss scores achieved by venetoclax-azacitidine in the drug screening approach (high responders [n = 2] vs low responders [n = 4]). Cells were sorted for high CD34 and moderate CD45 expression, and whole proteome was examined by mass spectrometry and compared. (B) Maximum venetoclax-azacitidine Bliss scores of FLT3 wild-type patient samples analyzed by proteomics. Bliss >5 was defined as high response. (C) Normalized enrichment score (NES) plot for FLT3 (left) and MAPK (right) signaling in AML samples with high ex vivo response vs low response to venetoclax-azacitidine. (D) Heat map of FLT3 signaling–associated (top) and MAPK signaling–associated (bottom) proteins differentially expressed in AML samples with high ex vivo response vs low response to venetoclax-azacitidine.
Proteomics of primary AML patient samples reveal upregulation of FLT3 and MAPK signaling in venetoclax-azacitidine–resistant samples. (A) Experimental setup of the proteomic experiments conducted with primary patient samples. FLT3 wild-type samples (n = 6) were divided into 2 groups according to Bliss scores achieved by venetoclax-azacitidine in the drug screening approach (high responders [n = 2] vs low responders [n = 4]). Cells were sorted for high CD34 and moderate CD45 expression, and whole proteome was examined by mass spectrometry and compared. (B) Maximum venetoclax-azacitidine Bliss scores of FLT3 wild-type patient samples analyzed by proteomics. Bliss >5 was defined as high response. (C) Normalized enrichment score (NES) plot for FLT3 (left) and MAPK (right) signaling in AML samples with high ex vivo response vs low response to venetoclax-azacitidine. (D) Heat map of FLT3 signaling–associated (top) and MAPK signaling–associated (bottom) proteins differentially expressed in AML samples with high ex vivo response vs low response to venetoclax-azacitidine.
Venetoclax-gilteritinib reduces viability and colony-formation capacity and induces apoptosis in FLT3 wild-type AML
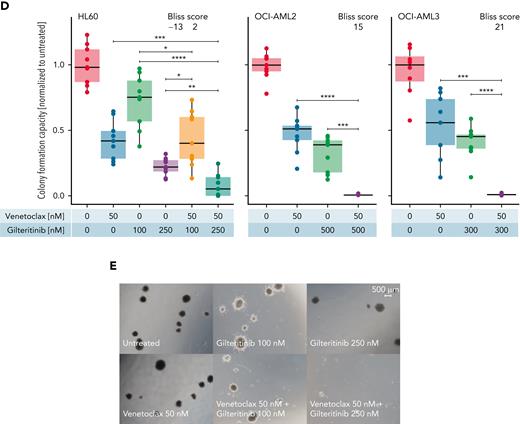
Next, we recapitulated the combinatorial effect of venetoclax-gilteritinib in FLT3 wild-type and FLT3 ITD cell lines. Ex vivo drug screens (supplemental Figure 6A-B) reflected the results obtained in primary samples. Upon the addition of 20 nM of venetoclax, we observed an increase in the RI in FLT3 wild-type cell lines, whereas only a minor difference was found for the FLT3 ITD samples (supplemental Figure 6C). In line with this, dose-response assays of venetoclax-gilteritinib in FLT3 wild-type HL60 and OCI-AML2 showed decreased cell viability after 48 hours in concentrations that can be reached in the plasma of patients (Figure 3A).25 The reduced cell viability was associated with increased apoptosis (Figure 3B-C). Furthermore, the drug combination synergistically reduced colony-formation capacity in 3 FLT3 wild-type AML cell lines (Figure 3D-E; supplemental Table 7).
Combination of venetoclax-gilteritinib reduces viability, increases apoptosis, and diminishes colony-formation capacity in FLT3 wild-type samples. (A) Dose-response assay for HL60 (left) and OCI-AML2 (right) cells treated with indicated concentrations of venetoclax and gilteritinib for 48 hours. Viability was assessed by staining with MTS reagent and was normalized to untreated controls. Data are presented as mean ± standard deviation (SD) of 3 technical replicates of 1 representative independent experiment (n = 3). (B) Percentage of apoptotic cells was measured by flow cytometry in HL60 and OCI-AML2 cells 24 hours after treatment with venetoclax (0, 10, and 100 nM) and gilteritinib (0 and 500 nM) upon staining with annexin V antibody and PI. Data are presented as mean ± SD of 2 independent experiments comprising 2 technical replicates each. Statistical significance was assessed using a 2-tailed Student unpaired t test. Bliss scores are given in a range of −100 to 100, with 100 as maximum Bliss score. (C) Representative images of fluorescence-activated cell sorting analysis of OCI-AML2 after 24 hours of treatment with gilteritinib (0 and 500 nM) and venetoclax (10 and 100 nM). (D) Effect of venetoclax and gilteritinib on colony-formation capacity of HL60, OCI-AML2, and OCI-AML3 cells was assessed by seeding cells in methylcellulose supplemented with the respective drugs for 10 days. Data of 3 independent experiments with 3 technical replicates each are presented. Bliss scores are given in a range of −100 to 100, with 100 as maximum Bliss score. (E) Representative microscopic images of colony-formation assays using HL60 cells treated with indicated concentrations of venetoclax and gilteritinib for 10 days. ∗P < .05, ∗∗P < .005, ∗∗∗P < .0005, ∗∗∗∗P < .00005.
