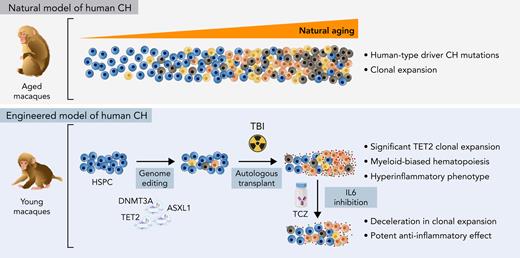
Key Points
Aging rhesus macaque HSPCs acquire clonal somatic mutations in the same set of genes as humans.
Engineered macaques demonstrate expansion of TET2 LOF clones and hyperinflammation and hold promise for testing interventions.
Abstract
Individuals with age-related clonal hematopoiesis (CH) are at greater risk for hematologic malignancies and cardiovascular diseases. However, predictive preclinical animal models to recapitulate the spectrum of human CH are lacking. Through error-corrected sequencing of 56 human CH/myeloid malignancy genes, we identified natural CH driver mutations in aged rhesus macaques matching genes somatically mutated in human CH, with DNMT3A mutations being the most frequent. A CH model in young adult macaques was generated via autologous transplantation of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9–mediated gene-edited hematopoietic stem and progenitor cells (HSPCs), targeting the top human CH genes with loss-of-function (LOF) mutations. Long-term follow-up revealed reproducible and significant expansion of multiple HSPC clones with heterozygous TET2 LOF mutations, compared with minimal expansion of clones bearing other mutations. Although the blood counts of these CH macaques were normal, their bone marrows were hypercellular and myeloid-predominant. TET2-disrupted myeloid colony-forming units isolated from these animals showed a distinct hyperinflammatory gene expression profile compared with wild type. In addition, mature macrophages purified from the CH macaques showed elevated NLRP3 inflammasome activity and increased interleukin-1β (IL-1β) and IL-6 production. The model was used to test the impact of IL-6 blockage by tocilizumab, documenting a slowing of TET2-mutated expansion, suggesting that interruption of the IL-6 axis may remove the selective advantage of mutant HSPCs. These findings provide a model for examining the pathophysiology of CH and give insights into potential therapeutic interventions.
Introduction
Clonal hematopoiesis (CH) refers to an expansion of hematopoietic stem and progenitor cells (HSPCs) carrying somatic mutations in individuals without evidence of hematological abnormalities.1,2 The prevalence of CH and clone size increase with age (age-related CH),3 and mutations in a specific set of genes previously associated with myeloid malignancies mainly drive CH. Particularly, loss-of-function (LOF) mutations in the epigenetic regulatory genes DNMT3A and TET2 account for more than half of the cases, followed by mutations in ASXL1.4 A variant allele frequency (VAF) of at least 2% is commonly considered to be a clinically relevant threshold for CH. These mutations provide an advantage that results in clonal expansion.5 CH has a broad range of physiological and prognostic consequences,6-8 including cardiovascular disease (CVD).9,10 However, the relationships between these mutations, clonal expansion, and clinical outcomes are not well understood due to difficulties in studying individuals with CH longitudinally in the absence of overt clinical abnormalities.
Murine models have revealed the hematopoietic roles of the common CH epigenetic regulators DNMT3A, TET2, and ASXL1 (DTA).11,12 A recent xenotransplant study documented selective expansion of human HSPCs with DTA LOF mutations in immunodeficient mice.13 These mouse models have provided critical insights into CH pathophysiology but have some limitations, including rapid progression to leukemic transformation in some models and differences in some underlying relevant properties between mice and humans, including HSC self-renewal rates and lifespans. Nonhuman primates, specifically noninbred rhesus macaques (RMs, Macaca mulatta), have characteristics that suggest relevance as a model for human CH, namely HSPC frequencies and cycling rates, telomere length, marrow architecture, lifespan, and size much closer to humans than rodent models, as well as aging phenotypes reported as similar to humans.14 Here, we report for the first time the natural occurrence of CH in aged RMs and describe the creation of a preclinical model for CH in young RMs via clustered regularly interspaced short palindromic repeats (CRISPR) engineering of DTA mutations in HSPCs, with relevance for testing therapies interrupting CH-related hyperinflammatory pathways.
Methods
Animals
Sixty aged macaques from 3 cohorts were included in the natural CH study, and 3 young adult macaques in the engineered CH studies (details in supplemental Methods, available on the Blood Web site). All procedures were approved by the Animal Care and Use Committees at National Heart, Lung, and Blood Institute (NHLBI), National Institute on Aging (NIA), and Oregon Health and Science University (OHSU).
Error-corrected panel sequencing for natural CH mutation detection
DNAs extracted from granulocytes and skin fibroblasts were sheared and libraries prepared for error-corrected sequencing using a custom RM target bait panel homologous to exons of 56 common human CH genes via the Agilent SureSelect XT HS hybridization protocol. Libraries were pooled and sequenced on the Illumina NovaSeq 150 bp paired-end platform with 5000× on-target depth.
HSPC autologous transplantation for engineered CH model creation
CD34+ HSPCs were mobilized, collected via apheresis, immunoselected, and electroporated with a guide RNA (gRNA) pool targeting DTA loci complexed to CRISPR-associated protein 9 (Cas9) ribonucleoprotein (Cas9 RNP) as described.17 Edited HSPCs were reinfused into the autologous animal following 10 Gy total body irradiation (TBI).
Cell purification
Mononuclear cells and granulocytes were separated from bone marrow (BM) and peripheral blood (PB) via density gradient centrifugation. CD34+ cells were immunoselected using magnetic beads. Hematopoietic lineage cells were purified via fluorescence-activated cell sorting following antibody staining (supplemental Table 1).
Targeted deep sequencing
On-target edited regions in DNA of RM samples were amplified using gene-specific primers (supplemental Table 2), ligated with customized indexes, and sequenced on Illumina MiSeq. Target sites (mean depth, ≥250 000) were aligned using CRISPResso (http://crispresso.rocks), and the number of reads for each unique indel across multiple time points was compiled via a custom pipeline. Heatmaps were generated via R.
Histopathologic analyses
BM biopsies from the iliac crests were fixed, decalcified, and embedded in paraffin. Serial sections were stained for immunohistochemical analyses.
Gene expression analyses
Myeloid colony-forming units (CFUs) derived from CD34+ cells were processed individually.18 Ultralow-input CFUs identified as wild type (WT) or TET2 uni-allelic mutant on Sanger sequencing were used for RNA sequencing (RNA-seq). Sequencing reads were analyzed using Salmon,19 Tximeta,20 DESeq2,21 and custom R code. Genome assembly Mmul_10 release 104 was used to build a transcript-level index along with macaque annotation from Ensembl. The differential gene expressions were visualized with heatmaps and clustered in an unsupervised manner using custom R code.
Enzyme-linked immunosorbent assays
Plasma was separated from cells by centrifugation at 4°C. Purified CD14+CD163+ macrophages were primed with lipopolysaccharide (LPS) for 4 hours and stimulated with adenosine triphosphate (ATP) for another 30 minutes, and cell culture supernatant was harvested. Interleukin-6 (IL-6), IL-6Ra, and IL-1β concentrations were measured in plasma or the supernatant using enzyme-linked immunosorbent assay kits (supplemental Table 1).
Quantitative reverse transcription polymerase chain reaction
Total RNA was extracted from the cultured macrophages with or without stimulation. Complementary DNA was synthesized using SuperScript III First-Strand Synthesis System (ThermoFisher) and expression of NLRP3, PYCARD, IL1b, IL18, IL6, IL6R, and TRAF6 measured using the QuantStudio 12K Flex Real-Time PCR system (ThermoFisher).
NLRP3 inflammasome activity
Either unstimulated or LPS/ATP-activated macrophages were stained with the FAM-FLICA Caspase-1 probe and analyzed for caspase-1 activity by flow cytometry. Cytospin slides prepared at a density of 1500 macrophages per slide were stained with a monoclonal antibody specific for Apoptosis-associated speck-like protein containing a CARD (ASC) (supplemental Table 1). ASC specks in each slide were imaged using a Leica SP8 confocal microscope.
TCZ infusions
Pharmaceutical-grade tocilizumab (TCZ) (Actemra) 10 mg/kg in a 52 mL solution was IV infused weekly over 1 hour for a total of 16 doses.
Results
RMs are a natural model for human age-related CH
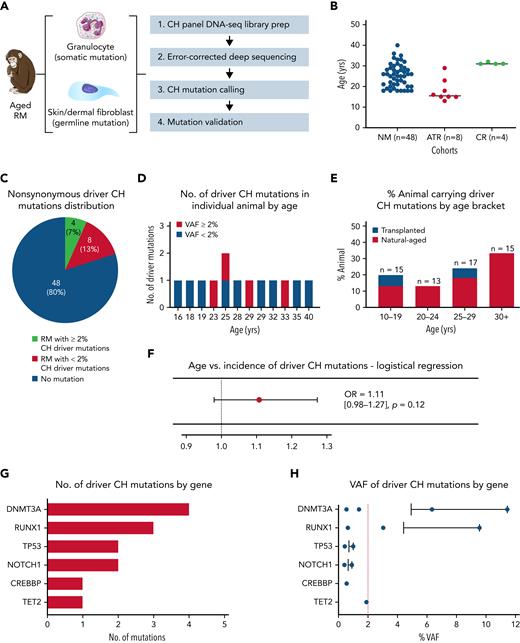
To determine the prevalence and mutational spectrum of CH in aged RMs, we performed error-corrected deep sequencing of 56 genes recurrently mutated in human CH (Figure 1A; supplemental Table 3) in a total of 60 animals with a median age of 25 years, obtained from 3 different cohorts (Figure 1B; supplemental Table 4), including nonmanipulated aged macaques (n = 48), autologous transplant recipients (n = 8), and calorie-restricted aged macaques (n = 4). RM age multiplied by 2.5 to 3 corresponds approximately to human age.22,23 Nonsynonymous somatic mutations (supplemental Tables 5 and 6) were identified in PB granulocytes via comparison with skin cells from the same animal to exclude germline polymorphisms. Greater than 5000× mean on-target coverage (supplemental Figure 1A; supplemental Table 7) and error correction permitted mutation calling down to 0.1%.
Natural CH in aged RMs. (A) Schematic workflow for error-corrected ultra-deep sequencing using a custom panel of primers covering RM homologs of 56 genes associated with human CH. Circulating granulocytes and dermal fibroblasts from aged RM (n = 60) were sequenced to detect somatic and germline mutations, respectively. (B) Cohort type and age distribution of the RMs enrolled in this analysis. (C) The number/percentage of aged RM carrying nonsynonymous driver CH somatic mutations identified by error-corrected deep sequencing (stratified by VAF ≥2%). (D) The number of driver CH mutations fitting conservative criteria in each of the 12 CH animals is depicted by age (red, VAF ≥2%; blue, VAF <2%). (E) The prevalence of aged RMs carrying CH somatic mutations stratified by age groups. Blue bars represent transplanted animals, and red bars represent nontransplanted naturally-aged animals. (F) Logistical regression analysis of age vs incidence of natural CH. Transplant recipients are excluded from this analysis. (G) The number and type of driver CH mutations are graphed by genes from the most to least frequent. (H) The VAF of each identified mutation are shown for each gene, from the most to least frequent. The bar represents the mean VAF of each gene, and the red dashed line indicates a 2% VAF. ATR, autologous transplant recipient; CR, calorie-restricted cohort; NM, nonmanipulated; OR, odds ratio.
Natural CH in aged RMs. (A) Schematic workflow for error-corrected ultra-deep sequencing using a custom panel of primers covering RM homologs of 56 genes associated with human CH. Circulating granulocytes and dermal fibroblasts from aged RM (n = 60) were sequenced to detect somatic and germline mutations, respectively. (B) Cohort type and age distribution of the RMs enrolled in this analysis. (C) The number/percentage of aged RM carrying nonsynonymous driver CH somatic mutations identified by error-corrected deep sequencing (stratified by VAF ≥2%). (D) The number of driver CH mutations fitting conservative criteria in each of the 12 CH animals is depicted by age (red, VAF ≥2%; blue, VAF <2%). (E) The prevalence of aged RMs carrying CH somatic mutations stratified by age groups. Blue bars represent transplanted animals, and red bars represent nontransplanted naturally-aged animals. (F) Logistical regression analysis of age vs incidence of natural CH. Transplant recipients are excluded from this analysis. (G) The number and type of driver CH mutations are graphed by genes from the most to least frequent. (H) The VAF of each identified mutation are shown for each gene, from the most to least frequent. The bar represents the mean VAF of each gene, and the red dashed line indicates a 2% VAF. ATR, autologous transplant recipient; CR, calorie-restricted cohort; NM, nonmanipulated; OR, odds ratio.
We applied very conservative criteria to define macaque somatic mutations as representing driver CH. The mutations had to fulfill criteria previously established for human driver mutations utilizing this sequencing panel (supplemental Table 5),24,25 and missense or in-frame mutations had to impact on homologous amino acids exactly matching human mutations reported in the Catalogue Of Somatic Mutations In Cancer (COSMIC) database (cancer.sanger.ac.uk).26 Thirteen driver CH mutations were detected in a total of 12 RMs (20%) with a median VAF of 1% (range, 0.40% to 11.45%), with 4 (7%) having a mutation with VAF ≥2% (Figure 1C; supplemental Table 6). Among those with CH mutations, a single mutation was detected in 11 (92%) animals, and 2 mutations were detected in 1 animal (8%) (Figure 1D). When the cohort was broken down by age brackets, the proportion of natural-aged animals with driver CH mutations showed a trend of increasing with age (Figure 1E), although this trend did not reach significance applying a logistical regression model accounting for sex, housing facility, and animal protocol, likely due to the limited sample size (odds ratio, 1.11; 95% confidence interval, 0.98-1.27; P = .12; Figure 1F).
We found a similar mutational spectrum in these aged RMs compared with human CH, with DNMT3A as the gene with the most frequent driver mutations and with the highest VAF (Figure 1G-H). Three animals were found to have the same DNMT3A frameshift mutation (p.S349X). Of note, RUNX1 frameshift mutations were found, despite having not been previously reported as a common human CH mutation, whereas no alterations were found to date in several human CH genes such as JAK2 and PPM1D, potentially due to cohort size.
The frameshift DNMT3A and RUNX1 mutations were validated by digital droplet PCR (ddPCR) on independently processed samples, and the overall results correlated closely with the VAF values from the panel sequencing (supplemental Figure 1B). In longitudinal samples collected over a time span of 1 to 5 years for the 3 living animals with DNMT3A-p.S349X mutations, clones with the mutations gradually rose in PB granulocytes and T cells but were undetectable in B cells. Two animals with RUNX1-p.TV114-115TX mutations showed evidence for clonal expansion in longitudinal samples collected over 1- to 2-year time periods. The other DNMT3A mutation (p.FF731F) was also validated by ddPCR in macaque M999, but only a single time point sample was available (supplemental Figure 1C). All animals had normal blood counts at the time of sampling, without evidence of hematologic neoplasms (supplemental Figure 2). These findings support aged RMs as natural models for human CH.
Creation of genetically engineered model of human CH in young macaques
Considering the difficulty in obtaining sufficient numbers of aged macaques in captivity for studies of naturally occurring CH, we sought to create a relevant preclinical model in younger RMs via autologous transplantation of HSPCs engineered with human LOF mutations, focusing on the commonly mutated DTA epigenetic regulator genes. At the time this study was initiated, ASXL1 human CH mutations were presumed LOF; however, subsequent investigation in murine models have cast doubt on this assumption.27 We designed candidate gRNAs targeting exons encoding 5′ and/or functional domains of the encoded protein for the 3 most commonly mutated genes in human CH. Exon sequences and structures of these epigenetic regulators are highly conserved between RMs and humans. Based on nonhomologous end joining efficiency of indel formation in RM primary CD34+ HSPCs, the most effective gRNAs for each gene were selected (supplemental Figure 3). Overall HSPC editing efficiency was targeted to be relatively low in order to analyze the competitive fitness of HSPCs with heterozygous mutations in a setting analogous to small human CH clones.
HSPCs from 3 young adult macaques aged 3 to 9 years were electroporated with Cas9 RNPs containing a gRNA pool targeting DTA and reinfused into the autologous animal following myeloablative TBI (Figure 2A; supplemental Table 8). In the first animal, ZL26, 20% of the HSPCs were edited with RNPs targeting the AAVS1 safe harbor locus, functioning as a predicted biologically inert comparator control cell population, and infused along with the DTA-edited cells (Figure 2B).17,28 All macaques engrafted promptly, with recovery of red blood cell, white blood cell, and platelet counts (supplemental Figure 4). Following engraftment, all animals maintained normal blood counts and distribution of circulating mature leukocytes for up to 55 months.
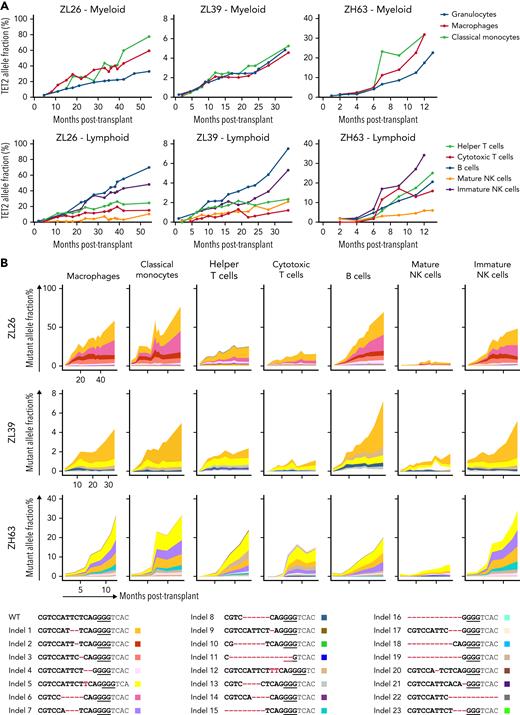
Longitudinal tracking of mutant clones in engineered CH macaques. (A) Schematic outline of RM HSPC gene editing for CH loci and subsequent autologous transplantation. (B) Summary of the gRNAs used for gene editing in each animal (n = 3; ZL26, ZL39, and ZH63). (C) Mutation frequencies at target sites are quantified from PB granulocytes collected at different time points from the 3 macaques using targeted deep sequencing. The percentage of reads containing indel mutations for each target site by individual macaque is plotted over time. (D) Indel frequencies at the TET2 target site in granulocytes collected over time are combined into a single graph for comparison. (E) Heatmaps on the right show the fractional contribution of each indel to total sequencing reads in granulocyte samples over time. Indels in HSPC measured before editing (Precut) and in infusion product (IP) are also shown in the first 2 columns. The most abundant indel types for the TET2 target site in each animal are listed on the left, with the WT sequence at the bottom. Indels are highlighted in red font, and the predicted Cas9 cut site is indicated by a red arrow. Changes in percent contribution of unique clones over time are shown stacked on the left, with each indel type marked with a distinct color across the 3 RMs. Td, doubling time; PAM, protospacer adjacent motif.
Longitudinal tracking of mutant clones in engineered CH macaques. (A) Schematic outline of RM HSPC gene editing for CH loci and subsequent autologous transplantation. (B) Summary of the gRNAs used for gene editing in each animal (n = 3; ZL26, ZL39, and ZH63). (C) Mutation frequencies at target sites are quantified from PB granulocytes collected at different time points from the 3 macaques using targeted deep sequencing. The percentage of reads containing indel mutations for each target site by individual macaque is plotted over time. (D) Indel frequencies at the TET2 target site in granulocytes collected over time are combined into a single graph for comparison. (E) Heatmaps on the right show the fractional contribution of each indel to total sequencing reads in granulocyte samples over time. Indels in HSPC measured before editing (Precut) and in infusion product (IP) are also shown in the first 2 columns. The most abundant indel types for the TET2 target site in each animal are listed on the left, with the WT sequence at the bottom. Indels are highlighted in red font, and the predicted Cas9 cut site is indicated by a red arrow. Changes in percent contribution of unique clones over time are shown stacked on the left, with each indel type marked with a distinct color across the 3 RMs. Td, doubling time; PAM, protospacer adjacent motif.
Clonal expansion of blood cells produced by edited HSPCs
To track the edited allele fractions of specific indels from tissues over time,13,29 we performed targeted deep sequencing focused on circulating granulocytes, a short-lived population that best reflects ongoing HSPC production.30 The frequency of granulocytes with TET2 indels increased markedly and steadily following infusion in all 3 macaques (Figure 2C). In ZL26, TET2 VAF in granulocytes reached 33% at 55 months posttransplantation, with a Td of 9.3 months (Figure 2D). This expansion consisted of cells carrying at least 6 different indel types, with 5 of the 6 unique indels resulting in predicted frameshift LOF mutations (Figure 2E). We found no detectable editing in the top 10 predicted off-target sites of the TET2 gRNA (supplemental Table 9). ZL39 and ZH63, followed for 34 and 13 months, respectively, also demonstrated clear expansion of granulocytes with similar indels at the TET2 locus. However, the TET2-mutated expansion rate measured in granulocytes among the 3 macaques varied markedly (Td: ZL26, 9.3 months; ZL39, 10.4 months; ZH63, 2.4 months). These results support the concept that specific intrinsic genetic factors and/or extrinsic environmental factors can regulate the expansion rate of TET2-mutated HSPCs.31,32
In contrast to TET2-mutated clones, granulocytes with DNMT3A mutations expanded very slowly only in ZL26 and ZL39. Those with ASXL1 mutations demonstrated no appreciable expansion in ZL39 and ZH63 (Figure 2C; supplemental Figure 5), confirming that LOF mutations in ASXL1 may not result in a competitive advantage. The overall low initial indel VAFs in these 3 genes in the infused CD34+ HSPCs as well as differences in expansion rates of cells containing indels in TET2 vs DNMT3A vs ASXL1 support that mutations in a single gene are sufficient for clonal expansion. VAFs of DTA mutations by targeted sequencing were validated by CH panel sequencing on granulocytes vs fibroblasts, with comparable values in all 3 macaques (supplemental Figure 6). Importantly, no additional somatic mutations were detected for any other genes in the panel.
We next examined TET2 mutation frequencies in cells of additional hematopoietic lineages (supplemental Figure 7). In general, several myeloid cell types (macrophages and classical monocytes) and CD56+ immature natural killer (NK) cells demonstrated higher TET2 mutant allele frequencies compared with T cells or mature CD16+ NK cells (Figure 3A). This supports the idea that TET2-mutated HSPCs may have a differentiation bias toward the monocytic/macrophage lineage, consistent with previous data from TET2-mutated human33 and murine HSPCs.34 The long-lived nature of T cells and mature NK cells35 may also contribute to slower clonal expansion within these compartments, as well as TBI affecting thymic function and delaying HSPC-derived T-cell reconstitution. When compared within each animal, the indel types across different lineages were similar in both identity and relative frequency (Figure 3B), suggesting that TET2-edited HSPCs maintain multipotency and are capable of giving rise to several different lineages.
Lineage analyses of output from edited HSPC.TET2 target site indel frequencies were analyzed in lineages of blood cells purified from ZL26, ZL39, and ZH63. (A) Indel frequencies in myeloid lineages, including macrophages (CD14+CD163+), classical monocytes (CD14+CD16−) and granulocytes (top), and lymphoid lineages, including helper T cells (CD4+), cytotoxic T cells (CD8+), B cells (CD20+), and mature (CD16+NKG2A+) and immature (CD56+) NK cells (bottom). (B) All indel types at the TET2 target site identified in any lineages from the 3 animals are listed on the bottom with unique colors. Indels are highlighted in red font, and the PAM sequence is underlined. The percent contribution of each indel in each cell type over time is shown in the graphs at the top.
Lineage analyses of output from edited HSPC.TET2 target site indel frequencies were analyzed in lineages of blood cells purified from ZL26, ZL39, and ZH63. (A) Indel frequencies in myeloid lineages, including macrophages (CD14+CD163+), classical monocytes (CD14+CD16−) and granulocytes (top), and lymphoid lineages, including helper T cells (CD4+), cytotoxic T cells (CD8+), B cells (CD20+), and mature (CD16+NKG2A+) and immature (CD56+) NK cells (bottom). (B) All indel types at the TET2 target site identified in any lineages from the 3 animals are listed on the bottom with unique colors. Indels are highlighted in red font, and the PAM sequence is underlined. The percent contribution of each indel in each cell type over time is shown in the graphs at the top.
Differentiation block and myeloid shift with TET2-mutated CH
In the absence of clinical indications for invasive marrow sampling, there is little available data regarding BM phenotype or morphology in humans with CH.7 We thus analyzed CFUs grown from the BM of CH macaques to evaluate the genotypes and characteristics of more primitive hematopoietic cells. Although there was no significant difference in total CFU potential among the 3 animals, ZL39, with a lower fraction of mutated alleles, exhibited a lower fraction of myeloid colonies compared with the other 2 macaques (Figure 4A). To delineate the frequency of BA vs UA target site mutations, we performed sequencing on individual myeloid CFUs from 3 animals (Figure 4B). Although AAVS1, DNMT3A, and ASXL1 indels were detected in <5% of individual CFUs, considerably more CFUs had mutations at TET2. The majority of edited CFUs contained UA indels, with a smaller portion of BA compound heterozygous colonies. No colonies had mutations in multiple genes. These findings support that this NHP model is representative of human CH, where the majority of expanded, mutated clones are heterozygous and involve only a single gene.1,2,36
Bone marrow characteristics of TET2-engineered CH macaques. BM CD34+ HSPCs obtained at 12 (ZL26 and ZH63) and 30 months (ZL39) posttransplantation were plated for CFU assays. Colonies were isolated individually on day 14 for analysis. (A) Total CFU potential and composition of erythroid and myeloid colonies are plotted. (B) WT, uniallelic (UA), and biallelic (BA) edits for each target site in individual CFUs were identified by deep sequencing. (C) Targeted deep sequencing was performed on BM granulocytes and purified CD34+ HSPCs from ZL26 at 6.5, 12, and 39 months, ZL39 at 30 and 34 months, and ZH63 at 12 months posttransplantation. The percentage of reads containing indels at the TET2, DNMT3A, and ASXL1 target sites is indicated. (D) Serial sections of BM core obtained from ZL26 and ZH63 were stained with hematoxylin and eosin and myeloperoxidase antibody, along with animals receiving lentivirally transduced (ZK48) or non-CH edited cells (ZI35) as controls, with BM samples collected at similar time points posttransplantation. Representative microscopic images at 50× magnification are shown. (E) The number of nucleated cells per field in at least 5 different fields was quantified by Image J software. Data are presented as mean plus or minus standard error of the mean, and statistical significances were calculated by One-way ANOVA followed by Tukey’s multiple comparisons test, ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
Bone marrow characteristics of TET2-engineered CH macaques. BM CD34+ HSPCs obtained at 12 (ZL26 and ZH63) and 30 months (ZL39) posttransplantation were plated for CFU assays. Colonies were isolated individually on day 14 for analysis. (A) Total CFU potential and composition of erythroid and myeloid colonies are plotted. (B) WT, uniallelic (UA), and biallelic (BA) edits for each target site in individual CFUs were identified by deep sequencing. (C) Targeted deep sequencing was performed on BM granulocytes and purified CD34+ HSPCs from ZL26 at 6.5, 12, and 39 months, ZL39 at 30 and 34 months, and ZH63 at 12 months posttransplantation. The percentage of reads containing indels at the TET2, DNMT3A, and ASXL1 target sites is indicated. (D) Serial sections of BM core obtained from ZL26 and ZH63 were stained with hematoxylin and eosin and myeloperoxidase antibody, along with animals receiving lentivirally transduced (ZK48) or non-CH edited cells (ZI35) as controls, with BM samples collected at similar time points posttransplantation. Representative microscopic images at 50× magnification are shown. (E) The number of nucleated cells per field in at least 5 different fields was quantified by Image J software. Data are presented as mean plus or minus standard error of the mean, and statistical significances were calculated by One-way ANOVA followed by Tukey’s multiple comparisons test, ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
We compared TET2 mutation frequencies between HSPCs and mature granulocytes purified from the same marrow aspirates in 3 animals. By 39 months in ZL26, the frequency of TET2 indels in HSPCs was nearly double that found in granulocytes (Figure 4C), suggesting a possible partial differentiation block for HSPCs edited at TET2. In morphologic analyses of BM, compared with age-matched transplanted control macaques, ZL26 and ZH63, the animals with the highest TET2 VAFs, exhibited increased cellularity and a myeloid shift. Neither animal showed dysplastic changes or any increase in blasts (Figure 4D-E; supplemental Figure 8).
Impact of TET2 disruption on DNA methylation and gene expression
Members of the TET protein family catalyze the oxidization of 5-mC to 5-hmC in genomic DNA.37 TET2 deficiency thus results in a decrease of 5-hmC relative to 5-mC, an effect likely to impact gene expression. We compared the 5-hmC to 5-mC ratio in peripheral blood mononuclear cells of the CH macaques to age-matched controls at the similar timepoints posttransplantation. The ratio was decreased in CH macaques, supporting that TET2 protein function is disrupted as a result of gene editing (supplemental Figure 9).
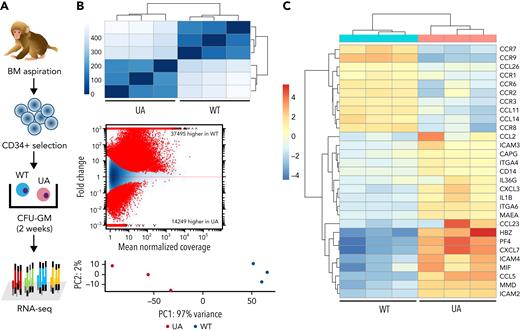
To investigate the impact of TET2 editing on gene expression in ZL26 at 15 months posttransplantation, we genotyped granulocyte-macrophage CFUs (CFU-GMs) to identify WT vs UA TET2 mutant colonies, then performed individual CFU RNA-seq (Figure 5A). By hierarchical Euclidian clustering, MA plotting based on log fold changes in both exonic regions and splice junctions, and principal component analysis, WT vs TET2+/− myeloid colonies showed distinct gene expression patterns (Figure 5B). Various inflammatory cytokine and chemokine genes, including CCL2, CXCL3, and IL1B, were upregulated in TET2-mutated colonies (Figure 5C), consistent with characteristics of Tet2-null myeloid cells in mouse models.9,38
Gene expression profile of hematopoietic clones with TET2 disruption. (A) Experimental scheme of ultralow-input CFU genotyping and RNA-seq. Enriched BM CD34+ cells collected at 15 months posttransplantation were plated at low density, and CFU-GMs were isolated individually on day 14 of culture. Three CFU-GMs identified as TET2 UA with predicted LOF mutations and 3 CFU-GMs identified as TET2 WT were analyzed via RNA-seq. (B) Heatmap of the Euclidean distance between WT and TET2 UA CFUs from RNA-seq genome-wide expression profiling (top). MA plot of differential usage in exonic regions and splice junctions based on the log fold changes between TET2 WT and UA CFU-GMs. Log fold expression change (M) is depicted on the y-axis and the average normalized coverage (A) generated by the DESeq2 Bioconductor package (middle) is depicted on the x-axis. Two-dimensional principal component analysis of TET2 WT and UA CFU-GMs (bottom). (C) Heatmap of select up- or downregulated genes between WT and TET2 UA-mutated CFUs from genome-wide expression profiling.
Gene expression profile of hematopoietic clones with TET2 disruption. (A) Experimental scheme of ultralow-input CFU genotyping and RNA-seq. Enriched BM CD34+ cells collected at 15 months posttransplantation were plated at low density, and CFU-GMs were isolated individually on day 14 of culture. Three CFU-GMs identified as TET2 UA with predicted LOF mutations and 3 CFU-GMs identified as TET2 WT were analyzed via RNA-seq. (B) Heatmap of the Euclidean distance between WT and TET2 UA CFUs from RNA-seq genome-wide expression profiling (top). MA plot of differential usage in exonic regions and splice junctions based on the log fold changes between TET2 WT and UA CFU-GMs. Log fold expression change (M) is depicted on the y-axis and the average normalized coverage (A) generated by the DESeq2 Bioconductor package (middle) is depicted on the x-axis. Two-dimensional principal component analysis of TET2 WT and UA CFU-GMs (bottom). (C) Heatmap of select up- or downregulated genes between WT and TET2 UA-mutated CFUs from genome-wide expression profiling.
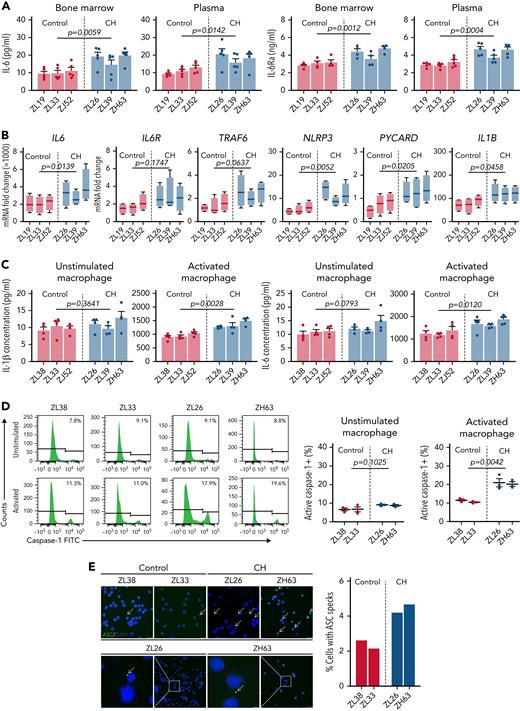
Hyperinflammation in TET2-deficient CH
Based on the CFU RNA-seq results, we assessed the in vivo systemic inflammatory status of CH macaques and controls. In both BM plasma and PB, CH macaques exhibited higher IL-6 and IL-6Ra levels than age-matched controls (Figure 6A). Overproduction of NLRP3 inflammasome-driven IL-1β and IL-6 results in the exacerbation of atherosclerotic CVD in Tet2-deficient mice, and elevated IL-6R production in TET2-disrupted myeloid cells is associated with further clonal expansion.9,31,39 We evaluated the transcription level of essential genes involved in the NLRP3 inflammasome and IL-6 signaling pathways within CD14+CD163+ macrophages in the presence or absence of immunostimulants. Although only subtle differences were observed in naïve cells, macrophages with TET2 indels showed significantly higher expression of NLRP3, IL1B, IL18, IL6, IL6R, and TRAF6 upon LPS/ATP activation compared with TET2-intact macrophages (Figure 6B). Notably, this was the case even for ZL39, despite a much lower fraction of mutated TET2 alleles, suggesting that even a small abnormal clone fraction may be enough to activate inflammatory pathways via paracrine events in WT myeloid cells.
Enhanced NLRP3 inflammasome and IL-6 signaling pathways in TET2-deficient myeloid cells. (A) Systemic IL-6 and IL-6Ra levels were quantified in both BM plasma and PB of CH macaques (ZL26, ZL39, and ZH63) and age-matched transplanted control macaques (ZL19, ZL33, and ZJ52). (B) CD14+CD163+ macrophages were purified from PB collected from the 3 CH-edited macaques and age-matched controls. The macrophages were incubated with LPS, followed by addition of ATP at the end of culture to activate the NLRP3 inflammasome. RNA from unstimulated or stimulated macrophages was analyzed by quantitative reverse transcription polymerase chain reaction. Fold changes of NLRP3 inflammasome-related genes (NLRP3, PYCARD, IL1B, and IL18) and IL-6-signaling genes (IL6, IL-6R, and TRAF6) calculated by δ-deltaCt relative to the unstimulated control are shown. (C) Concentrations of IL-1β and IL-6 were measured in the supernatants of macrophages cultured with or without specific stimulants (LPS and ATP for IL-1β; LPS for IL-6). Results from at least 3 independent experiments are graphed as mean plus or minus standard error of the mean. Statistical analysis was conducted using an unpaired t test between CH and controls group, and P values are directly marked on each panel. (D) Caspase-1 activity was measured by flow cytometry in unstimulated and LPS/ATP-activated macrophages from CH macaques (ZL26 and ZH63) and transplanted control macaques (ZL38 and ZL33). Representative histograms are shown on the left, and the results from the 3 independent experiments on the right. (E) The representative confocal microscopic images of cytoplasmic ASC specks (indicated by the arrow) in naïve macrophages from CH macaques and controls (left: scale bar, 25 μM). The percentage of nuclei with ASC specks from at least 10 different fields (right). FITC, fluorescein isothiocyanate.
Enhanced NLRP3 inflammasome and IL-6 signaling pathways in TET2-deficient myeloid cells. (A) Systemic IL-6 and IL-6Ra levels were quantified in both BM plasma and PB of CH macaques (ZL26, ZL39, and ZH63) and age-matched transplanted control macaques (ZL19, ZL33, and ZJ52). (B) CD14+CD163+ macrophages were purified from PB collected from the 3 CH-edited macaques and age-matched controls. The macrophages were incubated with LPS, followed by addition of ATP at the end of culture to activate the NLRP3 inflammasome. RNA from unstimulated or stimulated macrophages was analyzed by quantitative reverse transcription polymerase chain reaction. Fold changes of NLRP3 inflammasome-related genes (NLRP3, PYCARD, IL1B, and IL18) and IL-6-signaling genes (IL6, IL-6R, and TRAF6) calculated by δ-deltaCt relative to the unstimulated control are shown. (C) Concentrations of IL-1β and IL-6 were measured in the supernatants of macrophages cultured with or without specific stimulants (LPS and ATP for IL-1β; LPS for IL-6). Results from at least 3 independent experiments are graphed as mean plus or minus standard error of the mean. Statistical analysis was conducted using an unpaired t test between CH and controls group, and P values are directly marked on each panel. (D) Caspase-1 activity was measured by flow cytometry in unstimulated and LPS/ATP-activated macrophages from CH macaques (ZL26 and ZH63) and transplanted control macaques (ZL38 and ZL33). Representative histograms are shown on the left, and the results from the 3 independent experiments on the right. (E) The representative confocal microscopic images of cytoplasmic ASC specks (indicated by the arrow) in naïve macrophages from CH macaques and controls (left: scale bar, 25 μM). The percentage of nuclei with ASC specks from at least 10 different fields (right). FITC, fluorescein isothiocyanate.
On the protein level, macrophages from the edited macaques increased IL-1β and IL-6 secretion markedly upon inflammatory stimulation (Figure 6C). Moreover, these animals demonstrated hyperactivation of the NLRP3 inflammasome, with higher caspase-1 activity and cytoplasmic ASC speck deposition (Figure 6D-E). In aggregate, these data confirm that TET2 mutant macrophages possess increased NLRP3 inflammasome activity and IL-6 signaling in the CH macaques, consistent with findings from Tet2-null or haplo-insufficient mice and patients with myelodysplastic syndrome.39,40 Cardiac computed tomography scans performed at 12 months did not show coronary calcifications or other evidence for accelerated CVD due to hyperinflammation; however, follow-up was short, and these macaques were transplanted at a young adult age, and more time with CH may be required to observe any acceleration of overt CVD.
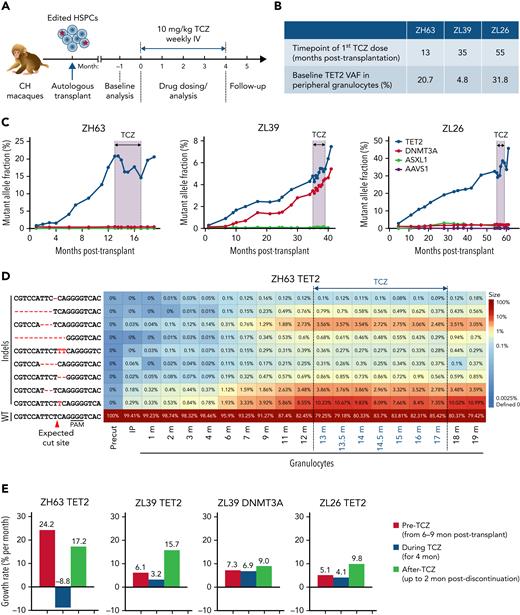
Response to pharmacological blockade of IL-6 signaling
Tet2 deficiency has been associated with both upregulation of IL-6 receptor expression and signaling as well as hypersecretion of IL-6 in murine models.31,32 This is theorized to result in a vicious cycle of clonal expansion simultaneously driven by and driving inflammation. We hypothesized that interrupting this cycle could halt clonal expansion. TCZ, a humanized monoclonal antibody that blocks IL-6 signaling via binding to the IL-6Ra, cross-reacts with macaque IL-6R41 and is used clinically in various human inflammatory diseases. All 3 CH macaques began a weekly dose of 10 mg/kg TCZ and continued on drug for 4 months. At initiation, all had active clonal expansion, although the starting VAF and time posttransplantation for TCZ initiation varied between animals (Figure 7A-B). Treatment was well tolerated, with no impact on blood counts or other clinical parameters (supplemental Figure 4).
Impact of IL-6 pathway inhibition on TET2 mutant clonal expansion. (A) Experimental protocol for TCZ administration. All 3 CH macaques were given a weekly dose of 10 mg/kg TCZ IV for 4 months. (B) Summary table of the status of 3 macaques at the time of initiation of TCZ. (C) Indel frequencies at the DTA target sites in PB granulocytes before, during, and after TCZ administration were quantified via targeted deep sequencing. The black line and shading indicate the period of TCZ administration. (D) Heatmap of specific TET2 indels over time before and after TCZ administration in ZH63 granulocytes. (E) Percent changes in growth rate per month of expanding clones in all 3 animals prior to TCZ, during TCZ, and after TCZ in the 3 animals, including TET2 clones in all 3 animals and DNMT3A clones in ZL39. The time periods used for pre-TCZ growth rate calculations began with samples collected 6 to 9 months posttransplantation, when stabilization of long-term hematopoiesis occurs in transplanted macaques, and until the first dose of TCZ. Negative values correspond to shrinkage of edited clones.
Impact of IL-6 pathway inhibition on TET2 mutant clonal expansion. (A) Experimental protocol for TCZ administration. All 3 CH macaques were given a weekly dose of 10 mg/kg TCZ IV for 4 months. (B) Summary table of the status of 3 macaques at the time of initiation of TCZ. (C) Indel frequencies at the DTA target sites in PB granulocytes before, during, and after TCZ administration were quantified via targeted deep sequencing. The black line and shading indicate the period of TCZ administration. (D) Heatmap of specific TET2 indels over time before and after TCZ administration in ZH63 granulocytes. (E) Percent changes in growth rate per month of expanding clones in all 3 animals prior to TCZ, during TCZ, and after TCZ in the 3 animals, including TET2 clones in all 3 animals and DNMT3A clones in ZL39. The time periods used for pre-TCZ growth rate calculations began with samples collected 6 to 9 months posttransplantation, when stabilization of long-term hematopoiesis occurs in transplanted macaques, and until the first dose of TCZ. Negative values correspond to shrinkage of edited clones.
In ZH63, the animal with the most rapid TET2 mutant clonal expansion pretreatment, the growth rate of mutated clones in granulocytes not only slowed but actually decreased by 30% by the end of the treatment period. Upon TCZ discontinuation, the TET2 VAF began to increase again and by 2 months postdiscontinuation rose back to the VAF present immediately before the start of TCZ treatment (Figure 7C; supplemental Figure 10A). Individual indel tracking data indicated that most of the indel types decreased, suggesting that the TCZ’s repressive activity is not limited to a subset of clones (Figure 7D). Although no actual sustained drop in VAF occurred in ZL39 and ZL26, the growth rate of mutated clones per month also showed a slowing during TCZ treatment in both animals and markedly rebounded following discontinuation (Figure 7E). Despite the differences in patterns and absolute effects between animals, these findings strongly support a biological effect of TCZ on CH. The frequency of ASXL1 and DNMT3A indels did not change markedly in any macaque, indicating that the repressive effect of TCZ is likely specific to the TET2 mutant phenotype.
The serum level of IL-6Ra rose equivalently in all animals, reflecting binding to similar TCZ concentrations. Additionally, the levels of tumor necrosis factor alpha (TNF-α) and IL-10 in ZH63 and ZL26 indicate systemic anti-inflammatory responses with TCZ administration that resolved with TCZ discontinuation (supplemental Figure 10B). There was no clear relationship between change in VAF and serum IL-6 concentration, ruling out compensatory increases in circulating IL-6.
Discussion
Previous small animal studies have revealed that loss of TET212,42 or DNMT3A11,43 results in increased HSPC self-renewal. However, rapid progression to myeloid neoplasia rather than a prolonged CH phase is seen in most models. Recently, Chin et al reported some natural human-type CH mutations in aged mice,44 although at a low incidence and VAF. This is perhaps due to differences in HSPC dynamics in short-lived rodents or the use of inbred strains maintained in a protective environment. We now report for the first time that aged RMs harbor a very similar spectrum of CH-associated mutations to humans, with VAFs reaching levels >2% in some animals, normal blood counts, and no evidence for hematologic malignancies, thus meeting criteria for clinically relevant CH. Furthermore, an upward trend of driver CH mutation incidence with age was noted in our cohort. The relative ages at which CH was documented in RMs vs humans correlates well with their relative lifespans.
Of interest, the DNMT3A in-frame deletion (p.FF731F) we retrieved from one animal is one of the very few indel hotspots previously reported in the human COSMIC cancer database.26 The mutation VAF in this animal was the highest in our cohort. Three animals were found to carry the same DNMT3A p.S349X frameshift mutation. We confirmed the presence of this mutation in multiple independently processed samples via both ddPCR and targeted sequencing. Bioinformatically, this mutation is predicted to cause a premature stop codon in the chromatin-associating PWWP domain, thus abolishing the function of DNMT3A and resulting in a competitive advantage. Although a homologous recurrent frameshift mutation at this site has not been reported in humans to date, other “hotspot” DNMT3A mutations have been reported in human CH samples (ie, R882).
Of note, DNMT3A mutations were almost absent in B cells and detected at a lower level in T cells in our naturally aged animals. Lower levels of these mutations in both B cells and T cells have also been documented in a proportion of lineage-sorted human samples,45,46 postulated to result from slower turnover of lymphoid cells, occurrence of these mutations at a different stem or progenitor cell stage, or an impact of some mutations on lineage bias. Although data are limited, in contrast to the findings in these macaques, most humans with low levels of DNMT3A mutations in B cells showed even lower levels in T cells. Further studies on expanded RM aging cohorts may provide additional insights into age associations and assess relationships to clinical outcomes, including hematologic malignancies and hyperinflammatory diseases.
Human CD34+ cells with DTA mutations are engraftable in immunodeficient mice, with expansion of LOF alleles over 5 months.13 However, prolonged follow-up and analysis of lineage differentiation is challenging in xenografts. Given the difficulty of maintaining sufficiently aged RMs in the long term to create large CH cohorts, we engineered HSPCs from young adult RMs to create CRISPR/Cas9-mediated LOF mutations in the 3 genes most frequently linked to human CH, tracking multiple mutated clones for more than 4 years. These animals developed CH expansions closely recapitulating TET2-associated human CH. Monoallelic LOF TET2 mutations alone were sufficient to drive striking clonal expansions while maintaining normal blood counts, aligning with human data.36,47 The doubling time for the TET2 mutant clones observed in our macaques fell within the ranges from human studies, demonstrating a relatively rapid rate of TET2 clonal expansion indicating robust clonal fitness.47-50
Prior human studies of CH in noncancer patients with normal blood counts have not analyzed the marrow compartment, given a lack of clinical indications for invasive BM biopsies. Serial marrow examinations are feasible in our longitudinal macaque model and showed consistently increased cellularity and a myeloid shift, despite a range of mutant TET2 VAFs in the marrow and blood. These changes could be explained by paracrine or microenvironmental effects on remaining WT HSPCs as well as intrinsic lineage-biased output from TET2 mutant cells.
DNMT3A clones with LOF mutations expanded more slowly in our model compared with TET2-mutated clones, suggesting innate differences in expansion ability for different gene targets. Of note, more rapid expansion rates for TET2- vs DNMT3A-mutated clones were reported in both a murine competitive repopulation model38,51 and a population study of very elderly women.47 It is also possible that transplantation differentially impacts expansion of TET2 vs DNMT3A clones given evidence that TET2 LOF directly increases HSPC self-renewal, which is critical during regeneration of the HSPC pool after transplantation.52 The lack of clear expansion of ASXL1 clones with LOF indels in exon 2 may be explained by recent evidence that CH expansions result from gain-of-function C-terminal protein truncations in this gene rather than LOF mutations.27,53
One of the most striking findings in our study was the marked difference in population doubling times for TET2 mutant HSPCs in individual macaques. The doubling rate was approximately constant over time in each animal and independent of the initial VAF. This suggest that differences may result from individual-specific genetic traits and/or extrinsic microenvironmental factors.54 We did not detect secondary somatic mutations in these animals to date. Expansion or transformation of somatically mutated HSPCs has been linked to inflammation in hematologic neoplasms,39,55 and aging itself is an inflammatory process.56 Aged murine HSPCs have distinct epigenetic changes resulting in myeloid lineage bias and resistance to cell death after DNA damage.57 In our CH model, the macaque with the most rapid clonal expansion (ZH63) was twice as old as the other 2 animals. Previously, we found that transplantation of barcoded aged HSPCs resulted in oligoclonal expansions and more lineage bias compared with young HSPCs, similar to findings in mice.58,59 However, our model demonstrates that aged HSPCs or an aged microenvironment are not required for TET2 mutant clonal expansion. In contrast, an aged/inflamed microenvironment may be required for DNTM3A expansion.54 Further study of aged macaques with natural mutations is warranted.
Inflammation and CH appear to share a reciprocal relationship: CH mutations lead to increased expression of inflammatory mediators, and an inflamed microenvironment stimulates further expansion of CH clones. Indeed, TET2 mutant HSPCs and myeloid progenitors are more sensitive to inflammatory cytokines via enhanced TNF-α and IL-6 receptor expression, resulting in a positive feedback loop driving further clonal expansion.31,32,60 Tet2-deficient murine macrophages have previously been shown to overproduce IL-6 in response to inflammatory stimuli and hypersecrete IL-1β and IL-6 via NLRP3 inflammasome activation.31,32,39 In our model, TET2-deficient myeloid CFUs exhibited increased expression of proinflammatory genes. We also observed hyperactivation of the NLRP3 inflammasome in macrophages from TET2-edited animals, as previously reported in human myelodysplastic syndrome.61 In our model, there was increased inflammasome activity and cytokine expression even when TET2-mutated VAFs were low. This is consistent with the concept of a vicious cycle of inflammatory cytokine release from mutant cells acting on WT cells and the microenvironment to release more inflammatory mediators, stimulating hypersensitive mutant HSPCs and potentially further increasing clone size.
Interrupting this vicious cycle would be predicted to disrupt the selective advantage of TET2-mutated HSPC. In mice, inhibition of IL-6 signaling attenuated both systemic inflammation and Tet2 clonal expansion.31,32 Furthermore, a cohort study linked the presence of an IL6R variant allele known to decrease IL-6 signaling to a lower risk of CVD in patients with CH.62 We administered an IL-6R blocking antibody and documented a drug-dependent decrease in the expansion rate for TET2-mutated HSPC in all animals; however, the absolute change and duration of the suppressive effect of TET2-mutated clones varied between the macaques, with an actual decrease in clone size observed in only 1 animal. This suggests that individual-specific factors may be involved in this noninbred species, in contrast to more homogeneous but possibly less relevant findings in inbred rodents. Although the small sample size precludes firm conclusions, our data suggest that there may be critical factors regulating clonal expansion, including overall clone size, systemic inflammatory state, age, gender, and/or underlying intrinsic genetic modifying factors. Even absent decreasing mutant clone size, anti-inflammatory interventions may mitigate the risk of CVD and other complications in individuals with CH.
In summary, the macaque model can test hypotheses regarding pathophysiology and clinical interventions as even a small number of macaques has yielded relevant insights, given the ability to study multiple clones in each animal. Utilization of this model may provide further insights into both intrinsic and extrinsic factors regulating clonal expansion and allow for testing of interventions to halt clonal expansion and reverse clinical consequences.
Acknowledgments
The authors thank the National Heart, Lung, and Blood Institute (NHLBI) DNA Sequencing and Genomics, Flow Cytometry, and Bioinformatics and Computational Biology Cores; Keyvan Keyvanfar, Stephanie Sellers, Lemlem Alemu, and Jun Zhu for technical assistance; Ilker Tunc for off-target prediction; Mehdi Pirooznia and Komudi Singh for bioinformatics discussions; Aylin Bonifacino for HSPC selection; and Allen Krouse, Nathaniel Linde, Teresa Engels, Justin Golomb, and the veterinary staff for care of the macaques.
This work was supported by the Basic Research Program through the National Research Foundation of Korea (NRF) funded by the Korean Government (2022R1C1C1009606, K.-R.Y.), a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare (HI14C1148, K.-R.Y.; H17C2039, T.-H.S.), the Oregon National Primate Research Center Core Grant OD011092, and the Intramural Research Programs of the National Institute of Aging and the National Heart, Lung, and Blood Institute, National Institutes of Health. Also, this research was funded in whole, or in part, by the Wellcome Trust [203151/Z/16/Z] and the UKRI Medical Research Council [MC_PC_17230].
Authorship
Contribution: K.-R.Y., Y.Z., and C.E.D. conceptualized the study; T.-H.S., Y.Z., S. Chen, M.Z.G., X.F., B.-C.L., A.A.A., M.A.F.C., J.-Y.M., H.N., K.L.M., and M.B. designed and performed the experiments; T.-H.S., Y.Z., S. Cordes, X.F., S.G.H., K.R.C., M.B., K.L.M., A.F.L., L.B., G.S.V., and K.-R.Y. analyzed data; K.L.V., J.A.M., S.G.K., M.A.F., N.U., S.D., and J.F.T. provided aged animal resources; and T.-H.S., Y.Z., S. Chen, K.-R.Y., and C.E.D. wrote and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kyung-Rok Yu, Animal Cell Biotechnology Laboratory, Department of Agricultural Biotechnology, Seoul National University, 1 Gwanak-ro, Bldg. 200, Room 4204, Gwanak-gu, Seoul 08826, Republic of Korea; e-mail: cellyu@snu.ac.kr; and Cynthia E. Dunbar, Translational Stem Cell Biology Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, 9000 Rockville Pike, Bldg. 10-CRC, Room 5E-3332, Bethesda, MD 20892; e-mail: dunbarc@nhlbi.nih.gov.
References
Author notes
∗T.-H.S., Y.Z., and S.C. contributed equally to this study.
The sequencing data from the natural CH study is available in the NCBI BioProject database (accession number PRJNA811244). The CFU RNA-seq datasets from the engineered macaques have been deposited to the Gene Expression Omnibus (GEO) database (accession number GSE148132).
Send data sharing requests via e-mail to the corresponding author.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal