Key Points
Exposure to thalidomide analogs, particularly lenalidomide, is associated with increased risk of TP53-mutated myeloid neoplasms.
Treatment with lenalidomide but not pomalidomide leads to expansion of preleukemic Trp53-mutant HSPCs due to selective degradation of Ck1α.
Abstract
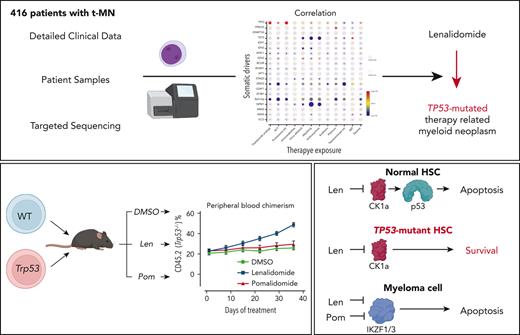
There is a growing body of evidence that therapy-related myeloid neoplasms (t-MNs) with driver gene mutations arise in the background of clonal hematopoiesis (CH) under the positive selective pressure of chemo- and radiation therapies. Uncovering the exposure relationships that provide selective advantage to specific CH mutations is critical to understanding the pathogenesis and etiology of t-MNs. In a systematic analysis of 416 patients with t-MN and detailed prior exposure history, we found that TP53 mutations were significantly associated with prior treatment with thalidomide analogs, specifically lenalidomide. We demonstrated experimentally that lenalidomide treatment provides a selective advantage to Trp53-mutant hematopoietic stem and progenitor cells (HSPCs) in vitro and in vivo, the effect of which was specific to Trp53-mutant HSPCs and was not observed in HSPCs with other CH mutations. Because of the differences in CK1α degradation, pomalidomide treatment did not provide an equivalent level of selective advantage to Trp53-mutant HSPCs, providing a biological rationale for its use in patients at high risk for t-MN. These findings highlight the role of lenalidomide treatment in promoting TP53-mutated t-MNs and offer a potential alternative strategy to mitigate the risk of t-MN development.
Introduction
Therapy-related myeloid neoplasms (t-MNs) represent one of the most devastating consequences of cancer therapy. These cancers arise from selective pressure introduced by chemo- and radiation therapies (CRTs) and are often treatment resistant with a median overall survival of 7 to 14 months and a 5-year overall survival of 10% to 20%.1-3 They typically present in a form of acute myeloid leukemia (t-AML) or myelodysplastic syndromes (t-MDS) and develop 3 to 7 years after treatment with CRTs. t-MNs are frequently associated with poor prognostic features, such as complex karyotypes.1 Exposure to certain types of chemotherapy has been associated with specific chromosomal alterations. For instance, prior exposures to topoisomerase II inhibitors have been linked to the development of t-MNs with KMT2A gene rearrangements at 11q23, whereas alkylating agents have been associated with chromosomes 5 and/or 7 abnormalities.4 With the advent of DNA sequencing technologies, the unique landscape of somatic mutations in t-MNs has become increasingly evident.5-7 However, little is known about how specific CRT exposures shape the somatic mutation profiles in t-MNs.
The model of t-MN development, in which CRT-induced mutations accumulate in hematopoietic stem and progenitor cells (HSPCs), has been challenged by increasing data suggesting that clonal selection of preexisting mutant HSPCs (ie, clonal hematopoiesis [CH] of indeterminate potential [CHIP]) occurs under the stress of cytotoxic therapy. For example, the mutational burden of de novo AML and t-AML are comparable,8 many somatic mutations in t-MNs are detectable years before CRT exposures,8-10 and somatic mutations in genes specifically involved in the DNA damage response, such as TP53 and PPM1D, are enriched in the blood of patients exposed to CRTs.11,12 Although this model may not be fully applicable to t-MNs with recurring gene rearrangements (eg, KMT2A translocation), these make up only a small fraction of all cases. Thus, understanding how individual therapies promote the outgrowth of specific mutant clones and the development of t-MNs and whether there are interventions or modifications in therapy that can decrease this risk is an important clinical problem. To address this question, we systematically analyzed the association between t-MN genotypes and prior CRT exposures in a large cohort of patients treated within a single institution.
Methods
Patients and samples
We retrospectively reviewed the clinical data of 416 patients with t-MNs who were diagnosed and treated at MD Anderson Cancer Center between November 2008 and February 2019. One hundred fifty-six of those were previously analyzed and reported in a study published elsewhere.6 The diagnosis of t-MNs was based on the 2016 World Health Organization classification.13 Cryopreserved diagnostic bone marrow (BM) or peripheral blood (PB) samples from the patients were used for mutation analysis by next-generation sequencing (NGS). As a comparison, 1021 patients with de novo MNs (611 with non–treatment-related AML and 410 with de novo MDS) who were diagnosed and treated during the same period in our institution were also analyzed (supplemental Table 1; available on the Blood website). All the patients provided informed consent under the Declaration of Helsinki guidelines and consented for sample collection, storage, and analysis.
DNA sequencing
Somatic mutations in BM or PB samples were detected by hybrid capture sequencing of coding regions of 300 genes (Agilent SureSelect XT, N = 156) or 81 genes (Agilent SureSelect XTHS, N = 260). A list of the genes targeted by the 2 panels are shown in supplemental Table 2. PPM1D was not covered by the 81-gene panel, and therefore; correlative analysis on PPM1D mutations was restricted to 156 patients analyzed by the 300-gene panel. For other cancer gene mutations covered by both panels (68 genes), correlative analysis was performed with 416 patients. Sequencing methods and bioinformatics pipelines to identify high-confidence cancer gene mutations were previously described.6
Multiplexed in vivo CRISPR knockout screen in mouse HSPCs
Concentrated lentiviruses encoding individual single guide RNAs (sgRNAs) (supplemental Table 3) and a tag-red fluorescent protein (tRFP) were prepared as previously described14 and titered on HEK293T cells. Live, lineagelo, Sca1+, c-Kit+ (LSK) cells were sorted from the BM of CRBNI391VMxCre Cas9+/−CD45.2+ mice15 and stimulated overnight at a concentration of 1 × 106 cells per mL in serum-free expansion medium (StemSpan SFEM, SemCell Technologies) supplemented with 50 ng/mL murine thrombopoietin (mTPO) and 50 ng/mL murine stem cell factor (mSCF, Peprotech). Lentiviral transduction was performed in an arrayed fashion, with LSK cells separated into individual wells and spinfected with individual viruses at 2200 revolutions per minute for 90 minutes at 37°C with the virus at a multiplicity of infection of ∼125 as determined by the titer in HEK293T cells. After spinfection, cells were allowed to rest for 2 hours, then an equal volume of fresh SFEM with TPO and SCF was added, and the cells were grown at 37°C for an additional 4 hours. Equal numbers of cells from each infection were then mixed, washed twice in phosphate buffer saline, and retro-orbitally injected into lethally irradiated (split dose 450 cGy × 2) Bl6.SJL CD45.1+ recipient mice. We chose retro-orbital injection over tail vein injection because we found it to be less traumatic for the mice. The remaining cells were grown for 2 days in SFEM with TPO and SCF, and DNA was purified for NGS. sgRNA guide frequencies were determined by purifying DNA from whole blood or BM and polymerase chain reaction (PCR) amplification of the lentiviral guide backbone followed by NGS. Quantitation was performed by counting the number of reads matching each specific guide sequence divided by the total number of reads matching all 8 guides. We have previously demonstrated that our sgRNAs induce deleterious indels adjacent to protospacer adjacent motif sites within our target genes and that the frequency of guide sequence correlates well with the presence of these indels.14,16
Generation of Hoxb8 cell lines and Hoxb8 cell line experiments
Immortalized myeloid progenitor cell lines were generated from a homozygous CrbnI391VMxCre Cas9+/− mouse and a CrbnwtMxCre Cas9+/− mouse via estrogen-regulated expression of Hoxb8 as previously described.15,17 c-KIT+ BM cells were infected with a murine stem cell virus retrovirus system, encoding ER-Hoxb8, then grown in vitro in RPMI supplemented with 10% fetal calf serum, penicillin/streptomycin, mSCF, and 1 μM estradiol (Sigma). After 1 month of culture, single-cell cloning was performed, and a representative clone with bilineage (neutrophil and macrophage) differentiation potential upon estrogen withdrawal was selected for all downstream applications. mSCF was generated from a Chinese hamster ovary cell line as previously described18 with conditioned media used at a final concentration of 2% (∼100 ng/mL). To generate cell lines carrying CH mutations, CrbnI391VMxCre Cas9+/− cells were spinfected with sgRNA-encoding lentiviruses. tRFP+ cells were sorted, and genotypes were confirmed by PCR amplification and sequencing of the targeted genes.
For drug sensitivity assays, cells were plated in triplicate in the presence of various concentrations of lenalidomide or pomalidomide prepared in dimethyl sulfoxide (DMSO). At 72 hours, cell viability was assessed using the CellTiterGlo (Promega) luminescent cell viability assay. For in vitro competition assays, blue fluorescent protein positive control cells and tRFP+ test cells were mixed in a 9:1 ratio and grown for 15 days. Every 72 hours, cells were harvested, with a portion analyzed by flow cytometry to assess blue fluorescent protein/tRFP percentages and a portion replated in fresh media with drug.
Competitive BM transplantation and thalidomide analog treatment
c-Kit+ cells were isolated from the BM of CrbnI391VTrp53−/−CD45.2, CrbnI391VvavCre Tet2fl/−CD45.2, CrbnI391VvavCre Csnk1a1−/+, and CRBNI391VCD45.1 mice using CD117 selection beads (Miltenyi). Cells were mixed at the indicated ratios in phosphate buffer saline and retro-orbitally injected into lethally irradiated (split dose 450 cGy × 2) Bl6.SJL CD45.1+ recipient mice. Stock solutions of lenalidomide, pomalidomide, and iberdomide were prepared in DMSO, stored at −80°C, and diluted to between 10% and 20% DMSO with sterile 0.9% sodium chloride, immediately before being administered by oral gavage. Retro-orbital bleeds were performed at the indicated time points and blood counts measured on a Hemavet (Drew Scientific). BM counts were performed on a Vi-CELL BLU automated cell analyzer (Beckman-Coulter). Flow cytometry was performed on a BD FACSCanto II.
Statistical analysis
Pearson chi-squared or Fisher exact tests were used to analyze the associations among categorical variables. We used odds ratio (OR) to assess the strength of associations between any 2 categorical variables. To avoid the infinity OR value, we used the Haldane correction. Mann-Whitney U test or Student t test was used to test the statistical difference between continuous variables. For comparison in continuous data with >2 groups, one-way analysis of variance (ANOVA) plus the Tukey honestly significant difference post hoc test was used. We performed a multivariate logistic regression to adjust for potential confounding factors in the analysis of association between mutations and prior exposures. The multicollinearity was evaluated, and variance inflation factor was used to evaluate the severity. The model was built by stepwise algorithm, and the Akaike information correction minimization method was used for the variable selection. A Firth logistic regression was applied instead, if the quasi-complete separation problem was detected. Where appropriate, adjustment for multiple testing was performed by the Benjamini-Hochberg method. The analysis was performed with Prism (version 9), SPSS (version 23), or statistical R software (version 4.0.2).
Results
The landscape of cancer gene mutations in t-MNs
To understand the landscape of cancer gene mutations in t-MNs, we searched our institutional medical database to identify newly diagnosed patients with t-MN whose diagnostic BM or PB specimens were available for sequencing. We identified 416 patients with t-MN, of whom 167 (40%) had t-AML and 249 (60%) had t-MDS. Patient characteristics are summarized in Table 1. About two-thirds (63%) of the patients had a primary diagnosis of solid tumors and the remainder (37%) had nonmyeloid hematologic cancers (Table 1; supplemental Figure 1A). Overall, 186 patients (45%) received prior treatment with chemotherapy alone, 69 patients (17%) had radiotherapy alone, and 161 patients (39%) had received both. Sixty-nine patients (17%) underwent autologous hematopoietic stem cell (HSC) transplant (supplemental Figure 1B). The median latency from initial CRT exposure to t-MN diagnosis was 6.2 years (interquartile range 3.0-12.0 years), with a significantly shorter latency for t-AML than t-MDS (t-AML vs t-MDS, median 5.0 years vs 6.4 years, P = .0283) (supplemental Figure 2). Cytogenetic profiles were consistent with those seen in high-risk MNs: 170 patients (41%) had a complex karyotype, 138 (34%) had −7/del(7q), 131 (32%) had −5/del(5q), and 29 (7%) had 11q23 rearrangements (Table 1). Although the prevalence of complex karyotype was similar between t-AML and t-MDS, −5/del(5q) and −7/del(7q) were more frequent in t-MDS. A small subset of the patients had favorable cytogenetic abnormalities including inv16/t(16;16) in 6 (1%) patients, t(15;17) in 4 (1%) patients, and t(8;21) in 3 (1%) patients.
Clinical characteristics of 416 patients with t-MNs
| . | Total . | t-AML . | t-MDS . | P value . |
|---|---|---|---|---|
| N | 416 | 167 | 249 | |
| Median age (range), y | 68 (17-91) | 65 (17-89) | 69 (22-91) | .002 |
| Male, n (%) | 221 (53) | 79 (47) | 143 (57) | .051 |
| Median latency (range), y | 6.0 (0.1-40) | 5.0 (0.1-40) | 6.4 (0.3-45) | .028 |
| Median Hgb (range), g/dL | 9.3 (5.2-14.8) | 9.3 (5.2-13.2) | 9.3 (5.9-14.8) | .079 |
| Median WBC (range), K/μL | 3.2 (0.1-267) | 3.8 (0.1-267) | 3 (0.2-85.7) | .000 |
| Median ANC (range), K/μL | 1.17 (0-62.5) | 0.98 (0-62.5) | 1.3 (0-50.6) | .170 |
| Median PLT (range), K/μL | 58 (3-895) | 41 (3-389) | 65 (6-895) | .002 |
| Previous malignancy, n (%) | .000 | |||
| Nonhematological | 285 (63) | 142 (78) | 143 (52) | |
| Hematologic | 169 (37) | 39 (22) | 130 (48) | |
| Active primary malignancy, n (%) | 63 (15) | 11 (7) | 52 (21) | .000 |
| Previous therapy, n (%) | .053 | |||
| Chemotherapy | 186 (45) | 66 (39) | 120 (48) | |
| Radiotherapy | 69 (16) | 36 (22) | 33 (13) | |
| Chemoradiotherapy | 161 (39) | 65 (39) | 96 (39) | |
| History of auto-HSCT, n (%) | 69 (17) | 32 (19) | 37 (15) | .282 |
| Cytogenetics∗, n (%) | ||||
| Diploid | 84 (21) | 34 (21) | 50 (21) | .941 |
| Complex† | 170 (41) | 67 (41) | 103 (43) | .622 |
| Del 5q/-5‡ | 131 (32) | 42 (25) | 89 (37) | .013 |
| Del 7q/-7 | 138 (34) | 33 (20) | 105 (43) | .000 |
| Inv 16/t(16;16) | 6 (1) | 6 (4) | 0 (0) | .003 |
| 11q23 | 29 (7) | 26 (16) | 3 (1) | .000 |
| t(15;17) | 4 (1) | 4 (2) | 0 (0) | .036 |
| t(8;21) | 3 (1) | 3 (2) | 0 (0) | .036 |
| -Y | 16 (4) | 9 (5) | 7 (3) | .201 |
| Del 20q | 38 (9) | 11 (7) | 27 (11) | .118 |
| Del 12p | 42 (10) | 14 (8) | 28 (12) | .297 |
| Trisomy 8 | 60 (15) | 25 (15) | 35 (14) | .887 |
| Inv3q/t(3;3) | 8 (2) | 2 (1) | 6 (2) | .358 |
| Trisomy 21 | 41 (11) | 18 (11) | 23 (10) | .673 |
| Del 17p/-17 | 54 (13) | 22 (13) | 32 (13) | .988 |
| . | Total . | t-AML . | t-MDS . | P value . |
|---|---|---|---|---|
| N | 416 | 167 | 249 | |
| Median age (range), y | 68 (17-91) | 65 (17-89) | 69 (22-91) | .002 |
| Male, n (%) | 221 (53) | 79 (47) | 143 (57) | .051 |
| Median latency (range), y | 6.0 (0.1-40) | 5.0 (0.1-40) | 6.4 (0.3-45) | .028 |
| Median Hgb (range), g/dL | 9.3 (5.2-14.8) | 9.3 (5.2-13.2) | 9.3 (5.9-14.8) | .079 |
| Median WBC (range), K/μL | 3.2 (0.1-267) | 3.8 (0.1-267) | 3 (0.2-85.7) | .000 |
| Median ANC (range), K/μL | 1.17 (0-62.5) | 0.98 (0-62.5) | 1.3 (0-50.6) | .170 |
| Median PLT (range), K/μL | 58 (3-895) | 41 (3-389) | 65 (6-895) | .002 |
| Previous malignancy, n (%) | .000 | |||
| Nonhematological | 285 (63) | 142 (78) | 143 (52) | |
| Hematologic | 169 (37) | 39 (22) | 130 (48) | |
| Active primary malignancy, n (%) | 63 (15) | 11 (7) | 52 (21) | .000 |
| Previous therapy, n (%) | .053 | |||
| Chemotherapy | 186 (45) | 66 (39) | 120 (48) | |
| Radiotherapy | 69 (16) | 36 (22) | 33 (13) | |
| Chemoradiotherapy | 161 (39) | 65 (39) | 96 (39) | |
| History of auto-HSCT, n (%) | 69 (17) | 32 (19) | 37 (15) | .282 |
| Cytogenetics∗, n (%) | ||||
| Diploid | 84 (21) | 34 (21) | 50 (21) | .941 |
| Complex† | 170 (41) | 67 (41) | 103 (43) | .622 |
| Del 5q/-5‡ | 131 (32) | 42 (25) | 89 (37) | .013 |
| Del 7q/-7 | 138 (34) | 33 (20) | 105 (43) | .000 |
| Inv 16/t(16;16) | 6 (1) | 6 (4) | 0 (0) | .003 |
| 11q23 | 29 (7) | 26 (16) | 3 (1) | .000 |
| t(15;17) | 4 (1) | 4 (2) | 0 (0) | .036 |
| t(8;21) | 3 (1) | 3 (2) | 0 (0) | .036 |
| -Y | 16 (4) | 9 (5) | 7 (3) | .201 |
| Del 20q | 38 (9) | 11 (7) | 27 (11) | .118 |
| Del 12p | 42 (10) | 14 (8) | 28 (12) | .297 |
| Trisomy 8 | 60 (15) | 25 (15) | 35 (14) | .887 |
| Inv3q/t(3;3) | 8 (2) | 2 (1) | 6 (2) | .358 |
| Trisomy 21 | 41 (11) | 18 (11) | 23 (10) | .673 |
| Del 17p/-17 | 54 (13) | 22 (13) | 32 (13) | .988 |
Bold denotes variables with P < .05.
ANC, absolute neutrophil counts; auto-HSCT, autologous hematopoietic stem cell transplant; Hgb, hemoglobin; PLT, platelet; WBC, white blood cell count.
Cytogenetic abnormalities were recorded either as a sole abnormality or with multiple other abnormalities; therefore, the total frequency exceeds 100%.
Among patients with complex karyotype, 40 (25%) did not have chromosome 5 or 7 abnormalities.
Seventy-nine patients (19%) had concurrent chromosome 5 and 7 abnormalities.
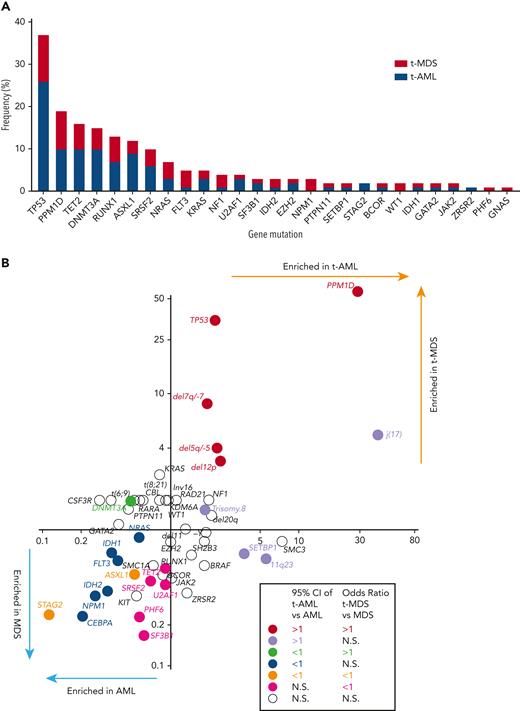
At least 1 gene mutation was detected in 352 of the 416 patients with t-MN (85%). Mutations in genes involved in the DNA damage response (TP53 [37%] and PPM1D [19%]) predominated, followed by TET2 (16%), DNMT3A (15%), RUNX1 (13%), ASXL1 (13%), and SRSF2 (10%) (Figure 1A). Consistent with a recent report,19 61% of TP53-mutated cases had multihit alterations (combination with del 17p or multiple TP53 mutations with total cancer-cell fraction exceeding 100%) with likely bi-allelic involvement (supplemental Figure 3). Although the median variant allele frequency (VAF) of TP53 mutations was 0.31, VAF of PPM1D mutations was predominantly subclonal (median VAF 0.045, supplemental Figure 4). A pair-wise analysis of mutation co-occurrence demonstrated that TP53 mutations co-occurred with aneuploid karyotype but were anticorrelated with many other gene mutations and with 11q23 rearrangements (supplemental Figure 5).5,20 The pattern of co-occurrence between inv16/t(16;16) and oncogenic RAS pathway mutations (NRAS, KRAS, and NF1), SRSF2 and IDH2, and NPM1 and FLT3 were consistent with the pattern observed in de novo MNs. By comparing the mutational frequency with that of AML and MDS without prior CRT exposures (N = 1021, supplemental Table 1), we determined which genes were more commonly mutated in each of the 4 disease subtypes (t-AML, t-MDS, AML, and MDS) (Figure 1B). As expected, TP53 and PPM1D mutations were significantly more frequent in t-MNs. By contrast, STAG2 and ASXL1 mutations were more common in AML/MDS without prior exposures. In addition, NPM1, IDH1/2, FLT3, CEBPA, and NRAS mutations were enriched in AML without exposures, whereas TET2, PHF6, and spliceosome gene mutations (SRSF2, SF3B1, and U2AF1) were found to be more common in MDS without prior exposures (Figure 1B).
Cancer gene mutations in t-MNs. (A) The frequency of cancer gene mutations detected in at least 1% of the studied cohort. Stacked bar graph shows the share of the mutations detected in t-AML and t-MDS each. (B) Mutation enrichment against 4 disease subtypes (t-AML, t-MDS, AML, and MDS). Enrichment is measured by OR for t-AML vs AML in x-axis and t-MDS vs MDS in y-axis. Statistically significant enrichment based on OR of 95% CI is highlighted by colors. CI, confidence interval.
Cancer gene mutations in t-MNs. (A) The frequency of cancer gene mutations detected in at least 1% of the studied cohort. Stacked bar graph shows the share of the mutations detected in t-AML and t-MDS each. (B) Mutation enrichment against 4 disease subtypes (t-AML, t-MDS, AML, and MDS). Enrichment is measured by OR for t-AML vs AML in x-axis and t-MDS vs MDS in y-axis. Statistically significant enrichment based on OR of 95% CI is highlighted by colors. CI, confidence interval.
Association between gene mutations and prior exposures
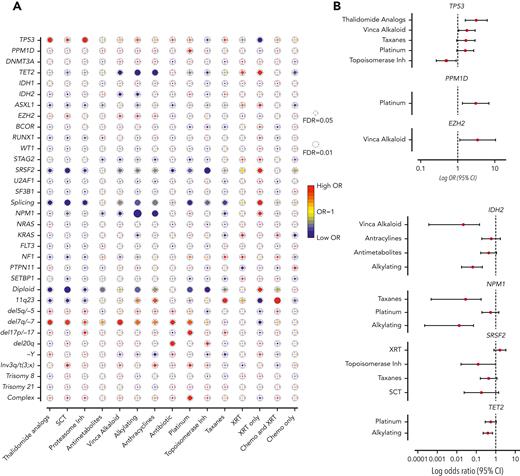
Next, we assessed the association between mutations and prior exposures (Figure 2A). As expected, we observed significant correlations between complex karyotype and platinum agents (OR, 1.88; 95% CI, 1.23-2.89; FDR = 0.052), and between chromosome 7 abnormalities with alkylating agents (OR, 1.64; 95% CI, 1.08-2.49; FDR = 0.057) and platinum drugs (OR, 1.65; 95% CI, 1.06-2.57; FDR = 0.057). Mutations that were associated with de novo AML/MDS (NPM1, IDH2, ASXL1, and splicing genes, Figure 1B) anticorrelated with agents that are commonly associated with t-MNs (alkylators, anthracyclines, and topoisomerase inhibitors), suggesting that t-MNs with these mutations may not have biological association with the prior exposures. In addition, patients who were exposed to radiation therapy alone had strong associations with NPM1, splicing genes, and normal karyotype, consistent with the previous study reporting the similar clinical outcomes of those patients with de novo MN.21
Association between cancer gene mutations and prior exposures. (A) The bubble plot showing univariate correlation between gene mutations and major chromosome abnormalities with prior exposures. Some of the genes are also grouped by their functional pathways. Splicing gene includes (SF3B1, SRSF2, U2AF1, and ZRSR2). The size of the bubbles represents the degree of statistical significance measured by FDR. The color of the bubble represents the degree of correlation based on the log OR; red color indicates positive correlation and green color indicates negative correlation. (B) Log OR (and 95% CI) with gene mutations and prior exposures after adjusting for confounding effect from multiple exposures with multilogistic regression analysis. Results of positive associations are shown in the top panel and negative associations are shown in the bottom panel. For full results, refer to supplemental Table 4. FDR, false discovery rate; SCT, stem cell transplant; XRT, radiotherapy.
Association between cancer gene mutations and prior exposures. (A) The bubble plot showing univariate correlation between gene mutations and major chromosome abnormalities with prior exposures. Some of the genes are also grouped by their functional pathways. Splicing gene includes (SF3B1, SRSF2, U2AF1, and ZRSR2). The size of the bubbles represents the degree of statistical significance measured by FDR. The color of the bubble represents the degree of correlation based on the log OR; red color indicates positive correlation and green color indicates negative correlation. (B) Log OR (and 95% CI) with gene mutations and prior exposures after adjusting for confounding effect from multiple exposures with multilogistic regression analysis. Results of positive associations are shown in the top panel and negative associations are shown in the bottom panel. For full results, refer to supplemental Table 4. FDR, false discovery rate; SCT, stem cell transplant; XRT, radiotherapy.
Notably, we found significant associations between TP53 mutations and proteasome inhibitors (OR, 3.06; 95% CI, 1.52-6.15; FDR = 0.025) and with thalidomide analogs (OR, 2.62; 95% CI, 1.36-5.05; FDR = 0.035). Multivariate logistic regression analysis to adjust for the confounding effect from multiple exposures in individual patients confirmed a significant association between TP53 mutations and prior exposures to thalidomide analogs (OR, 3.14; 95% CI, 1.60-6.18; P = .0009) and vinca alkaloids (OR, 1.76; 95% CI, 1.05-2.93; P = .031) and negative association with topoisomerase inhibitors (OR, 0.49; 95% CI, 0.26-0.91; P = .023) (Figure 2B; supplemental Table 4). The associations between gene mutations and prior malignancies were concordant with these findings (supplemental Figure 6). Correlation between gene mutations and prior malignancies revealed a significant association between TP53 mutations and history of multiple myeloma (MM) (FDR < 0.05) and ovarian cancer (FDR < 0.1), whereas breast cancer was associated with NF1 mutations (FDR < 0.05) and 11q23 rearrangements (FDR < 0.01) (supplemental Figure 6). Because thalidomide analogs are predominantly used for the treatment of MM, with prolonged administration as a maintenance therapy, we also analyzed the duration of exposure and t-MN genotype. Among the patients treated with lenalidomide, patients with TP53-mutated t-MN had significantly longer duration of exposure compared with patients with wild-type (WT) TP53 (supplemental Figure 7), suggesting that the t-MN genotype is also shaped by the duration of exposure. We also analyzed the secondary exposures in patients with MM who were treated with thalidomide analogs and had developed t-MNs (supplemental Figure 8). There were no significant differences in secondary exposures between patients who developed TP53-mutated or unmutated t-MN. These data suggest that specific CRT exposures promote the development of t-MNs with unique driver events.
Trp53 loss promotes resistance to lenalidomide but not pomalidomide
The significant association between TP53 mutations and prior exposures to thalidomide analogs (Figure 2) in t-MNs prompted us to directly investigate the effect of lenalidomide on TP53-mutant HSPCs because 92% of the thalidomide analog exposure in this cohort involved lenalidomide. TP53 mutations are associated with resistance to lenalidomide therapy in del(5q) MDS.22 Lenalidomide induces CK1α degradation leading to p53-dependent apoptosis in HSPCs.23 In addition, loss of p53 in Csnk1a1−/+ mouse HSPCs confers resistance to ex vivo treatment with lenalidomide.23 However, to the best of our knowledge, the effect of thalidomide analogs on preleukemic TP53-mutant HSPCs and their evolution to t-MN has never been assessed.
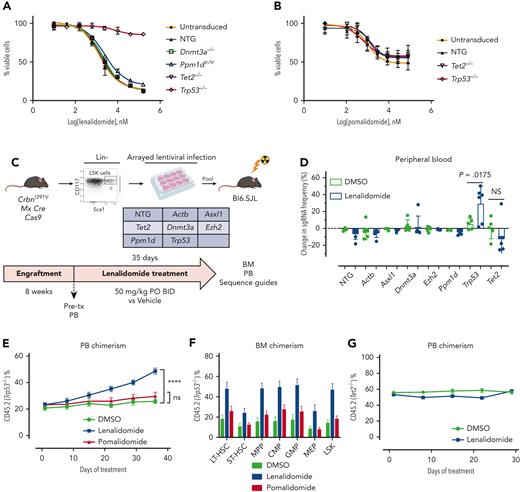
To address the hypothesis that thalidomide analogs promote evolution of TP53-mutated disease directly, we generated an immortalized mouse HSPC cell line with an estrogen-inducible Hoxb8 transgene (supplemental Figure 9A).17 Because mouse cells are inherently resistant to thalidomide analogs owing to a single amino acid difference from the human CRBN, we used a CrbnI391V knockin mouse model that we previously developed and characterized.15 Hoxb8 cells generated from the CrbnI391V mouse were sensitive to both lenalidomide and pomalidomide, whereas cells derived from a WT mouse were not (supplemental Figure 9B).
Hoxb8 CrbnI391V;Rosa26-Cas9 cells were engineered to carry mutations recurrently mutated in CH, including Trp53, using the CRISPR-Cas9 system, and then treated with lenalidomide. Only loss of Trp53 led to resistance to lenalidomide (Figure 3A), although it did not confer resistance to pomalidomide (Figure 3B). Similarly, in long-term in vitro competition assays, Trp53 mutant cells rapidly outcompeted control Hoxb8 cells in the presence of lenalidomide, but with pomalidomide, showed only a mild proliferative advantage at high drug doses (supplemental Figure 9C-D). Because almost all patients with MM were exposed to both thalidomide analogs and proteasome inhibitors, it was difficult to separate the effect of the 2 drugs from clinical analysis. Therefore, we also treated Trp53-mutated and WT Hoxb8 immortalized mouse HSPCs with bortezomib or carfilzomib. However, compared with lenalidomide, Trp53-mutated clones showed only mild resistance to the proteasome inhibitors, further suggesting that lenalidomide is the main driver of clonal selection of TP53-mutated clones (supplemental Figure 10).
Clonal selection of Trp53-mutant HSPCs with lenalidomide therapy but not with pomalidomide therapy. Hoxb8 cells derived from the CrbnI391V Rosa26-Cas9 mouse and transduced with sgRNAs targeting the labeled genes were grown in the presence of serially diluted lenalidomide (A) or pomalidomide (B) for 72 hours and cell density measured using CellTiter-Glo. Data are normalized to the vehicle (DMSO) control and shown as the mean ± standard deviation (n = 3 replicates). Curves represent the logistic regression. (C) Schematic for the generation of pooled parallel CRISPR-Cas9 in vivo mouse screen. LSK cells were lentivirally transduced with individual guides, pooled at equal ratios, and then transplanted into lethally irradiated Bl6.SJL recipient mice. (D) PB was collected before and after treatment of mice from pooled CRISPR-Cas9 experiment, DNA was harvested, sgRNA sequence was PCR amplified, and then subjected to NGS. The proportions of read mapping to each sgRNA sequence was determined and the percentage change in read frequency for each sgRNA between pre- and posttreatment samples was determined. Shown is the mean ± standard deviation (n = 6 mice per group). P values are from unpaired single-sided t tests. (E) PB was collected weekly and chimerism was measured using fluorescence-assisted cell sorting. (F) BM was harvested and chimerism measured within each of the following cellular compartments: LSK, long-term HSCs (LT-HSC: LSK, CD150+, and CD48−), short-term–HSC (ST-HSC: LSK, CD150−, and CD48−), multipotent progenitor (MPP: LSK, CD150−, and CD48+), common myeloid progenitor (CMP: LK, CD16/32−, and CD34+), granulocyte-monocyte progenitor (GMP: LK, CD16/32+, and CD34+), megakaryocyte-erythroid progenitor (MEP: LK, CD16/32−, and CD34−). Shown is the mean ± the standard error of the mean (n = 9-10 mice per group). ∗∗∗∗P < .0001, one-way ANOVA. (G) After hematopoietic reconstitution, chimeric transplants containing 50% Tet2−/−, CrbnI391V (mutant, CD45.2), and 50% CrbnI391V (WT, CD45.1) were treated with vehicle (DMSO) or lenalidomide 50 mg/kg, twice daily (BID). PB was collected weekly and chimerism was measured using fluorescence-assisted cell sorting. Shown is the mean ± the standard error of the mean (n = 15 mice per group). Actb, cutting guide targeting intronic sequence within β-actin gene; NTG, nontargeting guide; ns, not significant; PO, postoperative; tx, treatment.
Clonal selection of Trp53-mutant HSPCs with lenalidomide therapy but not with pomalidomide therapy. Hoxb8 cells derived from the CrbnI391V Rosa26-Cas9 mouse and transduced with sgRNAs targeting the labeled genes were grown in the presence of serially diluted lenalidomide (A) or pomalidomide (B) for 72 hours and cell density measured using CellTiter-Glo. Data are normalized to the vehicle (DMSO) control and shown as the mean ± standard deviation (n = 3 replicates). Curves represent the logistic regression. (C) Schematic for the generation of pooled parallel CRISPR-Cas9 in vivo mouse screen. LSK cells were lentivirally transduced with individual guides, pooled at equal ratios, and then transplanted into lethally irradiated Bl6.SJL recipient mice. (D) PB was collected before and after treatment of mice from pooled CRISPR-Cas9 experiment, DNA was harvested, sgRNA sequence was PCR amplified, and then subjected to NGS. The proportions of read mapping to each sgRNA sequence was determined and the percentage change in read frequency for each sgRNA between pre- and posttreatment samples was determined. Shown is the mean ± standard deviation (n = 6 mice per group). P values are from unpaired single-sided t tests. (E) PB was collected weekly and chimerism was measured using fluorescence-assisted cell sorting. (F) BM was harvested and chimerism measured within each of the following cellular compartments: LSK, long-term HSCs (LT-HSC: LSK, CD150+, and CD48−), short-term–HSC (ST-HSC: LSK, CD150−, and CD48−), multipotent progenitor (MPP: LSK, CD150−, and CD48+), common myeloid progenitor (CMP: LK, CD16/32−, and CD34+), granulocyte-monocyte progenitor (GMP: LK, CD16/32+, and CD34+), megakaryocyte-erythroid progenitor (MEP: LK, CD16/32−, and CD34−). Shown is the mean ± the standard error of the mean (n = 9-10 mice per group). ∗∗∗∗P < .0001, one-way ANOVA. (G) After hematopoietic reconstitution, chimeric transplants containing 50% Tet2−/−, CrbnI391V (mutant, CD45.2), and 50% CrbnI391V (WT, CD45.1) were treated with vehicle (DMSO) or lenalidomide 50 mg/kg, twice daily (BID). PB was collected weekly and chimerism was measured using fluorescence-assisted cell sorting. Shown is the mean ± the standard error of the mean (n = 15 mice per group). Actb, cutting guide targeting intronic sequence within β-actin gene; NTG, nontargeting guide; ns, not significant; PO, postoperative; tx, treatment.
Trp53 mutation confers a selective advantage on HSPCs in the presence of lenalidomide therapy
To further assess the effect of lenalidomide on clonal outgrowth in vivo, we virally transduced LSK cells harvested from CrbnI391V;Rosa26-Cas9 mice with sgRNAs targeting 6 recurrently mutated genes in CH (Dnmt3a, Tet2, Asxl1, Trp53, Ppm1d, and Ezh2) and 2 control sgRNAs in parallel, pooled the mutant cells at equal ratios, and then transplanted them into lethally irradiated WT recipient mice (Figure 3C).14,24 NGS of the input cells after 2 days in culture demonstrated an equal distribution of sgRNAs across the 6 genes and 2 controls, and flow cytometry demonstrated efficient engraftment of the fluorescently labeled mutant hematopoietic cells in the mice (supplemental Figure 11A-B). After hematopoietic reconstitution, we treated the mice with lenalidomide or vehicle for 5 weeks and collected PB and BM for both flow cytometric analysis and NGS. As expected, treatment with lenalidomide led to selective loss of CrbnI391V donor cells (supplemental Figure 11C). Sequencing of the PB before and after treatment with lenalidomide demonstrated a 23% average increase in cells carrying the sgRNA targeting Trp53 (P = .0175) (Figure 3D). We also observed a trend toward a reduction in cells carrying the sgRNA targeting Tet2 (Figure 3D). Similar results were seen in the BM (supplemental Figure 11D).
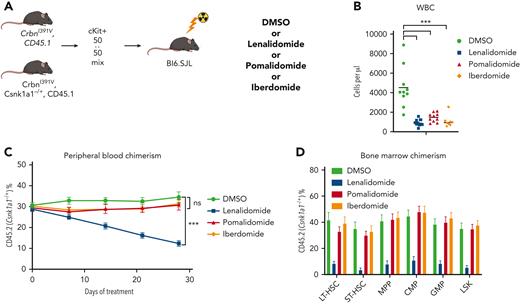
To further study the impact of thalidomide analogs on the outgrowth of Trp53-mutant HSPCs in vivo, we crossed the CrbnI391V mouse to a Trp53−/− knockout mouse.15,25 We transplanted c-Kit+ cells with donor marrow consisting of 20% CD45.2;Trp53−/−;CrbnI391V (mutant) cells mixed with 80% CD45.1;CrbnI391V (WT) cells (supplemental Figure 12A). After hematopoietic reconstitution at 8 weeks, we treated mice with vehicle (10% DMSO), lenalidomide 50 mg/kg, or pomalidomide 20 mg/kg. Treatment with both lenalidomide and pomalidomide led to a similar degree of PB leukopenia and decreased BM cellularity (supplemental Figure 12B-C). Other PB parameters showed only mild differences, consistent with prior work (supplemental Figure 12C).15 In general, whereas both agents led to equal levels of PB leukopenia and decreased BM cellularity, lenalidomide led to further decrease in long-term HSCs and myeloid progenitors than did pomalidomide (supplemental Figure 12D). Treatment with lenalidomide, but not pomalidomide, led to outgrowth of Trp53−/−-mutant cells in all blood cell lineages, suggesting selection in HSPCs (Figure 3E; supplemental Figure 12E). Analysis of the BM demonstrated significant preferential expansion of the Trp53−/−-mutant cells in all stem cell and progenitor populations, only in the lenalidomide treated mice (Figure 3F; supplemental Figure 12F). Competitive transplantation of Tet2−/− HSPCs failed to demonstrate any selective depletion of Tet2-mutant cells after treatment with lenalidomide (Figure 3G; supplemental Figure 13A-D). Thus, we conclude that treatment with lenalidomide but not pomalidomide leads to the selective outgrowth of Trp53-mutant HSPCs.
Differential degradation of CK1α defines the toxicity of thalidomide analogs in HSPCs
Although lenalidomide and pomalidomide both facilitate the degradation of IKZF1 and IKZF3, only lenalidomide promotes degradation of CK1α23,26,27 (supplemental Figure 14A). Because suppression of CK1α induces p53-mediated apoptosis,15,28 we hypothesized that lenalidomide treatment would be more toxic to normal HSPCs compared with pomalidomide and could explain the competitive advantage seen for TP53-mutated HSPCs. Consistent with this hypothesis, we observed that treatment with lenalidomide led to a more significant reduction in long-term HSCs and myeloid progenitors compared with pomalidomide (supplemental Figure 12D).
To further characterize the role of CK1α degradation in the toxicity of thalidomide analogs in normal HSPCs, we performed a competitive BM transplant experiment with Csnk1a1 heterozygous knockout (Csnk1a1−/+) mouse cells and WT cells in the Crbn1391V background, and treated the transplanted mice with lenalidomide, pomalidomide, and iberdomide.29 Iberdomide is a next-generation thalidomide analog currently in clinical development.30 We found that, similar to pomalidomide, it degrades IKZF1 and ZFP91 but not CK1α (supplemental Figure 14A). Although all 3 drugs led to a similar degree of leukopenia, only lenalidomide treatment selectively depleted Csnk1a1−/+ cells in the PB and hematopoietic stem and progenitor populations (Figure 4C-D; supplemental Figure 14). These data strongly suggest that the HSPC toxicity seen during lenalidomide treatment is driven by the degradation of CK1α, that this effect is specific to lenalidomide, and that CK1α degradation provides a mechanistic basis for selection of Trp53-mutant HSPCs.
Lenalidomide toxicity is enhanced by haploinsufficiency of Csnk1a1. (A) Schematic for the Csnk1a1 competitive transplant experiment. After hematopoietic reconstitution, chimeric transplants containing 50% Csnk1a1−/+, CrbnI391V (mutant, CD45.2), and 50% CrbnI391V (WT, CD45.1) were treated with vehicle (DMSO), lenalidomide 50 mg/kg BID, pomalidomide 20 mg/kg BID, or iberdomide 20 mg/kg BID. (B) Total peripheral white blood cell count was determined using fluorescence-assisted cell sorting. (C) PB was collected weekly and chimerism was measured using fluorescence-assisted cell sorting. (D) BM was harvested and chimerism measured within each of the following cellular compartments: LSK, LT-HSC (LSK, CD150+, and CD48−), ST-HSC (LSK, CD150−, and CD48−), MPP (LSK, CD150−, and CD48+), CMP (LK, CD16/32−, and CD34+), and GMP (LK, CD16/32−, and CD34−). Shown is the mean ± the standard error of the mean (n = 8-10 mice per group). ∗∗∗P < .0001, one-way ANOVA.
Lenalidomide toxicity is enhanced by haploinsufficiency of Csnk1a1. (A) Schematic for the Csnk1a1 competitive transplant experiment. After hematopoietic reconstitution, chimeric transplants containing 50% Csnk1a1−/+, CrbnI391V (mutant, CD45.2), and 50% CrbnI391V (WT, CD45.1) were treated with vehicle (DMSO), lenalidomide 50 mg/kg BID, pomalidomide 20 mg/kg BID, or iberdomide 20 mg/kg BID. (B) Total peripheral white blood cell count was determined using fluorescence-assisted cell sorting. (C) PB was collected weekly and chimerism was measured using fluorescence-assisted cell sorting. (D) BM was harvested and chimerism measured within each of the following cellular compartments: LSK, LT-HSC (LSK, CD150+, and CD48−), ST-HSC (LSK, CD150−, and CD48−), MPP (LSK, CD150−, and CD48+), CMP (LK, CD16/32−, and CD34+), and GMP (LK, CD16/32−, and CD34−). Shown is the mean ± the standard error of the mean (n = 8-10 mice per group). ∗∗∗P < .0001, one-way ANOVA.
Discussion
Here, we have analyzed the mutational profiles of 416 patients with t-MN and their associated clinical characteristics including prior cancer therapy exposures. We identified a significant association between TP53-mutated t-MN and prior exposure to thalidomide analogs, especially lenalidomide. Using in vitro and in vivo mouse models, we demonstrate that lenalidomide, but not pomalidomide, treatment promotes selective outgrowth of Trp53-mutant HSPCs, which likely forms the basis of t-MN development in this context.
Mutations in TP53 within both leukemic cells and otherwise normal HSPCs lead to resistance to a variety of cytotoxic agents.8,31 This likely explains their predominance in the setting of t-MN, where the disease has evolved in the setting of this selective pressure. Loss of p53 is likely to allow tolerance for additional genomic instability and karyotypic abnormalities, promoting the development of the high-risk cytogenetic abnormalities that have been associated with a poor prognosis.32 Thus, it appears that TP53-CHIP may represent a unique high-risk lesion with increased preleukemic potential33,34 and risk for transformation, especially in the setting of cytotoxic therapy.35
Mutations in TP53 have also been proposed to mediate resistance to lenalidomide in del(5q) MDS and secondary AML based on data that these mutations can often be found in patients who progress on treatment.22,36 We demonstrate that TP53-mutated t-MNs are particularly enriched in patients with MM treated with thalidomide analogs. We have also previously observed that although there was no increased risk of t-MN among patients with CH at the time of autologous stem cell transplant for MM, those that did develop t-MN had universally been exposed to thalidomide analogs and most had a preexisting TP53-mutant clone.37 Because MM is a clonal plasma-cell disorder, one could postulate that patients with MM are more likely to carry mutant HSCs and hence are at increased risk of t-MN development. However, previous studies have found a similar prevalence of CHIP in patients with MM and solid tumors.12,35,37 In addition, the risk of t-MN in patients with MM has evolved significantly over time, accompanied by the changes in treatment modalities.38 These observations argue against the idea that patients with MM have inherently higher risk of having abnormal HSCs that predispose them for t-MN, but rather, treatment exposures are driving the risk. Our multiplexed CRISPR-Cas9 experiments demonstrate that the selective advantage is exclusive to Trp53-mutant HSPCs and is not conferred on HSPCs carrying other mutations that are commonly found in CHIP (eg, Dnmt3a, Tet2, Asxl1, Ppm1d, and Ezh2). Although this model may not fully recapitulate the setting in patients, these data suggest a direct role for lenalidomide in driving the development of TP53-mutant t-MN.
We also demonstrate that pomalidomide does not exert the same selective pressure on Trp53-mutated HSPCs. The primary difference between the activities of lenalidomide and pomalidomide is the degradation of CK1α. Both drugs bind CRBN and induce degradation of Ikaros and Aiolos; however, pomalidomide is a significantly less potent degrader of CK1α.23,27 CK1α is essential for HSPC survival.29 Thus, pomalidomide may have less impact on WT HSPCs, limiting the selective advantage of TP53-mutant clones, a finding we observed in competitive transplant assays. Using competitive transplantation with Csnk1a1−/+ cells, we confirmed that the differential toxicity of lenalidomide and pomalidomide is largely driven by the lenalidomide-specific degradation of CK1α. This finding in mouse HSPCs will require further prospective validation in patients but suggests the possibility that future therapeutic regimens using thalidomide analogs with a narrower target degradation profile (eg, pomalidomide or iberdomide) could mitigate the risk of t-MN while maintaining their antimyeloma activity. At this point, because the clinical use of pomalidomide is restricted to patients previously treated with lenalidomide, it is difficult to identify a clinical cohort that has only been treated with pomalidomide but not lenalidomide. Future and ongoing clinical trials investigating the efficacy of newer thalidomide analogs (eg, iberdomide) might offer the opportunities to investigate the difference in t-MN risk based on the choice of thalidomide analogs.
Although it is important to identify the etiologic agents and genetic hallmarks of t-MN, it is equally important to define those patients with mutations more commonly seen in AML/MDS without prior exposures and thus likely to represent t-MNs that are biologically unrelated to the prior therapy and instead represent a second primary AML/MDS. Mutations in NPM1, IDH2, ASXL1, and spliceosome genes were anticorrelated with agents that are commonly associated with t-MNs (alkylators, anthracyclines, and topoisomerase inhibitors). These exact mutations are also more commonly identified in AML/MDS without prior exposures. Our data suggest that genetic profiling may distinguish true t-MN from second primary AML/MDS with a coincident history of CRT exposure. Such distinctions are important for improved risk stratification and trial allocation because many patients with t-MN are currently excluded from participating in clinical trials.
Our data add to the growing understanding of the interactions between specific clonal somatic mutations in the blood, exogenous stressors, and the development of clinical disease.6,8,35,39-41 Although CHIP, as an entity, is associated with adverse outcomes including increased cardiovascular disease and risk of myeloid malignancy, the specific risk of any disease outcome is likely to involve a complex interplay between genotype and environmental perturbagens. Comprehensive understanding of the heterogeneous interactions between CHIP and exogenous stressors will help advance a personalized approach to risk reduction and early intervention.
Acknowledgments
The authors thank D. Sykes and D. Scadden for the kind gift of the Hoxb8 viral vector and CHO-SCF cells. The authors also thank Koji Sasaki for helping chart review.
This study was supported by the National Institutes of Health (NIH), National Cancer Institute grants K12CA087723 and K08CA252174 (A.S.S.); an Evans Foundation Young Investigator Award (P.G.M.); an American Society of Hematology Research Training Award for Fellows (P.G.M.); NIH National Heart, Lung, and Blood Institute grant R01HL082945 (B.L.E.); NIH National Cancer Institute grants R01CA237291 (K.T.), P01CA265748 (K.T.), and P01CA066996 (B.L.E.); the Howard Hughes Medical Institute grant (B.L.E.); the Edward P. Evans Foundation grant (B.L.E.); the Leukemia and Lymphoma Society grant (B.L.E. and K.T.); the Adelson Medical Research Foundation grant (B.L.E.); the Cancer Prevention and Research Institute of Texas grant R120501 (P.A.F.); the Welch Foundation grant G-0040 (P.A.F.); the University of Texas System STARS award (grant PS100149) (P.A.F.); Physician Scientist Program at MD Anderson (K.T.); Andrew Sabin Family Foundation award (K.T.); American Society of Hematology Scholar Award (K.T.); Dresner Foundation Early Investigator award (K.T.); Lyda Hill Foundation grant (P.A.F.); the Charif Souki Cancer Research fund (H.K.); the MD Anderson Cancer Center Leukemia SPORE grant (NIH P50 CA100632) (H.K. and K.T.); the MD Anderson Cancer Center Support grant (NIH/NCI P30 CA016672); and generous philanthropic contributions to MD Anderson Moon Shot Program grant (P.A.F., K.T., G.G.-M., and H.K.).
Authorship
Contribution: A.S.S., J.A.K., A.T.N., P.G.M., M.E.M., V.A.Q.B., and B.L.E. designed and performed in vitro and in vivo experiments; V.A.G. collected data and wrote the manuscript; Y.Y. performed statistical analysis; J.I.H. and L.Z. collected data; F.W., K.F., and J.Z. performed bioinformatics analysis; L.L. and C.G. performed DNA sequencing; N.D., R.K.-S., C.D.D., T.K., F.R., H.K., and G.G.-M. collected samples and data; A.S.S., P.G.M., B.L.E., and K.T. wrote the manuscript; A.S.S., B.L.E., P.A.F., and K.T. provided leadership and managed the study team; and all authors read and approved the manuscript.
Conflict-of-interest disclosure: B.L.E. received research funding from Celgene, Deerfield, Novartis, and Calico and consulting fees from GRAIL and serves on the scientific advisory board for and is a shareholder in Neomorph Therapeutics, Skyhawk Therapeutics, TenSixteen Bio, and Exo Therapeutics. P.G.M. received consulting fees from Foundation Medicine, Inc. F.R. received honoraria from Celgene and BMS as an advisory board member. C.D.D. received research support (to institution) from AbbVie, Agios, Bayer, Calithera, Cleave, BMS/Celgene, Daiichi-Sankyo, and ImmuneOnc and is a consultant/advisory board member at AbbVie, Agios, Celgene/BMS, Daiichi-Sankyo, ImmuneOnc, Novartis, Takeda, and Notable Labs. H.K. received research grants from AbbVie, Amgen, Ascentage, BMS, Daiichi-Sankyo, Immunogen, Jazz, Novartis, Pfizer, and Sanofi and honoraria from AbbVie, Actinium, Adaptive Biotechnologies, Amgen, Apptitude Health, BioAscend, Daiichi-Sankyo, Delta Fly, Janssen Global, Novartis, Oxford Biomedical, Pfizer and, Takeda. K.T. received consulting fees from Celgene, GSK, and Novartis; received honoraria from Mission Bio and Illumina; and serves on the scientific advisory board for Symbio Pharmaceuticals. The remaining authors declare no competing financial interests.
Correspondence: Benjamin L. Ebert, Department of Medical Oncology, Dana-Farber Cancer Institute, 450 Brookline Ave, D1610A, Boston, MA 02115; e-mail: benjamin_ebert@dfci.harvard.edu; and Koichi Takahashi, Department of Leukemia, Unit 428, The University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: ktakahashi@mdanderson.org.
References
Author notes
∗A.S.S., V.A.G., J.A.K., and Y.Y. contributed equally to this study.
Data are available on request from the corresponding authors, Benjamin L. Ebert (benjamin_ebert@dfci.harvard.edu) and Koichi Takahahsi (ktakahashi@mdanderson.org). The list of all high-confidence somatic mutations detected by the 300-gene panel sequencing is provided in supplemental Table 5.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

Comments
Lenalidomide – From a Friend (in Short Term) to a Foe (in Long Term)
Abbreviations -
CK1α-Casein kinase 1α, HSCT – Hematopoietic Stem Cell Transplant, MRD – Minimal Residual Disease, TP53 - Tumor Protein p53
References –
1. Sperling AS, Guerra VA, Kennedy JA, et al. Lenalidomide promotes the development of TP53-mutated therapy-related myeloid neoplasms. Blood. 2022;140(16):1753-1763. doi:10.1182/blood.2021014956
2. Fink EC, Ebert BL. The novel mechanism of lenalidomide activity. Blood. 2015 Nov 19;126(21):2366-9. doi: 10.1182/blood-2015-07-567958.
3. Kumar L, Chellapuram SK, Sahoo R, et al. VRd versus VCd as induction therapy for newly diagnosed multiple myeloma: A Phase III, randomised study. Clin Lymphoma Myeloma Leuk. 2019; 19(10):E361. https://doi.org/10.1016/j.clml.2019.09.597
4. Richardson PG, Jacobus SJ, Weller EA, et al. Triplet Therapy, Transplantation, and Maintenance until Progression in Myeloma. N Engl J Med. 2022;387(2):132-147. doi:10.1056/NEJMoa2204925
5. Jones JR, Cairns D, Pawlyn C, et al. Myeloma XI Trial for Newly Diagnosed Multiple Myeloma (NDMM); Long Term Second Primary Malignancy (SPM) Incidence in the Context of Lenalidomide Maintenance. Blood. 2019; 134 (Supplement_1): 3132
https://ashpublications.org/blood/article/134/Supplement_1/3132/423895/Title-Myeloma-XI-Trial-for-Newly-Diagnosed
6. Jones JR, Cairns D, Menzies T, et al. Second Primary Malignancy Incidence in Patients Receiving Lenalidomide at Induction and Maintenance; Long-Term Follow up of 4358 Patients Enrolled to the Myeloma XI Trial. Oral Abstract #754 64th American Society of Hematology Annual Meeting and Exposition. Dec 12, 2022; New Orleans, US
https://ash.confex.com/ash/2022/webprogram/Paper168723.html
7. Attal M, Lauwers-Cances V, Hulin C, et al. Lenalidomide, Bortezomib, and Dexamethasone with Transplantation for Myeloma. N Engl J Med. 2017;376(14):1311-1320. doi:10.1056/NEJMoa1611750
8. Facon T, Kumar S, Plesner T, et al. Daratumumab plus Lenalidomide and Dexamethasone for Untreated Myeloma. N Engl J Med. 2019;380(22):2104-2115. doi:10.1056/NEJMoa1817249
9. Samur MK, et al. High-Dose Melphalan Significantly Increases Mutational Burden in Multiple Myeloma Cells at Relapse: Results from a Randomized Study in Multiple Myeloma. Blood 2020;136:4-5
10. Costa LJ, Chhabra S, Medvedova E, et al. Daratumumab, Carfilzomib, Lenalidomide, and Dexamethasone With Minimal Residual Disease Response-Adapted Therapy in Newly Diagnosed Multiple Myeloma. J Clin Oncol. 2022;40(25):2901-2912. doi:10.1200/JCO.21.01935
Dr Sumeet Mirgh
Email - drsumeetmirgh@gmail.com