Combination of venetoclax-gilteritinib reduces viability, increases apoptosis, and diminishes colony-formation capacity in FLT3 wild-type samples. (A) Dose-response assay for HL60 (left) and OCI-AML2 (right) cells treated with indicated concentrations of venetoclax and gilteritinib for 48 hours. Viability was assessed by staining with MTS reagent and was normalized to untreated controls. Data are presented as mean ± standard deviation (SD) of 3 technical replicates of 1 representative independent experiment (n = 3). (B) Percentage of apoptotic cells was measured by flow cytometry in HL60 and OCI-AML2 cells 24 hours after treatment with venetoclax (0, 10, and 100 nM) and gilteritinib (0 and 500 nM) upon staining with annexin V antibody and PI. Data are presented as mean ± SD of 2 independent experiments comprising 2 technical replicates each. Statistical significance was assessed using a 2-tailed Student unpaired t test. Bliss scores are given in a range of −100 to 100, with 100 as maximum Bliss score. (C) Representative images of fluorescence-activated cell sorting analysis of OCI-AML2 after 24 hours of treatment with gilteritinib (0 and 500 nM) and venetoclax (10 and 100 nM). (D) Effect of venetoclax and gilteritinib on colony-formation capacity of HL60, OCI-AML2, and OCI-AML3 cells was assessed by seeding cells in methylcellulose supplemented with the respective drugs for 10 days. Data of 3 independent experiments with 3 technical replicates each are presented. Bliss scores are given in a range of −100 to 100, with 100 as maximum Bliss score. (E) Representative microscopic images of colony-formation assays using HL60 cells treated with indicated concentrations of venetoclax and gilteritinib for 10 days. ∗P < .05, ∗∗P < .005, ∗∗∗P < .0005, ∗∗∗∗P < .00005.
Venetoclax-gilteritinib combination suppresses ERK and GSK3A/B phosphorylation and induces proteasomal degradation of MCL-1
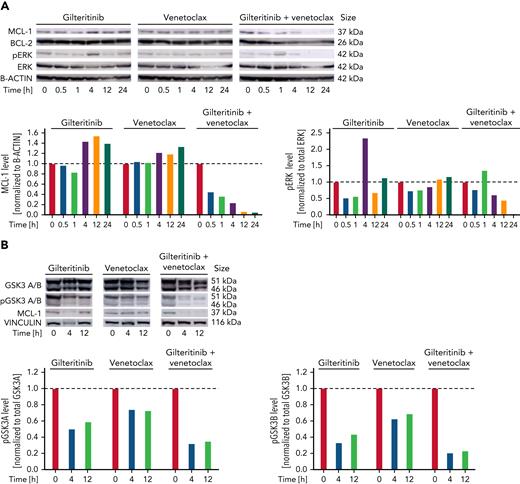
MCL-1 confers resistance to venetoclax.22 We thus examined protein expression of MCL-1 in HL60 cells after exposure to gilteritinib, venetoclax, or the combination. For gilteritinib, a concentration of 1 μM, which is slightly higher than usual drug plasma concentrations in patients,25 was chosen for optimal visualization of drug effects. MCL-1 protein expression decreased in venetoclax-gilteritinib–exposed cells compared with nontreated or single agent–treated cells (Figure 4A). Besides other kinases, gilteritinib predominantly inhibits FLT3 and AXL and thereby affects downstream signaling pathways, such as PI3K/AKT and Ras/Raf/ERK. Activation of most of these pathways occurs in FLT3 wild-type and mutant cells.26 The venetoclax-gilteritinib combination inhibited phosphorylation of ERK1/2 in FLT3 wild-type HL60 cells (Figure 4A). MCL-1 has a short half-life and is constantly degraded by the proteasome.27 Degradation of MCL-1 is mediated by various mechanisms that are not yet entirely understood.28 In addition to decreasing MCL-1 levels under venetoclax-gilteritinib, we observed a decrease of GSK3A/B phosphorylation (Figure 4B). Phosphorylation of GSK3 by pERK has been shown to reduce kinase activity.29 We further observed that MCL-1 T163 phosphorylation levels decreased upon exposure to the venetoclax-gilteritinib combination (Figure 4C). T163 phosphorylation induced by pERK stabilizes MCL-1.28 In addition, we found that venetoclax-gilteritinib increased MCL-1 S159 phosphorylation, whereas the single drugs did not (Figure 4C). Additional phosphorylation at S159 has been shown to be mediated by GSK3A/B and increased MCL-1 proteasomal degradation.29 In line, decreased pT163 and increased pS159 levels were associated with reduced levels of total MCL-1 in HL60 cells treated with venetoclax-gilteritinib (Figure 4C) and suggested proteasomal degradation of MCL-1. We performed venetoclax, gilteritinib, and combination treatment of HL60 cells in the presence and absence of the proteasome inhibitors carfilzomib and ixazomib, respectively. MCL-1 levels increased upon proteasome inhibition in cells treated with gilteritinib monotherapy or the drug combination (Figure 4D; supplemental Figure 7A). To evaluate whether the inactivation (ie, dephosphorylation) of ERK by venetoclax-gilteritinib is important for drug efficiency, we treated HL60 cells with venetoclax, gilteritinib, venetoclax-gilteritinib, or the combination of the ERK inhibitor SCH772984 with venetoclax. As venetoclax-gilteritinib, the combination of ERK inhibition with venetoclax reduced HL60 viability (supplemental Figure 7B). Immunoblotting of treated cells further demonstrated that direct ERK inhibition in combination with venetoclax strongly decreased pMCL-1 T163 and thus reduced total MCL-1 levels comparable to venetoclax-gilteritinib (supplemental Figure 7C). After overexpression of ERK, levels of pMCL-1 T163 increased (supplemental Figure 7D), and total MCL-1 levels remained stable upon treatment with venetoclax-gilteritinib (supplemental Figure 7E). Therefore, reduction of pERK-mediated phosphorylation of MCL-1 at T163 is crucial for the mode of action of venetoclax-gilteritinib. The inhibition of GSK3, in contrast, reduced the combination effect of venetoclax-gilteritinib and partially prevented the downregulation of MCL-1 (supplemental Figure 7F-G).
Venetoclax-gilteritinib combination reduces ERK and GSK3B phosphorylation and MCL-1 protein levels via proteasomal degradation. (A) Protein expression of MCL-1, BCL-2, pERK, and total ERK in HL60 cells treated for indicated timespan with 1 μM of gilteritinib, 20 nM of venetoclax, or the combination of both as analyzed by western blotting. B-actin levels are given as loading control. All western blot images have been cropped for improved clarity and conciseness. Quantification was performed using ImageJ. Data are representative of 3 independent experiments. (B) Protein expression of pGSK3A and B, total GSK3A and B, and MCL-1 in HL60 cells treated for indicated timespan with 1 μM of gilteritinib, 20 nM of venetoclax, or the combination of both as analyzed by western blotting. Vinculin levels are given as loading control. Quantification was performed using ImageJ. Data are representative of 3 independent experiments. (C) Protein expression of MCL-1, pMCL-1 S159, and pMCL-1 T163 in HL60 cells treated for indicated time points with 1 μM of gilteritinib, 20 nM of venetoclax, or the combination of both as analyzed by western blotting. B-actin levels are given as loading control. Quantification was performed using ImageJ. Data were obtained from the same biologic replicate as data shown in panel A and are representative of 3 independent experiments. (D) Protein expression of MCL-1, BCL-2, pERK, and total ERK in HL60 cells treated with 1 μM of gilteritinib, 20 nM of venetoclax, or the combination of both with or without the addition of the proteasome inhibitor carfilzomib for 4 hours. B-actin levels are given as loading control. Quantification was performed using ImageJ. Data are representative of 3 independent experiments.
Venetoclax-gilteritinib combination reduces ERK and GSK3B phosphorylation and MCL-1 protein levels via proteasomal degradation. (A) Protein expression of MCL-1, BCL-2, pERK, and total ERK in HL60 cells treated for indicated timespan with 1 μM of gilteritinib, 20 nM of venetoclax, or the combination of both as analyzed by western blotting. B-actin levels are given as loading control. All western blot images have been cropped for improved clarity and conciseness. Quantification was performed using ImageJ. Data are representative of 3 independent experiments. (B) Protein expression of pGSK3A and B, total GSK3A and B, and MCL-1 in HL60 cells treated for indicated timespan with 1 μM of gilteritinib, 20 nM of venetoclax, or the combination of both as analyzed by western blotting. Vinculin levels are given as loading control. Quantification was performed using ImageJ. Data are representative of 3 independent experiments. (C) Protein expression of MCL-1, pMCL-1 S159, and pMCL-1 T163 in HL60 cells treated for indicated time points with 1 μM of gilteritinib, 20 nM of venetoclax, or the combination of both as analyzed by western blotting. B-actin levels are given as loading control. Quantification was performed using ImageJ. Data were obtained from the same biologic replicate as data shown in panel A and are representative of 3 independent experiments. (D) Protein expression of MCL-1, BCL-2, pERK, and total ERK in HL60 cells treated with 1 μM of gilteritinib, 20 nM of venetoclax, or the combination of both with or without the addition of the proteasome inhibitor carfilzomib for 4 hours. B-actin levels are given as loading control. Quantification was performed using ImageJ. Data are representative of 3 independent experiments.
Taken together, our data show that the combination of venetoclax-gilteritinib reduced levels of pERK, pGSK3A/B, and pMCL-1 T163 and induced S159 phosphorylation, which was associated with proteasomal degradation of MCL-1.
MCL-1 S159A mutant cells do not respond to venetoclax-gilteritinib
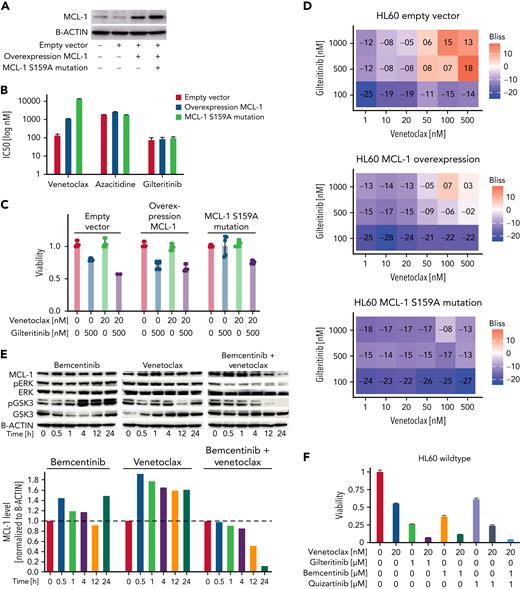
We sought to confirm the crucial role of S159 phosphorylation for efficacy of venetoclax-gilteritinib by lentiviral overexpression of MCL-1 with a phosphorylation-resistant S159A mutation (Figure 5A). MCL-1 S159A-overexpressing cells showed increased resistance toward venetoclax (Figure 5B). The venetoclax-gilteritinib combination was not synergistic, and the MCL-1 S159A cells were resistant to the drug combination (Figure 5C-D), underlining the importance of MCL-1 degradation for the effect of venetoclax-gilteritinib.
Overexpression of MCL-1 with S159A mutation inducesresistance to venetoclax, and cotargeting of FLT3 and AXL kinase was crucial for combination effect. (A) Protein expression of MCL-1 in parental HL60, empty vector–transduced, MCL-1–overexpressing, and MCL-1 S159A–mutated HL60 cells as analyzed by western blotting. B-actin levels are given as loading control. Data are representative of 3 independent experiments. (B) Fifty percent inhibitory concentrations (IC50s) for venetoclax, azacitidine, and gilteritinib in empty vector–transduced, MCL-1–overexpressing, and MCL-1 S159A–mutated HL60 cells were measured by treating cells in triplicate with the drugs in 7 concentrations (1 nM, 10 nM, 50 nM, 100 nM, 500 nM, 1 μM, and 10 μM) for 48 hours and staining with MTS reagent. IC50 was calculated at grcalculator.org, and representative results of 3 independent experiments are shown. (C) Effect of 20 nM of venetoclax, 500 nM of gilteritinib, or the combination of both on viability of empty vector–transduced, MCL-1–overexpressing, and MCL-1 S159A–mutated HL60 cells. Viability was assessed by staining with MTS reagent and was normalized to untreated cells. Data are presented as mean ± standard deviation (SD) of 3 technical replicates of 1 representative independent experiment (n = 3). (D) Venetoclax-gilteritinib Bliss scores calculated from effect of the drug combination on viability of respectively transduced HL60 cells. Bliss scores are given in a range of −100 to 100, with 100 as maximum Bliss score. Bliss scores of ≥0 indicate synergy. (E) Protein expression of MCL-1, pERK, ERK, pGSK3, and GSK3 in HL60 cells treated for indicated time span with 1 μM of bemcentinib, 20 nM of venetoclax, or the combination of both as analyzed by western blotting. B-actin levels are given as loading control. Quantification was performed using ImageJ. Data are representative of 3 independent experiments. (F) Effect of 20 nM of venetoclax, 1 μM of gilteritinib, 1 μM of bemcentinib, 1 μM of quizartinib, or various combinations of the drugs on viability of HL60 cells. Viability was assessed by staining with MTS reagent and was normalized to untreated cells. Data are presented as mean ± SD of 3 technical replicates of 1 representative independent experiment (n = 3).
Overexpression of MCL-1 with S159A mutation inducesresistance to venetoclax, and cotargeting of FLT3 and AXL kinase was crucial for combination effect. (A) Protein expression of MCL-1 in parental HL60, empty vector–transduced, MCL-1–overexpressing, and MCL-1 S159A–mutated HL60 cells as analyzed by western blotting. B-actin levels are given as loading control. Data are representative of 3 independent experiments. (B) Fifty percent inhibitory concentrations (IC50s) for venetoclax, azacitidine, and gilteritinib in empty vector–transduced, MCL-1–overexpressing, and MCL-1 S159A–mutated HL60 cells were measured by treating cells in triplicate with the drugs in 7 concentrations (1 nM, 10 nM, 50 nM, 100 nM, 500 nM, 1 μM, and 10 μM) for 48 hours and staining with MTS reagent. IC50 was calculated at grcalculator.org, and representative results of 3 independent experiments are shown. (C) Effect of 20 nM of venetoclax, 500 nM of gilteritinib, or the combination of both on viability of empty vector–transduced, MCL-1–overexpressing, and MCL-1 S159A–mutated HL60 cells. Viability was assessed by staining with MTS reagent and was normalized to untreated cells. Data are presented as mean ± standard deviation (SD) of 3 technical replicates of 1 representative independent experiment (n = 3). (D) Venetoclax-gilteritinib Bliss scores calculated from effect of the drug combination on viability of respectively transduced HL60 cells. Bliss scores are given in a range of −100 to 100, with 100 as maximum Bliss score. Bliss scores of ≥0 indicate synergy. (E) Protein expression of MCL-1, pERK, ERK, pGSK3, and GSK3 in HL60 cells treated for indicated time span with 1 μM of bemcentinib, 20 nM of venetoclax, or the combination of both as analyzed by western blotting. B-actin levels are given as loading control. Quantification was performed using ImageJ. Data are representative of 3 independent experiments. (F) Effect of 20 nM of venetoclax, 1 μM of gilteritinib, 1 μM of bemcentinib, 1 μM of quizartinib, or various combinations of the drugs on viability of HL60 cells. Viability was assessed by staining with MTS reagent and was normalized to untreated cells. Data are presented as mean ± SD of 3 technical replicates of 1 representative independent experiment (n = 3).
Dual inhibition of FLT3 and AXL is essential for venetoclax-gilteritinib synergism
Gilteritinib affects kinases beyond FLT3, including the AXL receptor tyrosine kinase. We used bemcentinib, a specific AXL inhibitor, to analyze involvement of AXL in the synergistic effects of venetoclax-gilteritinib. MCL-1, pERK, and pGSK3 levels were evaluated after venetoclax-bemcentinib treatment (Figure 5E). In contrast to gilteritinib, the FLT3 inhibitors quizartinib and midostaurin, which do not target AXL, did not reduce levels of MCL-1, pERK, or pGSK3 in combination with venetoclax (supplemental Figure 7H-I). In line with this, treatment with quizartinib alone or in combination with venetoclax did not reduce the viability of HL60 wild-type cells, as did gilteritinib with or without venetoclax (Figure 5F). However, bemcentinib added to venetoclax-quizartinib mimicked the effects of venetoclax-gilteritinib (Figure 5F). These findings suggest that dual targeting of FLT3 and AXL is required for the synergistic effects of venetoclax-gilteritinib.
Gilteritinib combined with venetoclax reduces engraftment of FLT3 wild-type TP53-mutated PDX model in vivo
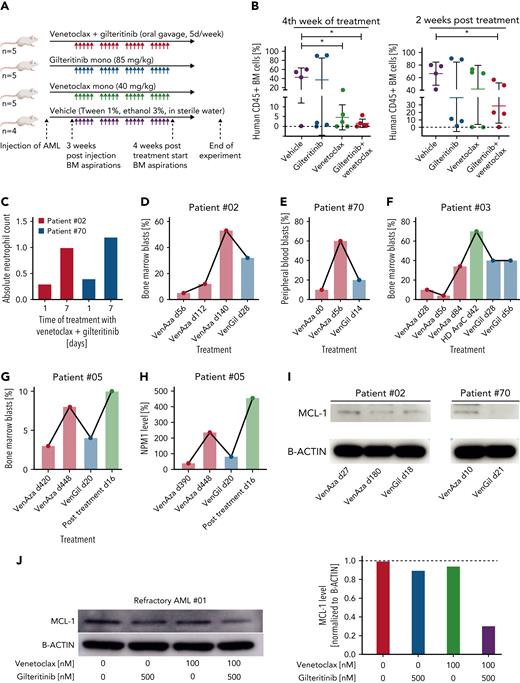
Next, we injected NSG mice with a highly aggressive PDX model (AML 372) to analyze in vivo efficiency of venetoclax-gilteritinib (supplemental Table 8). An FLT3 wild-type model with TP53 mutation was chosen because the drug screen suggested the highest activity in AML with a TP53 mutation. Mice were divided into 4 treatment groups: venetoclax, gilteritinib, venetoclax-gilteritinib, and vehicle (Figure 6A). Within the fourth week of treatment, engraftment was analyzed. The lowest engraftment was observed in the venetoclax-gilteritinib group, which was significantly different from engraftment levels in the control group (Figure 6B). Two weeks posttreatment, mice were euthanized. Again, the percentage of CD45+ blasts was lowest in the venetoclax-gilteritinib group. Interestingly, only engraftment levels within the venetoclax-gilteritinib group remained significantly lower compared with the vehicle group (P = .0308; Figure 6B). If only samples with a bone marrow engraftment >0.3% were included, engraftment within the venetoclax-gilteritinib group was significantly reduced compared with the venetoclax group (P = .0326; supplemental Figure 8A).
Primary AML from patients with venetoclax-azacitidine–refractory disease respond to venetoclax-gilteritinib in vitro and in vivo. (A) Overview of PDX model experiment. Treatment with gilteritinib (n = 5 mice), venetoclax (n = 5 mice), combination of both (n = 5 mice), or vehicle (n = 4 mice) started 3 weeks postinjection and lasted 4 weeks. Two weeks after end of treatment, mice were euthanized, and bone marrow (BM) was analyzed. (B) Percentage of CD45+ cells in BM obtained from PDX-transformed mice in the fourth week of treatment (left) or after euthanasia (right). (C) Absolute neutrophil count for patients 02 and 70 upon treatment with venetoclax-gilteritinib. (D-G) Percentage of BM (D, F, G) or peripheral blood (E) blasts of 4 venetoclax-azacitidine–refractory patients treated with venetoclax-gilteritinib. Blast percentage was analyzed at indicated time points upon start of the respective treatment condition. (H) Percentage of NPM1 level of patient 05. NPM1 level was analyzed at indicated time points upon start of the respective treatment condition. (I) Blasts from patients 02 and 70 obtained at different stages of therapy were lysed and analyzed for levels of MCL-1 and B-actin. (J) Primary AML blasts from patient 01 were treated for 12 hours with 100 nM of venetoclax, 500 nM of gilteritinib, or the combination of both. MCL-1 and B-actin levels were detected by western blotting and compared with untreated cells. ∗P < .05.
Primary AML from patients with venetoclax-azacitidine–refractory disease respond to venetoclax-gilteritinib in vitro and in vivo. (A) Overview of PDX model experiment. Treatment with gilteritinib (n = 5 mice), venetoclax (n = 5 mice), combination of both (n = 5 mice), or vehicle (n = 4 mice) started 3 weeks postinjection and lasted 4 weeks. Two weeks after end of treatment, mice were euthanized, and bone marrow (BM) was analyzed. (B) Percentage of CD45+ cells in BM obtained from PDX-transformed mice in the fourth week of treatment (left) or after euthanasia (right). (C) Absolute neutrophil count for patients 02 and 70 upon treatment with venetoclax-gilteritinib. (D-G) Percentage of BM (D, F, G) or peripheral blood (E) blasts of 4 venetoclax-azacitidine–refractory patients treated with venetoclax-gilteritinib. Blast percentage was analyzed at indicated time points upon start of the respective treatment condition. (H) Percentage of NPM1 level of patient 05. NPM1 level was analyzed at indicated time points upon start of the respective treatment condition. (I) Blasts from patients 02 and 70 obtained at different stages of therapy were lysed and analyzed for levels of MCL-1 and B-actin. (J) Primary AML blasts from patient 01 were treated for 12 hours with 100 nM of venetoclax, 500 nM of gilteritinib, or the combination of both. MCL-1 and B-actin levels were detected by western blotting and compared with untreated cells. ∗P < .05.
In conclusion, the venetoclax-gilteritinib combination is capable of effectively reducing FLT3 wild-type TP53-mutated AML.
Patients with AML relapsed/refractory to venetoclax-azacitidine respond to venetoclax-gilteritinib
Patients relapsing after venetoclax-azacitidine treatment do not respond to any known treatment.30 We treated 4 patients with relapsed or refractory FLT3 wild-type AML for whom additional treatment options had been exhausted with venetoclax-gilteritinib in off-label use. All patients had previously undergone allogeneic stem cell transplantation. Two (02 and 70) relapsed after treatment with venetoclax-azacitidine. One patient (03) was upfront refractory to venetoclax-azacitidine as well as to a treatment combination including high-dose cytarabine after the second relapse. For patient 02, treatment with venetoclax-gilteritinib led to a rise in absolute neutrophil count (Figure 6C) and peripheral blast clearance and a bone marrow blast reduction from 53% to 30% (Figure 6D). Unfortunately, this patient experienced infectious complications, with no further blast reduction, and died 4 weeks later. For patient 70, treatment with venetoclax-gilteritinib led to a peripheral blast reduction (Figure 6E) and a rise in absolute neutrophil count to >1.0/nL (Figure 6C). In patient 03, the combination approach led to a reduction in bone marrow blasts from 70% to 40% (Figure 6F). However, pancytopenia resulting from heavy pretreatment could not be resolved. Because the blast count was still at 40% 8 weeks after the start of venetoclax-gilteritinib treatment, this palliative treatment approach was stopped. Patient 05 relapsed after 15 courses of venetoclax-azacitidine. Bone marrow aspiration displayed blast counts of 8%, whereas the NPM1 level rose to 286%. After 3 weeks of treatment with venetoclax-gilteritinib, bone marrow aspiration displayed cytologic complete response with incomplete count recovery, and NPM1 levels dropped to 81% (Figure 6G-H). The treatment was discontinued because of grade 3 neutropenia (by Common Terminology Criteria for Adverse Events) and infectious complications. The treatment was not reinitiated based on patient decision and was changed to best supportive care.
We analyzed the expression of MCL-1 in bulk bone marrow blasts obtained from patients 02 and 70 receiving venetoclax-azacitidine and venetoclax-gilteritinib treatment, respectively. Notably, MCL-1 was strongly depleted in blasts from patient 70 and slightly decreased in blasts from patient 02 during venetoclax-gilteritinib treatment (Figure 6I).
To replicate the venetoclax-gilteritinib treatment findings in vitro, we treated blasts from patients 02 and 70 with venetoclax-gilteritinib in cell culture. Samples from patient 70 were also included in our ex vivo drug screen (supplemental Table 1), and here venetoclax-gilteritinib had additive effects, whereas response to venetoclax-azacitidine was weak (supplemental Figure 8B). No material for in vitro analysis was available from patients 03, 04, and 05. Therefore, we included material from an additional FLT3 wild-type patient (01) with a second relapse 20 months after allogeneic stem cell transplantation at progressive disease who was upfront refractory to venetoclax-azacitidine (supplemental Table 1).
In vitro, the venetoclax-gilteritinib combination reduced cell viability synergistically (supplemental Figure 8C), whereas venetoclax-azacitidine did not (supplemental Figure 8D). Furthermore, the combination significantly reduced colony-formation capacity in a synergistic manner (supplemental Figure 8E). MCL-1 protein levels were suppressed upon venetoclax-gilteritinib treatment in bone marrow blasts from patient 01 (Figure 6J), from whom sufficient material was available for western blotting.
Venetoclax-azacitidine resistance correlates with upregulation of MCL-1 and FLT3 downstream pathways
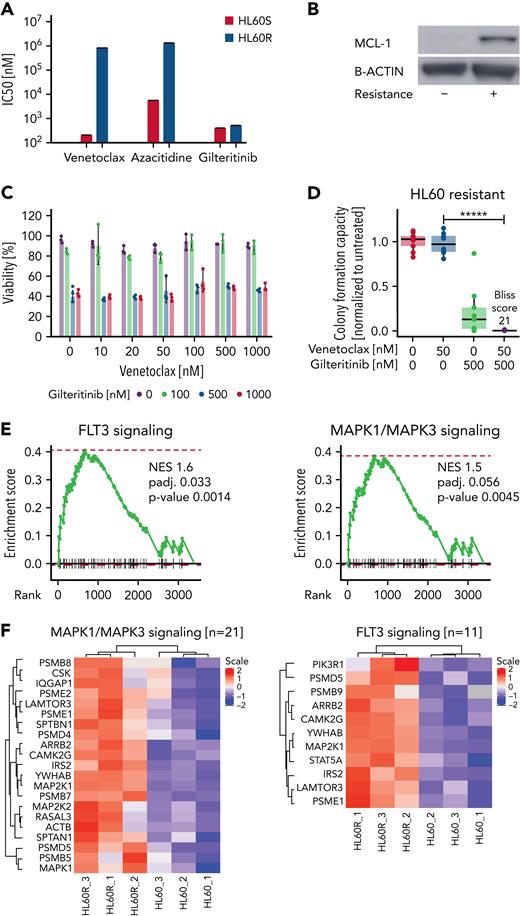
We generated resistant HL60 cells (HL60R) by constant exposure to increasing concentrations of venetoclax-azacitidine (Figure 7A). HL60R cells expressed high levels of MCL-1 (Figure 7B). Nonetheless, gilteritinib and venetoclax-gilteritinib decreased cell viability and inhibited the colony-formation capacity of HL60R (Figure 7C-D). Proteome analyses of parental HL60 and HL60R cells revealed that several FLT3 downstream signaling pathways were upregulated in venetoclax-azacitidine–resistant cells. FLT3 signaling –as well as MAPK signaling–associated proteins were enriched in HL60R (Figure 7E-F; supplemental Tables 9 and 10).
Venetoclax-azacitidine resistanceisassociated with upregulation of MCL-1 and FLT3 signaling and could be partly overcome by gilteritinib. (A) Fifty percent inhibitory concentrations (IC50s) for venetoclax, azacitidine, and gilteritinib in sensitive and venetoclax-azacitidine–resistant HL60 cells were measured by treating cells in triplicate with the drugs in 7 concentrations (1 nM, 10 nM, 50 nM, 100 nM, 500 nM, 1 μM, and 10 μM) for 48 hours and staining with MTS reagent. IC50 was calculated at grcalculator.org, and representative results of 3 independent experiments are shown. (B) MCL-1 levels in sensitive and resistant HL60 cells as estimated by western blotting. Blot is representative of 3 independent experiments. (C) Resistant HL60 cells were treated in technical triplicate with venetoclax (0, 10, 20, 50, 100, 500, and 1000 nM) and gilteritinib (0, 100, 500, and 1000 nM) for 48 hours. Viability was assessed by staining with MTS reagent. Data are presented as mean ± standard deviation from 1 of 3 independent experiments. (D) Effect of venetoclax-gilteritinib combination on colony-formation capacity of resistant HL60 cells was assessed by seeding cells in methylcellulose supplemented with the respective drugs for 10 days. Data of 3 independent experiments with 3 technical replicates each are shown. Bliss scores are given in a range of −100 to 100, with 100 as maximum Bliss score. (E) Normalized enrichment score (NES) plot for FLT3 (left) and MAPK (right) signaling in sensitive vs venetoclax-azacitidine–resistant HL60 cells. (F) Heat map of MAPK (left) and FLT3 (right) signaling–associated proteins differentially expressed in sensitive vs venetoclax-azacitidine–resistant HL60 cells. ∗∗∗∗∗P < .000005.
Venetoclax-azacitidine resistanceisassociated with upregulation of MCL-1 and FLT3 signaling and could be partly overcome by gilteritinib. (A) Fifty percent inhibitory concentrations (IC50s) for venetoclax, azacitidine, and gilteritinib in sensitive and venetoclax-azacitidine–resistant HL60 cells were measured by treating cells in triplicate with the drugs in 7 concentrations (1 nM, 10 nM, 50 nM, 100 nM, 500 nM, 1 μM, and 10 μM) for 48 hours and staining with MTS reagent. IC50 was calculated at grcalculator.org, and representative results of 3 independent experiments are shown. (B) MCL-1 levels in sensitive and resistant HL60 cells as estimated by western blotting. Blot is representative of 3 independent experiments. (C) Resistant HL60 cells were treated in technical triplicate with venetoclax (0, 10, 20, 50, 100, 500, and 1000 nM) and gilteritinib (0, 100, 500, and 1000 nM) for 48 hours. Viability was assessed by staining with MTS reagent. Data are presented as mean ± standard deviation from 1 of 3 independent experiments. (D) Effect of venetoclax-gilteritinib combination on colony-formation capacity of resistant HL60 cells was assessed by seeding cells in methylcellulose supplemented with the respective drugs for 10 days. Data of 3 independent experiments with 3 technical replicates each are shown. Bliss scores are given in a range of −100 to 100, with 100 as maximum Bliss score. (E) Normalized enrichment score (NES) plot for FLT3 (left) and MAPK (right) signaling in sensitive vs venetoclax-azacitidine–resistant HL60 cells. (F) Heat map of MAPK (left) and FLT3 (right) signaling–associated proteins differentially expressed in sensitive vs venetoclax-azacitidine–resistant HL60 cells. ∗∗∗∗∗P < .000005.
Discussion
Venetoclax combination therapy constitutes a major breakthrough in AML. However, relapses occur frequently, and few if any patients with high-risk AML are cured by the currently approved venetoclax combinations. Identification of the best synergistic combinations might improve patient responses and remission duration. However, these efforts are hampered by the fact that venetoclax is rather ineffective as monotherapy in AML. In addition, synergism drug screens with primary cancer cells are challenging.
In our high-throughput drug screening with primary high-risk AML specimens, gilteritinib was identified as a promising combination partner for venetoclax therapy in FLT3 wild-type specimens. Synergistic activity of venetoclax-gilteritinib has already been reported in FLT3-mutated AML in vitro and in vivo.31-35 Prior in vitro studies also detected synergistic effects of venetoclax-gilteritinib in FLT3 wild-type samples, but these initial findings were not further investigated,31,32 and the mechanism of action is unknown. An antileukemic effect of gilteritinib in patients with FLT3 wild-type AML was also observed in clinical studies.35 This effect might be explained by an autocrine activation of the nonmutated FLT3 kinase in patients with AML.36 The high synergism with venetoclax was specific for gilteritinib and was not found for other FLT3 inhibitors like midostaurin or quizartinib in our drug screen. Gilteritinib also inhibits AXL, a kinase that is significantly upregulated in AML samples.37,38 In our study, effects of either AXL inhibition with venetoclax or FLT3 inhibition (quizartinib and midostaurin) with venetoclax were lower than venetoclax-gilteritinib effects, respectively. Accordingly, combined AXL and FLT3 targeting is the likely mechanism for venetoclax-gilteritinib synergism in FLT3 wild-type AML.
Induction of MCL-1 was identified as a major mechanism of resistance against therapy with the BCL-2 inhibitor venetoclax.22 FLT3 downstream signaling was found to induce upregulation of MCL-1.39,40 AXL inhibition has been demonstrated to reduce MCL-1 levels in chronic lymphocytic leukemia samples.41 Both receptor tyrosine kinases, FLT3 and AXL, support proliferation and survival of AML cells through PI3K/AKT, Ras/Ref/MEK/ERK, and JAK/STAT signaling pathways.42 We identified ERK inhibition with subsequent activation of GSK3A/B, increased pS159, and decreased pT163, leading to reduced MCL-1 levels as mechanisms of action of the venetoclax-gilteritinib combination. In our experiments, higher concentrations of gilteritinib even reestablished venetoclax sensitivity in MCL-1–overexpressing cells.
MCL-1 is known to be regulated by various pathways.28 AML clones selected during venetoclax-azacitidine treatment have been described as more monocytic with higher expression of MCL-1.11 A reasonable treatment approach could be to add gilteritinib early to venetoclax in order to abolish the formation of resistant MCL-1–expressing clones.
Overall, venetoclax-gilteritinib was synergistic in FLT3 wild-type primary high-risk specimens. Of note, the combination reduced blast counts in several heavily pretreated FLT3 wild-type patients who lacked other treatment options. Given the late-stage disease of the patients and the individualized concepts, no additional conclusions can be drawn at this time. A clinical trial is required to assess the venetoclax-gilteritinib combination in patients with FLT3 wild-type AML.
Taken together, our study results show that rational in vitro drug testing opens new avenues to further improve venetoclax-based treatment options in AML.
Acknowledgments
The authors thank V. Eckstein for support with cell sorting. The authors also thank the teams of the biomaterial banks of Medical Department V, Heidelberg University Hospital; Heidi Altmann at the Department of Internal Medicine I, University Hospital of Dresden Carl Gustav Carus, Germany; and all patients for providing primary human cells.
M.J. received funding from the Physician Scientist Program of the University of Heidelberg. C.S. received funding from the Mildred Scheel Doctoral Program of German Cancer Aid. M.B. was supported in part by German Cancer Aid (Mildred-Scheel Junior Research Centre). C.M.-T. received funding from German Cancer Aid (grant BW70113908), the German Research Foundation (grant MU1328/18-1), the Federal Ministry of Education and Research (BMBF; Projektträger Jülich 031L0212A [SMART-CARE]), and the Wilhelm Sander Foundation (2021.145.1). S.D. received funding from the German Research Foundation (Sonderforschungsbereich 873 grants 122491522 and 464596535) and the BMBF (grant 464596535SYMPATY; project 01ZX1506).
Authorship
Contribution: M.J. designed the study, performed experiments and analyses, and wrote the manuscript. C.S. performed experiments and analyses and wrote the manuscript. P.-M.B. performed experiments and analyses and edited the manuscript. M.F.B. performed proteomic experiments and analyses. C.R. performed computational analyses. A.W. performed experiments and analyses and edited the manuscript. D.H. performed experiments and analyses. S.G. performed analyses and wrote the manuscript. S.R. performed experiments and analyses and edited the manuscript. S.A.H. performed experiments. M.K. performed experiments. C.K. performed experiments. L.V., B.B., A.P., K.W., and A.K.L. performed experiments. M.F. performed experiments and analyses. M.G. performed experiments and computational analyses. R.F.S. edited the manuscript. F.S., M.B., C.R., U.P, C.B., H.S., T.S., and S.R. edited the manuscript. C.P. supervised the project and edited the manuscript. G.V. supervised experiments and data analyses. B.V. performed experiments and analyses, supervised the project, and edited the manuscript. I.J., A.T., and J.K. supervised the project and edited the manuscript. C.M.-T. and S.D. designed the study, supervised the project, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Maike Janssen, University Hospital Heidelberg, Medical Department V, Hematology, Oncology, Rheumatology, Im Neuenheimer Feld 410, 69120 Heidelberg, Germany; e-mail: maike.janssen@med.uni-heidelberg.de.
References
Author notes
For original data, please contact maike.janssen@med.uni-heidelberg.de. Comprehensive visualization of results for viability, synergy, and relative inhibition for all samples from the drug screen is available in a shiny app (https://shiny-portal.embl.de/shinyapps/app/07_drug-screen).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
M.J. and C.S. as well as C.M.-T. and S.D. contributed equally to this work.



![Proteomics of primary AML patient samples reveal upregulation of FLT3 and MAPK signaling in venetoclax-azacitidine–resistant samples. (A) Experimental setup of the proteomic experiments conducted with primary patient samples. FLT3 wild-type samples (n = 6) were divided into 2 groups according to Bliss scores achieved by venetoclax-azacitidine in the drug screening approach (high responders [n = 2] vs low responders [n = 4]). Cells were sorted for high CD34 and moderate CD45 expression, and whole proteome was examined by mass spectrometry and compared. (B) Maximum venetoclax-azacitidine Bliss scores of FLT3 wild-type patient samples analyzed by proteomics. Bliss >5 was defined as high response. (C) Normalized enrichment score (NES) plot for FLT3 (left) and MAPK (right) signaling in AML samples with high ex vivo response vs low response to venetoclax-azacitidine. (D) Heat map of FLT3 signaling–associated (top) and MAPK signaling–associated (bottom) proteins differentially expressed in AML samples with high ex vivo response vs low response to venetoclax-azacitidine.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/140/24/10.1182_blood.2021014241/4/m_blood_bld-2021-014241-gr2.jpeg?Expires=1767702950&Signature=07YIUvheOcJUrt00~AF5YA6VfKtsgQlwrqe16nSlETchUAKnPaNPLsWL7y-qBoQO2UfPNQWZG1xTw62kO5JsM4Adbwd90OKP1GE-AhShZH~hHDpemQ9DFCN~v5Ww7XaDL~7vHY0cK-g4SirYn7eGjjFdMpQauG91TClljtsYzKecMxICLlj-UQItXabh50qsvRHhQsWjupfniziyko~yLjERUkcA4QS54pv-6mGUcoFuBnPXv3rGexgBqzwxB~Rjma1kbRVA4yTaWetzPUUBC9M5uoyaE9iCImavlg7BTa2fP4HTT26ieHjjcS5Zk2gpHYfQ-SuUKT~qSVMqhU5oNg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal