Key Points
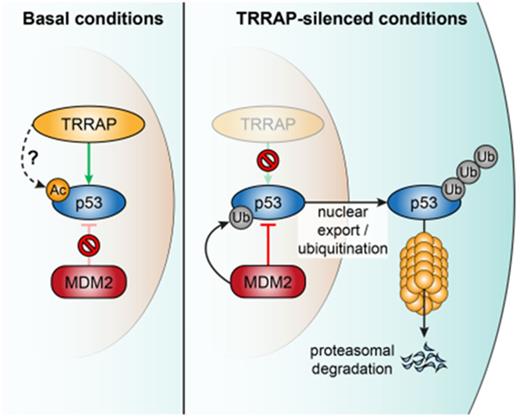
The HAT complex member TRRAP is vital for maintaining high p53 levels by shielding it against the natural p53 degradation machinery.
Acetylation-modifying complexes regulate p53 protein stability, which may provide a basis for therapeutic targeting of mutant p53.
Abstract
Tumors accumulate high levels of mutant p53 (mutp53), which contributes to mutp53 gain-of-function properties. The mechanisms that underlie such excessive accumulation are not fully understood. To discover regulators of mutp53 protein accumulation, we performed a large-scale RNA interference screen in a Burkitt lymphoma cell line model. We identified transformation/transcription domain-associated protein (TRRAP), a constituent of several histone acetyltransferase complexes, as a critical positive regulator of both mutp53 and wild-type p53 levels. TRRAP silencing attenuated p53 accumulation in lymphoma and colon cancer models, whereas TRRAP overexpression increased mutp53 levels, suggesting a role for TRRAP across cancer entities and p53 mutations. Through clustered regularly interspaced short palindromic repeats (CRISPR)–Cas9 screening, we identified a 109-amino-acid region in the N-terminal HEAT repeat region of TRRAP that was crucial for mutp53 stabilization and cell proliferation. Mass spectrometric analysis of the mutp53 interactome indicated that TRRAP silencing caused degradation of mutp53 via the MDM2-proteasome axis. This suggests that TRRAP is vital for maintaining mutp53 levels by shielding it against the natural p53 degradation machinery. To identify drugs that alleviated p53 accumulation similarly to TRRAP silencing, we performed a small-molecule drug screen and found that inhibition of histone deacetylases (HDACs), specifically HDAC1/2/3, decreased p53 levels to a comparable extent. In summary, here we identify TRRAP as a key regulator of p53 levels and link acetylation-modifying complexes to p53 protein stability. Our findings may provide clues for therapeutic targeting of mutp53 in lymphoma and other cancers.
Introduction
Aggressive lymphoma subtypes such as Burkitt lymphoma (BL), a B-cell non-Hodgkin lymphoma caused by c-Myc translocation,1 are characterized by a high incidence of p53 mutations (∼30% to 40%).2,3 Gain-of-function (GOF) mutations equip p53 with novel properties that actively contribute to tumorigenesis.4 Such mutations predict a poor response to chemotherapy and short survival in many different cancer entities,5 including lymphoma.6
Wild-type p53 (wtp53) is tightly controlled on the posttranslational level. In physiological conditions, wtp53 levels are low due to constitutive degradation by the E3 ubiquitin ligase MDM2, which is a transcriptional target of wtp53.7 In contrast, tumors harboring mutant p53 (mutp53) are typically characterized by substantial accumulation of p53 protein.4,5 Knockin studies have revealed that mutp53 is not intrinsically stable in normal cells but is reversibly stabilized in response to stress.8-12 It has been suggested that disruption of the MDM2-p53 loop is responsible for mutp53 accumulation.4 However, tumors often show residual MDM2 expression12,13 and, more importantly, MDM2 is capable of mutp53 binding and ubiquitination.14,15 Therefore, alternative mechanisms must contribute to the ability of mutp53 to evade degradation in cancer cells. Similar to wtp53, posttranslational modifications (eg, acetylation) of mutp53 may contribute to its stability.16
Numerous studies have shown that elevated levels of mutp53 are a prerequisite for its GOF properties: mutp53 silencing results in suppression of tumor growth, attenuated invasion and metastasis formation, and increased chemosensitivity in vitro and in vivo.4,5,17 This suggests that tumors may become “addicted” to constitutively high mutp53 levels. Thus, interference with mutp53 accumulation may be exploitable for cancer therapy.
Since our understanding of regulators of mutp53 accumulation is presently still limited, we performed an RNA interference (RNAi) screen targeting ∼5000 genes in the BL cell line Raji. The screen revealed a central role of transformation/transcription domain-associated protein (TRRAP), a histone acetyltransferase (HAT) complex constituent, in stabilizing both mutp53 and wtp53. Furthermore, we found that small-molecule histone deacetylase (HDAC) inhibitors decreased mutp53 levels to a similar extent as TRRAP silencing. Thus, we link acetylation-modifying complexes to the regulation of p53 protein stability, which may provide a basis for therapeutic targeting of mutp53 in lymphoma.
Materials and methods
Detailed materials and methods are available in the supplement (available on the Blood Web site). Oligonucleotide sequences are provided in supplemental Tables 1-6.
Patient samples/tissue microarray
Samples were investigated as part of the Molecular Mechanisms in Malignant Lymphomas (MMML) Network Project18 or were described previously.19 The MMML protocols have been approved centrally by the institutional review board of the coordination center in Göttingen, Germany. p53 tissue microarray was performed according to standard protocols with anti-p53 clone DO-7 (Dako, Santa Clara, CA).
p53 flow cytometry
The staining protocol was adapted from Mohr et al20 and showed a high correlation with p53 western blot (data not shown). Cells were stained with Zombie UV (BioLegend, San Diego, CA) for live/dead cell discrimination and fixed in either (1) 2% paraformaldehyde/phosphate-buffered saline for 30 minutes at 4°C and permeabilized in 80% EtOH/phosphate-buffered saline for 30 min at −20°C or (2) ice-cold MeOH for 30 minutes at −20°C. Cells were stained with anti-p53 clone DO-7 (554298; BD Biosciences, San Jose, CA) for 1 hour at 4°C and measured using an LSRFortessa or LSRII (BD Biosciences). Unstained and isotype control samples were used to evaluate specificity of the staining. Median fluorescence intensities (MFIs) were used for quantification.
RNAi and clustered regularly interspaced short palindromic repeats (CRISPR) Cas9
The RNAi screen was performed as described previously21 with modifications using the DECIPHER Pooled Lentiviral short hairpin RNA (shRNA) Library Human Module I (Cellecta, Mountain View, CA). For single knockdowns, shRNAs were expressed in the vectors pRSI12-U6-(sh)-UbiC-TagRFP-2A-Puro (constitutive) or pLKO_IPTG_3xLacO (isopropyl β-D-1-thiogalactopyranoside [IPTG] inducible). For single knockouts, single guide RNAs (sgRNAs) were expressed in the vectors lentiCRISPRv2 or pLKO5.sgRNA.EFS.tRFP. CRISPR-Cas9 screening of TRRAP protein domains was performed as described previously.22 TRRAP sgRNAs were designed based on rules defined by Doench et al,23 aiming for comparable on-target scores. TagRFP657 expression and p53 levels were monitored by flow cytometry.
Immunoblotting
Cells were lysed in RIPA buffer (Merck, Darmstadt, Germany) containing protease inhibitors (Roche Diagnostics, Mannheim, Germany). Lysates were transferred to polyvinylidene difluoride membranes (Bio-Rad Laboratories, Hercules, CA) and probed with antibodies against TRRAP (3966; Cell Signaling Technology, Danvers, MA), MDM2 (sc-965; Santa Cruz Biotechnology, Dallas, TX), p53 (HAF1355; R&D Systems, Minneapolis, MN), GAPDH (G9295; Sigma-Aldrich, St. Louis, MO), p21 (556431; BD Biosciences), PARP (9542; Cell Signaling Technology), XPO1 (sc-74454, Santa Cruz Biotechnology), or EIF3F (638201, BioLegend).
Immunoprecipitation/mass spectrometry
Namalwa cells with IPTG-inducible knockdowns were induced for 48 to 96 hours before they were harvested. For immunoprecipitation, lysates were precleared twice and protein complexes were captured for 4 hours at 4°C using antibody-coupled agarose beads (p53: sc-126 AC; immunoglobulin G [IgG]: sc-2343, Santa Cruz Biotechnology) and eluted with 0.1 M glycine (pH 2.5). Eluents were neutralized with 1.5 M Tris-HCl (pH 8.8). Samples were labeled with TMT10 reagents24 and analyzed on a QExactive plus (Thermo Fisher Scientific, Waltham, MA). Raw data were processed by IsobarQuant,25 and differential total protein expression was assessed using the DEP package in R.26 The experiment was performed in biological triplicates.
Gene expression analysis
Total DNase-digested RNA was purified using the RNeasy Mini Kit (Qiagen, Hilden, Germany). 500 ng RNA was used for cDNA synthesis using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed with the Power SYBR Green Master Mix (Thermo Fisher Scientific) and run on a cobas z 480 Analyzer (Roche Diagnostics). Gene expression was quantified using the ΔΔCt method and normalized to GAPDH. Global gene expression changes after TRRAP silencing were determined using an Illumina HumanHT-12 v4 BeadChip (data are accessible at the National Center for Biotechnology Information [NCBI] Gene Expression Omnibus [GEO] database under accession number GSE103915). Gene set enrichment analysis was performed using parametric analysis of gene set enrichment (PAGE).27
Results
mutp53 accumulates in lymphoma
Consistent with prior findings of high mutp53 levels in tumors, we found in a cohort of primary B-cell lymphoma samples that cases with p53 mutation showed significantly higher p53 protein levels than wild-type cases (diffuse large B-cell lymphoma: P = 1.40 × 10−10, n = 134; BL: P = .016, n = 20; supplemental Figure 1A). This finding was also confirmed in a panel of 14 lymphoma cell lines (8 mutp53, 6 wtp53) using flow cytometry to quantify intracellular p53 levels (supplemental Figure 1B). Consistent with a posttranscriptional mechanism of mutp53 accumulation,28,29 we found no correlation between p53 mRNA and protein expression (supplemental Figure 1C). MDM2 inhibition with Nutlin-3a further elevated the mutp53 level in eight lymphoma cell lines (supplemental Figure 1D), suggesting that mutp53 was still subject to MDM2-mediated degradation.
RNAi screen identifies TRRAP as a regulator of mutp53 accumulation
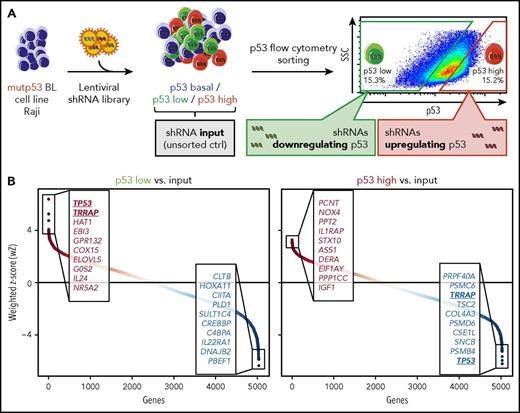
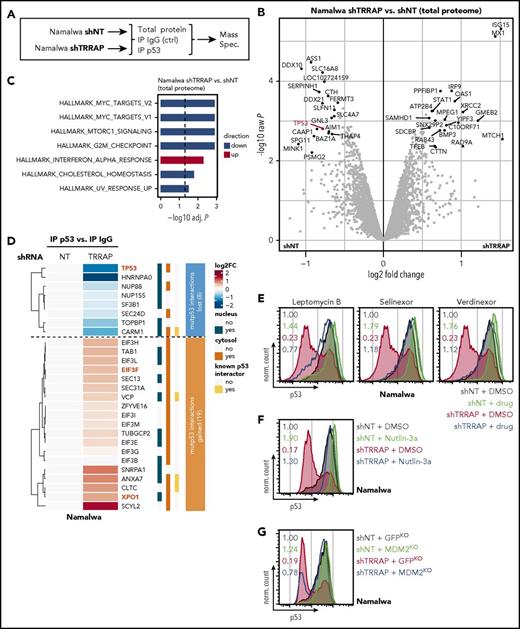
To identify regulators of mutp53 protein levels, we performed an RNAi screen in the Raji cell line, a model for mutp53 BL (Figure 1A). We used a shRNA library targeting ∼5000 genes with 5 or 6 independent shRNAs per gene. Eight days after transduction, cells were sorted by flow cytometry into a p53-low and a p53-high population (supplemental Figure 2A). shRNA barcodes of sorted and input (nonsorted) samples and the initial shRNA plasmid library were sequenced to identify enriched and depleted shRNAs (supplemental Table 7). The counts of shRNA barcodes were reproducible between technical replicates (r = 0.53-0.87; supplemental Figure 2B). As expected, shRNAs targeting essential genes were depleted in the input samples (supplemental Figure 2C).30 Notably, shRNAs against CDK6 (cyclin-dependent kinase 6) were the most depleted, consistent with the essential function of CDK6 in BL (supplemental Table 7).31
RNAi screen for regulators of mutp53 accumulation. (A) Screen layout. After lentiviral transfer of a pooled shRNA library, cells were sorted by flow cytometry into p53-low and p53-high cells. shRNA barcodes were amplified and their abundance was determined by high-throughput sequencing. The experiment was performed in technical duplicates. (B) Screen results. Data from multiple shRNAs per gene were combined using the wZ method, which assigns a high score to genes targeted by multiple enriched shRNAs. wZs were calculated for p53-low and p53-high cell populations by comparison against the input samples. Boxes highlight the top and bottom 10 shRNA-targeted candidate genes enriched or depleted in the respective population. SSC, side scatter.
RNAi screen for regulators of mutp53 accumulation. (A) Screen layout. After lentiviral transfer of a pooled shRNA library, cells were sorted by flow cytometry into p53-low and p53-high cells. shRNA barcodes were amplified and their abundance was determined by high-throughput sequencing. The experiment was performed in technical duplicates. (B) Screen results. Data from multiple shRNAs per gene were combined using the wZ method, which assigns a high score to genes targeted by multiple enriched shRNAs. wZs were calculated for p53-low and p53-high cell populations by comparison against the input samples. Boxes highlight the top and bottom 10 shRNA-targeted candidate genes enriched or depleted in the respective population. SSC, side scatter.
To combine data from multiple shRNAs per gene, we calculated weighted z-scores (wZ) of the p53-low and p53-high cell populations compared with input samples (Figure 1B; supplemental Table 7). TP53 shRNAs were the most enriched (rank 1/5033, wZ = 6.38) in the p53-low population and the most depleted in the p53-high population (rank 5033/5033, wZ = −6.21), a strong validation of our experimental approach. Next, we focused on shRNAs behaving similarly to those targeting TP53 (ie, enriched in the p53-low population while depleted in the p53-high population). Among the top/bottom 10 shRNA-targeted genes, TRRAP was the only candidate meeting this stringent criterion (Figure 1B), with pronounced shRNA enrichment in the p53-low population (rank 2/5033, wZ = 5.24) and strong shRNA depletion in the p53-high population (rank 5026/5033, wZ = −4.97). Notably, when we considered the top/bottom 100 shRNA-targeted genes, there were 11 additional genes that appeared in both lists (supplemental Table 7). This included TNF (p53 low: rank 12/5033, wZ = 3.64; p53 high: rank 4980/5033, wZ = −3.90), which positively regulates the nuclear mutp53 level.32
TRRAP stabilizes mutp53 across cancer entities and p53 mutations
TRRAP is a member of the phosphatidylinositol 3-kinase–related kinase (PIKK) family, which includes the well-characterized p53 regulators ATM, ATR, and DNA-PKcs. As a component of multiple HAT complexes, the most characterized function of TRRAP is the recruitment of HATs to chromatin during transcription and DNA repair.33 Although TRRAP is highly expressed in healthy B cells and many lymphomas,34 mutations in TRRAP are very rare in lymphoma (supplemental Figure 3).35
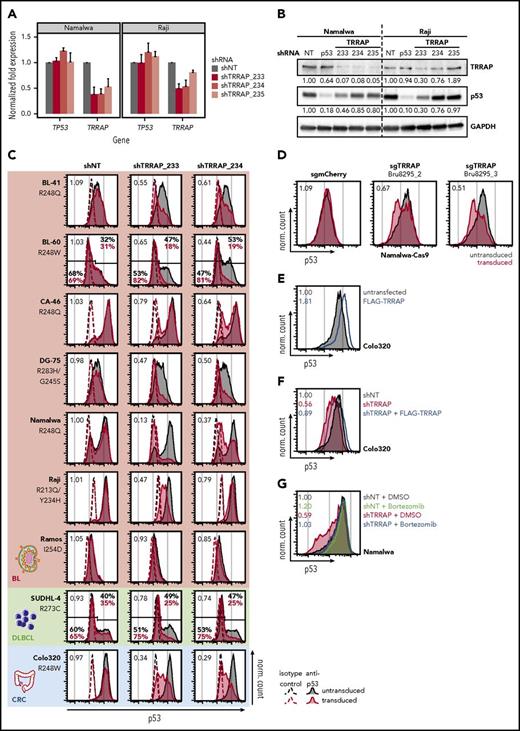
To confirm the effect of TRRAP silencing on mutp53 levels, we studied three independent shRNAs and verified the on-target effect for 2 shRNAs across cell lines (shTRRAP_233 and _234; Figure 2A-B). These 2 shRNAs also reduced mutp53 to 30% to 85% of its basal level (Figure 2B), with no consistent change in TP53 mRNA expression (Figure 2A), suggesting posttranscriptional regulation of p53. TRRAP silencing decreased mutp53 levels in a panel of 8 lymphoma cell lines (Figure 2C); however, we observed variable downregulation, which was independent of the specific p53 mutation. Notably, TRRAP silencing also reduced the mutp53 level in the colorectal cancer cell line Colo320 (Figure 2C). To confirm the RNAi effects, we employed CRISPR-Cas9 genome editing and observed that the mutp53 level was approximately halved upon TRRAP knockout (Figure 2D). These data suggest that TRRAP positively regulates mutp53 accumulation in a diverse set of cancers.
TRRAP stabilizes mutp53 across cancer entities and p53 mutations. (A) mRNA level of TRRAP and TP53 in Namalwa and Raji cells transduced with shRNAs against TRRAP. Cells were selected with puromycin for 48 hours and harvested 3 days after transduction. Expression values (mean + SD, n = 2) were determined by qRT-PCR and normalized to GAPDH and to cells transduced with a nontargeting shRNA (NT). (B) Protein level of TRRAP and p53 in Namalwa and Raji cells transduced with shRNAs against TRRAP and p53. Cells were selected with puromycin for 48 hours and harvested 7 days after transduction. Expression was determined by western blot and normalized to GAPDH and to cells transduced with a nontargeting shRNA (NT). (C) p53 flow cytometry 7 or 8 days after shRNA-mediated knockdown of TRRAP in mutp53 cancer cell lines: BL, diffuse large B-cell lymphoma (DLBCL), and colorectal cancer (CRC). The p53 mutation is specified for each cell line. Values denote the ratio of the p53 MFI between transduced (red) and untransduced cells (gray). For cell lines with a bimodal p53 level, gates and the corresponding percentages for the p53-low and p53-high population are indicated. Dashed lines indicate isotype control stainings, and filled colored histograms indicate samples stained with anti-p53. shNT, nontargeting shRNA. (D) p53 flow cytometry 7 days after sgRNA-mediated knockout of TRRAP in Namalwa-Cas9 cells. Values denote the ratio of the p53 MFI between transduced (red) and untransduced cells (gray). sgmCherry, negative control. (E) p53 flow cytometry 72 hours after transfection with FLAG-TRRAP in Colo320 cells. Values denote the ratio of the p53 MFI between transfected (blue) and untransfected cells (gray). (F) Molecular rescue of the TRRAP knockdown phenotype by FLAG-TRRAP overexpression in Colo320 cells. Cells were transduced with either a nontargeting shRNA (shNT; gray) or with a shRNA against TRRAP (shRNA #233; red). 24 hours after transduction, part of the TRRAP-silenced cells was transfected with FLAG-TRRAP (blue). Cells were analyzed by p53 flow cytometry 4 days after transduction. Values denote p53 MFI normalized to cells transduced with the NT. (G) Rescue of the TRRAP-silencing–mediated mutp53 degradation by treatment with the proteasome inhibitor bortezomib. Namalwa cells were transduced with an IPTG-inducible shRNA against TRRAP (shRNA #233) or a nontargeting control (NT). 24 hours after induction, cells were treated with dimethyl sulfoxide (DMSO; 0.1%) or bortezomib (100 nM) for 14 hours before being subjected to p53 flow cytometry. Values denote p53 MFI normalized to DMSO-treated cells transduced with the NT.
TRRAP stabilizes mutp53 across cancer entities and p53 mutations. (A) mRNA level of TRRAP and TP53 in Namalwa and Raji cells transduced with shRNAs against TRRAP. Cells were selected with puromycin for 48 hours and harvested 3 days after transduction. Expression values (mean + SD, n = 2) were determined by qRT-PCR and normalized to GAPDH and to cells transduced with a nontargeting shRNA (NT). (B) Protein level of TRRAP and p53 in Namalwa and Raji cells transduced with shRNAs against TRRAP and p53. Cells were selected with puromycin for 48 hours and harvested 7 days after transduction. Expression was determined by western blot and normalized to GAPDH and to cells transduced with a nontargeting shRNA (NT). (C) p53 flow cytometry 7 or 8 days after shRNA-mediated knockdown of TRRAP in mutp53 cancer cell lines: BL, diffuse large B-cell lymphoma (DLBCL), and colorectal cancer (CRC). The p53 mutation is specified for each cell line. Values denote the ratio of the p53 MFI between transduced (red) and untransduced cells (gray). For cell lines with a bimodal p53 level, gates and the corresponding percentages for the p53-low and p53-high population are indicated. Dashed lines indicate isotype control stainings, and filled colored histograms indicate samples stained with anti-p53. shNT, nontargeting shRNA. (D) p53 flow cytometry 7 days after sgRNA-mediated knockout of TRRAP in Namalwa-Cas9 cells. Values denote the ratio of the p53 MFI between transduced (red) and untransduced cells (gray). sgmCherry, negative control. (E) p53 flow cytometry 72 hours after transfection with FLAG-TRRAP in Colo320 cells. Values denote the ratio of the p53 MFI between transfected (blue) and untransfected cells (gray). (F) Molecular rescue of the TRRAP knockdown phenotype by FLAG-TRRAP overexpression in Colo320 cells. Cells were transduced with either a nontargeting shRNA (shNT; gray) or with a shRNA against TRRAP (shRNA #233; red). 24 hours after transduction, part of the TRRAP-silenced cells was transfected with FLAG-TRRAP (blue). Cells were analyzed by p53 flow cytometry 4 days after transduction. Values denote p53 MFI normalized to cells transduced with the NT. (G) Rescue of the TRRAP-silencing–mediated mutp53 degradation by treatment with the proteasome inhibitor bortezomib. Namalwa cells were transduced with an IPTG-inducible shRNA against TRRAP (shRNA #233) or a nontargeting control (NT). 24 hours after induction, cells were treated with dimethyl sulfoxide (DMSO; 0.1%) or bortezomib (100 nM) for 14 hours before being subjected to p53 flow cytometry. Values denote p53 MFI normalized to DMSO-treated cells transduced with the NT.
To investigate the impact of TRRAP and mutp53 silencing on cell proliferation, we transduced BL cell lines at an efficiency of ∼50% with shRNAs together with RFP and monitored the proportion of transduced cells over time (supplemental Figure 4A-B). Cells with knockdown of mutp53 were outgrown by their untransduced counterparts, indicating that silencing impaired cell proliferation and thus providing evidence for mutp53 GOF. TRRAP knockdown impaired proliferation of mutp53 and wtp53 BL cells equally, suggesting that TRRAP’s essential function for cell survival36 is p53 independent. Both TRRAP and mutp53 silencing did not induce apoptosis but elicited a severe arrest in G0/G1 phase (supplemental Figure 4C-D).
To further understand the role of TRRAP in regulating mutp53 accumulation, we tested the impact of TRRAP overexpression on mutp53 levels in Colo320 cells. Transfection of FLAG-TRRAP elevated mutp53 protein levels (1.81-fold; Figure 2E) without changing TP53 mRNA abundance (supplemental Figure 5). To verify the on-target effect of TRRAP knockdown, we performed TRRAP silencing in the context of TRRAP overexpression and observed a rescue of mutp53 levels (Figure 2F). To test for a potential association between p53 and TRRAP protein levels, we performed western blot analysis in lymphoma cell lines and primary samples, and observed no significant correlation (supplemental Figure 6).
To assess whether proteasomal degradation was involved in TRRAP-dependent regulation of mutp53, we silenced TRRAP, treated the cells with the proteasome inhibitor bortezomib, and quantified mutp53 levels by flow cytometry (Figure 2G). Bortezomib treatment mildly upregulated mutp53 (1.20-fold) in control knockdown samples. Notably, the reduced mutp53 levels in TRRAP-silenced cells were restored to basal control levels by proteasome inhibition. This was also confirmed by western blot analysis (supplemental Figure 7A). Correspondingly, we observed increased ubiquitination of mutp53 upon TRRAP knockdown (supplemental Figure 7B).
Together, these results suggest that by inhibiting mutp53 ubiquitination and proteasomal degradation, TRRAP acts as a positive posttranscriptional regulator of mutp53 levels across different cancer entities and p53 mutations.
Identification of the TRRAP domain crucial for mutp53 stabilization and cell proliferation
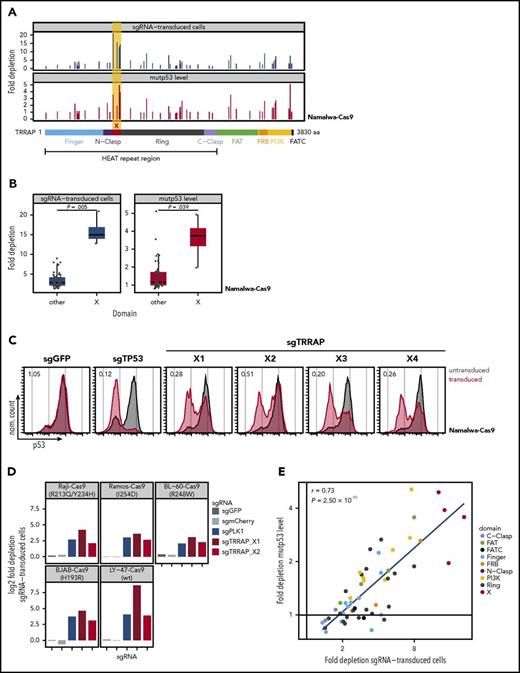
To identify functional regions of TRRAP that mediate mutp53 stabilization and cell proliferation, we performed a CRISPR-Cas9 mutagenesis screening. This approach is based on the fact that cells transduced with sgRNAs targeting functionally important protein domains will be preferentially outcompeted by untransduced cells, when compared with cells infected with sgRNAs targeting irrelevant protein domains.22 Namalwa-Cas9 cells were transduced at an efficiency of ∼25% in an arrayed format with 55 sgRNAs spanning the entire TRRAP coding region together with TagRFP657. The proportion of transduced cells and mutp53 levels were monitored by flow cytometry (Figure 3A). In line with our previous RNAi results, cells transduced with any TRRAP sgRNA were outcompeted by untransduced cells. Targeting of a 109-amino-acid region (residues 1050-1158) within TRRAP’s HEAT repeat region (between the N-clasp and ring)37 drove significantly stronger depletion of transduced cells (mean: 15.9-fold vs 3.5-fold, P = .005) and mutp53 (3.6-fold vs 1.5-fold, P = .039) than targeting of any other region of TRRAP (Figure 3A-C, “X”). We validated the effects of the newly identified TRRAP domain in 5 additional BL cell lines (Figure 3D). Of note, we identified a strong positive correlation between cell depletion and mutp53 downregulation (r = 0.73, P = 2.50 × 10−10, Figure 3E), suggesting that the ability of TRRAP to support cell proliferation and regulate mutp53 levels may be mediated by a common mechanism.
Identification of the TRRAP domain crucial for mutp53 stabilization and cell proliferation. (A) CRISPR-Cas9 screening of TRRAP protein domains. Namalwa-Cas9 cells were transduced in an arrayed format with 55 sgRNAs targeting different regions of TRRAP protein, covering all annotated domains according to Diaz-Santin et al.37 The proportion of transduced cells and the mutp53 level was quantified by flow cytometry. Negative cell selection (top, blue) and mutp53 depletion (bottom, red) is shown as fold depletion of transduced cells or mutp53 protein after 21 days or 7 days in culture, respectively. Every bar represents an independent sgRNA, and the location of each sgRNA relative to TRRAP protein is indicated along the x-axis. FAT, focal adhesion targeting domain; FATC, FRAP, ATM, TRRAP C-terminal domain; FRB, FKBP12-rapamycin binding domain; PI3K, phosphatidylinositol 3-kinase domain. (B) Comparison of fold depletion of cells (blue) and mutp53 protein (red) after transduction with sgRNAs targeting either domain X or any other region of TRRAP. P values were calculated with Student t test. Data reproduced from panel A. (C) p53 flow cytometry 7 days after TRRAP knockout in Namalwa-Cas9 cells. Values denote the ratio of the p53 MFI between transduced (red) and untransduced cells (gray). sgGFP, negative control; sgTP53, positive control. (D) Quantification of selection against sgTRRAP-transduced cells in BL-Cas9 cell lines. Bars indicate log2 fold depletion of sgRNA-transduced cells after 24 days in culture. The p53 status is specified for each cell line. sgGFP and sgmCherry, negative controls; sgPLK1, positive control. (E) Correlation of toxicity (depletion of sgRNA-transduced cells) and mutp53 depletion after TRRAP knockout using sgRNAs targeting different protein domains. Each data point represents a single sgRNA from panel A. Colors indicate the targeted domains. The Pearson correlation coefficient (r) and the corresponding P value are indicated.
Identification of the TRRAP domain crucial for mutp53 stabilization and cell proliferation. (A) CRISPR-Cas9 screening of TRRAP protein domains. Namalwa-Cas9 cells were transduced in an arrayed format with 55 sgRNAs targeting different regions of TRRAP protein, covering all annotated domains according to Diaz-Santin et al.37 The proportion of transduced cells and the mutp53 level was quantified by flow cytometry. Negative cell selection (top, blue) and mutp53 depletion (bottom, red) is shown as fold depletion of transduced cells or mutp53 protein after 21 days or 7 days in culture, respectively. Every bar represents an independent sgRNA, and the location of each sgRNA relative to TRRAP protein is indicated along the x-axis. FAT, focal adhesion targeting domain; FATC, FRAP, ATM, TRRAP C-terminal domain; FRB, FKBP12-rapamycin binding domain; PI3K, phosphatidylinositol 3-kinase domain. (B) Comparison of fold depletion of cells (blue) and mutp53 protein (red) after transduction with sgRNAs targeting either domain X or any other region of TRRAP. P values were calculated with Student t test. Data reproduced from panel A. (C) p53 flow cytometry 7 days after TRRAP knockout in Namalwa-Cas9 cells. Values denote the ratio of the p53 MFI between transduced (red) and untransduced cells (gray). sgGFP, negative control; sgTP53, positive control. (D) Quantification of selection against sgTRRAP-transduced cells in BL-Cas9 cell lines. Bars indicate log2 fold depletion of sgRNA-transduced cells after 24 days in culture. The p53 status is specified for each cell line. sgGFP and sgmCherry, negative controls; sgPLK1, positive control. (E) Correlation of toxicity (depletion of sgRNA-transduced cells) and mutp53 depletion after TRRAP knockout using sgRNAs targeting different protein domains. Each data point represents a single sgRNA from panel A. Colors indicate the targeted domains. The Pearson correlation coefficient (r) and the corresponding P value are indicated.
These results suggest that a portion of the TRRAP HEAT domain is crucial for promoting cell proliferation and mutp53 stabilization.
TRRAP silencing destabilizes mutp53 via the MDM2-proteasome axis
To obtain a global view on proteins regulated by TRRAP, we profiled the total proteome by mass spectrometry after TRRAP depletion (Figure 4A). As expected, TRRAP and mutp53 expression were reduced in the total proteome upon TRRAP silencing (supplemental Figure 8A). TRRAP knockdown significantly altered the abundance of 40 proteins (19 downregulated, 21 upregulated; Figure 4B; supplemental Table 8). TRRAP silencing suppressed proteins associated with the G2/M checkpoint36,38,39 and MYC targets,40,41 whereas it induced proteins associated with interferon α signaling (Figure 4C).
TRRAP silencing destabilizes mutp53 via the MDM2-proteasome axis. (A) Experimental design. Namalwa cells were transduced with an IPTG-inducible shRNA against TRRAP (shRNA #233) or a nontargeting control (NT). 48 to 96 hours after induction, cells were lysed and either collected immediately (total protein) or subjected to immunoprecipitation (IP) with antibodies against p53 or IgG (negative control) before mass spectrometry. The experiment was performed in biological triplicates. (B) Volcano plot visualizing differential total protein expression 96 hours after TRRAP knockdown compared with a nontargeting control (NT). Proteins denoted in black were significantly differentially expressed (adjusted P < .05, |log2FC| > 0.5). p53 is highlighted. P values were calculated using empirical Bayes statistics on protein-wise linear models. (C) Gene set enrichment analysis of differentially expressed proteins from panel B. Upregulated gene sets are denoted in red, downregulated in blue. Analysis was performed using PAGE with the MSigDB gene set collection “Hallmark.” Only significantly altered gene sets are shown (adjusted P < .05). (D) Heatmap of mutp53-bound proteins (proteins significantly enriched in p53 IPs compared with IgG control IPs). Values denote hierarchically clustered log2 fold changes of protein abundances in TRRAP-silenced samples compared with control knockdown (NT) samples. The cellular localization44 and known p53 interactions42 are indicated. Numbers within the clusters indicate the number of proteins. TP53, EIF3F, and XPO1 are highlighted. (E) Rescue of the TRRAP-silencing–mediated mutp53 degradation by treatment with nuclear export inhibitors. Namalwa cells were transduced with an IPTG-inducible shRNA against TRRAP (shRNA #233) or a nontargeting control (NT). 48 hours after induction, cells were treated with DMSO (0.005%), leptomycin B (20 nM), selinexor (500 nM), or verdinexor (500 nM) for 14 hours before they were subjected to p53 flow cytometry. Values denote p53 MFI normalized to DMSO-treated cells transduced with the NT. (F) Rescue of the TRRAP-silencing–mediated mutp53 degradation by treatment with the MDM2 inhibitor Nutlin-3a. Namalwa cells were transduced with a shRNA against TRRAP or a non-targeting control (NT). 4 days after transduction, cells were treated with DMSO (0.1%) or Nutlin-3a (10 µM) for 48 hours before they were subjected to p53 flow cytometry. Values denote p53 MFI normalized to DMSO-treated cells transduced with the NT. (G) Rescue of the TRRAP-silencing–mediated mutp53 degradation by CRISPR-Cas9–mediated knockout (KO) of MDM2. KO cells were transduced with a shRNA against TRRAP or a nontargeting control (NT) and subjected to p53 flow cytometry 4 days after transduction. Values denote p53 MFI normalized to control (GFP) KO cells transduced with the NT. KOs were generated by transduction of Namalwa cells with constructs harboring both Cas9 and sgRNAs against MDM2 or GFP (negative control).
TRRAP silencing destabilizes mutp53 via the MDM2-proteasome axis. (A) Experimental design. Namalwa cells were transduced with an IPTG-inducible shRNA against TRRAP (shRNA #233) or a nontargeting control (NT). 48 to 96 hours after induction, cells were lysed and either collected immediately (total protein) or subjected to immunoprecipitation (IP) with antibodies against p53 or IgG (negative control) before mass spectrometry. The experiment was performed in biological triplicates. (B) Volcano plot visualizing differential total protein expression 96 hours after TRRAP knockdown compared with a nontargeting control (NT). Proteins denoted in black were significantly differentially expressed (adjusted P < .05, |log2FC| > 0.5). p53 is highlighted. P values were calculated using empirical Bayes statistics on protein-wise linear models. (C) Gene set enrichment analysis of differentially expressed proteins from panel B. Upregulated gene sets are denoted in red, downregulated in blue. Analysis was performed using PAGE with the MSigDB gene set collection “Hallmark.” Only significantly altered gene sets are shown (adjusted P < .05). (D) Heatmap of mutp53-bound proteins (proteins significantly enriched in p53 IPs compared with IgG control IPs). Values denote hierarchically clustered log2 fold changes of protein abundances in TRRAP-silenced samples compared with control knockdown (NT) samples. The cellular localization44 and known p53 interactions42 are indicated. Numbers within the clusters indicate the number of proteins. TP53, EIF3F, and XPO1 are highlighted. (E) Rescue of the TRRAP-silencing–mediated mutp53 degradation by treatment with nuclear export inhibitors. Namalwa cells were transduced with an IPTG-inducible shRNA against TRRAP (shRNA #233) or a nontargeting control (NT). 48 hours after induction, cells were treated with DMSO (0.005%), leptomycin B (20 nM), selinexor (500 nM), or verdinexor (500 nM) for 14 hours before they were subjected to p53 flow cytometry. Values denote p53 MFI normalized to DMSO-treated cells transduced with the NT. (F) Rescue of the TRRAP-silencing–mediated mutp53 degradation by treatment with the MDM2 inhibitor Nutlin-3a. Namalwa cells were transduced with a shRNA against TRRAP or a non-targeting control (NT). 4 days after transduction, cells were treated with DMSO (0.1%) or Nutlin-3a (10 µM) for 48 hours before they were subjected to p53 flow cytometry. Values denote p53 MFI normalized to DMSO-treated cells transduced with the NT. (G) Rescue of the TRRAP-silencing–mediated mutp53 degradation by CRISPR-Cas9–mediated knockout (KO) of MDM2. KO cells were transduced with a shRNA against TRRAP or a nontargeting control (NT) and subjected to p53 flow cytometry 4 days after transduction. Values denote p53 MFI normalized to control (GFP) KO cells transduced with the NT. KOs were generated by transduction of Namalwa cells with constructs harboring both Cas9 and sgRNAs against MDM2 or GFP (negative control).
Next, we profiled the mutp53 interactome after TRRAP silencing (Figure 4A,D). Overall, mutp53-bound proteins were enriched for known p53 interactors (4/27, 14.8%, P = .034).42 For example, the histone-arginine methyltransferase CARM1 has been shown to physically interact with wtp53 and to regulate p53-dependent transcription via histone methylation.43 TRRAP knockdown induced major alterations in mutp53-interacting proteins; 8 mutp53 interactions were lost and 19 interactions were gained upon TRRAP silencing (Figure 4D). Remarkably, while the great majority of proteins whose interactions were lost upon TRRAP knockdown were nuclear (7/8 [87.5%]), almost all gained interactions involved cytosolic proteins (18/19 [94.7%]).44 Specifically, exportin 1 (XPO1/CRM1), the key facilitator of protein export from the nucleus to the cytosol, was recruited to mutp53 after TRRAP silencing. We validated the interaction of mutp53 with XPO1 and EIF3F, a cytosolic eIF3 complex member,45 upon TRRAP silencing via coimmunoprecipitation (supplemental Figure 8B).
We hypothesized that a subset of mutp53 was exported from the nucleus to the cytosol following TRRAP knockdown and that export might be associated with mutp53 degradation. To test this, we silenced TRRAP and treated cells with the XPO1 inhibitors leptomycin B, selinexor, or verdinexor and quantified mutp53 levels (Figure 4E). Treatment with the inhibitors mildly upregulated mutp53 levels (1.44- to 1.79-fold) in control samples. Importantly, the reduction in mutp53 levels upon TRRAP silencing was rescued by cotreatment with each nuclear export inhibitor. This indicates that nuclear export is required to mediate the effect of TRRAP silencing on mutp53 levels.
The nuclear export and consequent proteasomal degradation of mutp53 observed upon TRRAP silencing alluded to the involvement of MDM2. To test this, we silenced TRRAP in the context of the MDM2 inhibitor Nutlin-3a and quantified mutp53 levels. Indeed, the reduction in mutp53 levels upon TRRAP silencing was rescued by cotreatment with Nutlin-3a (Figure 4F). To further confirm the involvement of MDM2, we generated MDM2 knockout cells using CRISPR-Cas9 (supplemental Figure 8C) and found that shTRRAP-mediated mutp53 degradation was almost completely abrogated in these cells (Figure 4G). Notably, we observed no consistent changes in MDM2 mRNA and protein expression after TRRAP silencing (supplemental Figure 8D-E), indicating that mutp53 destabilization occurred independent of changes in MDM2 levels.
In summary, these data show that TRRAP silencing targets mutp53 to the MDM2-dependent p53 degradation machinery.
TRRAP silencing attenuates stabilization and activity of wtp53 upon genotoxic stress
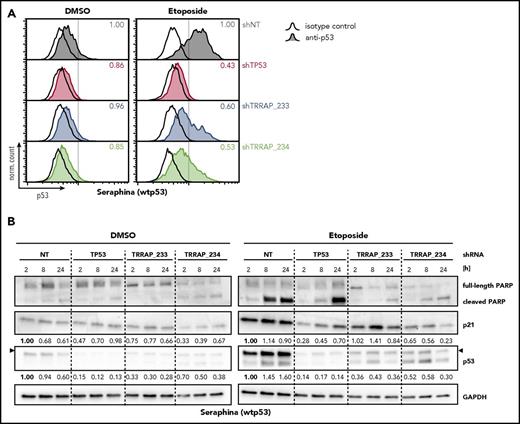
The mechanisms driving mutp53 stabilization are believed to mirror, to a large extent, the mechanisms responsible for wtp53 activation in response to stress. To explore the function of TRRAP in wtp53 stabilization upon genotoxic stress, we treated the wtp53 BL cell line Seraphina with the DNA-damaging agent etoposide and quantified p53 levels in the context of TRRAP silencing (Figure 5A). Etoposide treatment strongly stabilized wtp53, which was decreased upon TRRAP knockdown. To examine whether this also led to attenuated wtp53 activity, we monitored expression of p21 and PARP cleavage. As expected, etoposide treatment induced wtp53 activity in control cells, as reflected by increased p53 and p21 levels and PARP cleavage (Figure 5B). In line with the flow cytometry results, p53 induction was attenuated in TRRAP-silenced cells. One TRRAP shRNA impaired p21 induction, whereas both shRNAs suppressed PARP cleavage, consistent with diminished proapoptotic activity of p53. This suggests that TRRAP also regulates the stability and activity of wtp53 upon exposure to genotoxic stress.
TRRAP silencing attenuates stabilization and activity of wtp53 upon genotoxic stress. (A) Seraphina wtp53 BL cells were transduced with shRNAs against TRRAP, p53, or a nontargeting control (NT). 5 days after transduction, cells were treated with DMSO (0.1%) or etoposide (25 µM) for 8 hours before they were subjected to p53 flow cytometry. Values denote p53 MFI normalized to cells transduced with the NT. Hollow histograms indicate isotype control stainings, and filled histograms indicate samples stained with anti-p53. (B) Protein level of p53, p21, and PARP in Seraphina wtp53 BL cells transduced with shRNAs against TRRAP, p53, or a nontargeting control (NT). Cells were selected with puromycin for 72 hours. 6 days after transduction, cells were treated with DMSO (0.1%) or etoposide (25 µM) and harvested at the indicated time points. Expression was determined by western blot and normalized to GAPDH and to cells transduced with the NT. Arrows indicate the specific bands for p53.
TRRAP silencing attenuates stabilization and activity of wtp53 upon genotoxic stress. (A) Seraphina wtp53 BL cells were transduced with shRNAs against TRRAP, p53, or a nontargeting control (NT). 5 days after transduction, cells were treated with DMSO (0.1%) or etoposide (25 µM) for 8 hours before they were subjected to p53 flow cytometry. Values denote p53 MFI normalized to cells transduced with the NT. Hollow histograms indicate isotype control stainings, and filled histograms indicate samples stained with anti-p53. (B) Protein level of p53, p21, and PARP in Seraphina wtp53 BL cells transduced with shRNAs against TRRAP, p53, or a nontargeting control (NT). Cells were selected with puromycin for 72 hours. 6 days after transduction, cells were treated with DMSO (0.1%) or etoposide (25 µM) and harvested at the indicated time points. Expression was determined by western blot and normalized to GAPDH and to cells transduced with the NT. Arrows indicate the specific bands for p53.
HDAC inhibition decreases p53 levels similarly to TRRAP silencing
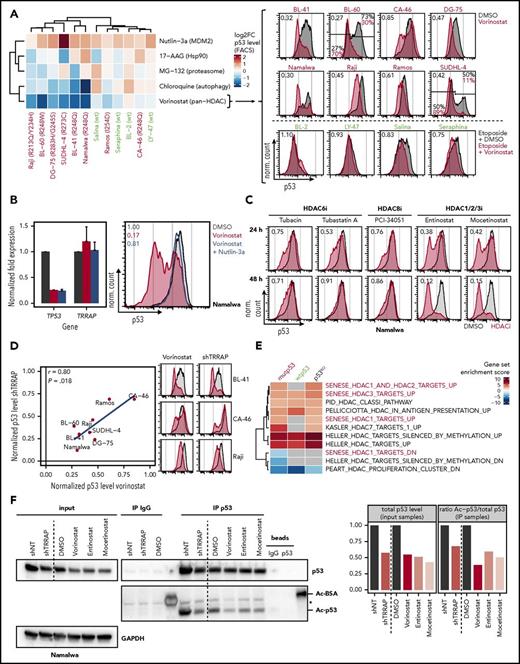
To identify small-molecule inhibitors able to modulate p53 levels similarly to TRRAP knockdown, we quantified p53 levels in a panel of 12 lymphoma cell lines treated with various inhibitors (wtp53 cells were additionally exposed to etoposide to stabilize p53) (Figure 6A). The pan-HDAC inhibitor vorinostat depleted mutp53 across all cell lines in a dose-dependent manner (Figure 6A; supplemental Figure 9A).46-49 This was confirmed using the pan-HDAC inhibitor panobinostat (supplemental Figure 9B) and in a subset of primary lymphoma samples (supplemental Figure 9C). In addition, vorinostat mildly interfered with induction of wtp53 upon genotoxic stress (Figure 6A). This suggests that the activity of HDACs is important for the accumulation of both mutp53 and wtp53 proteins.
HDAC inhibition decreases p53 levels similarly to TRRAP silencing. (A) Impact of different small-molecule inhibitors on p53 levels. Lymphoma cell lines were treated with 0.1% DMSO or the indicated inhibitors. 24 hours after treatment, cells were subjected to p53 flow cytometry. wtp53 cell lines were additionally treated with 25 µM etoposide for 5 hours before harvest to stabilize p53. The p53 mutation status is specified for each cell line (green, wtp53; red, mutp53). Values in heatmap denote log2 fold changes of p53 MFI normalized to DMSO-treated or etoposide-treated cells, respectively. Values in fluorescence-activated cell sorter (FACS) plots denote the ratio of the p53 MFI between vorinostat-treated (red) and DMSO-treated cells (gray). For cell lines with a bimodal p53 level, gates and the corresponding percentages for the p53-low and p53-high population are indicated (10 µM Nutlin-3a, MDM2 inhibitor; 5 µM 17-AAG, Hsp90 inhibitor; 5 µM MG-132, proteasome inhibitor; 10 µM chloroquine, autophagy inhibitor; 5 µM vorinostat, HDAC inhibitor). (B) Effect of vorinostat on mutp53 expression and evaluation of MDM2-dependency. Namalwa cells were treated with DMSO (0.1%, gray), vorinostat (5 µM, red), or with a combination of vorinostat and Nutlin-3a (10 µM, blue) for 24 hours before they were harvested. mRNA expression values (mean + SD, n = 2) determined by qRT-PCR and normalized to GAPDH and to DMSO-treated cells (left). p53 flow cytometry (right). Values denote p53 MFI normalized to DMSO-treated cells. (C) p53 flow cytometry of Namalwa cells after treatment with 0.1% DMSO (gray) or different HDAC inhibitors (red). The target proteins of the different inhibitors are indicated. Values denote p53 MFI normalized to DMSO-treated cells (7.5 µM tubacin; 7.5 µM tubastatin A; 10 µM PCI-34051; 500 nM entinostat; 500 nM mocetinostat). (D) Correlation of the impact of TRRAP silencing (8 days after transduction) and Vorinostat treatment (5 µM, 24 hours) on mutp53 levels determined by flow cytometry. Values denote p53 MFIs normalized to either untransduced or DMSO-treated cells, respectively. FACS plots in parts reproduced from panel A. The Pearson correlation coefficient (r) and the corresponding P value are indicated. (E) Gene set enrichment analysis of differentially expressed genes after TRRAP knockdown (shRNA #233) in mutp53 Namalwa, wtp53 Seraphina, and p53KO Seraphina cells (compared with nontargeting control). Cells were selected with puromycin for 48 hours and harvested 6 to 8 days after transduction. Analysis was performed using PAGE with the MSigDB gene set collection “c2” filtered for the term “HDAC.” Only gene sets that were significantly altered in at least 1 cell line are shown (adjusted P < .05), and nonsignificant enrichments are shown in gray. HDAC1/2/3-related pathways are highlighted.50 (F) Impact of HDAC inhibition and TRRAP silencing on mutp53 acetylation. Namalwa cells were treated with DMSO (0.05%) or different HDAC inhibitors before they were subjected to p53 or control (IgG) immunoprecipitation (IP). Silencing experiments were performed using Namalwa cells with IPTG-inducible shRNAs against TRRAP (shRNA #233) or a nontargeting control (NT). Cells were harvested 18 hours after drug exposure or 4 days after shRNA induction and were treated with the deacetylase inhibitors trichostatin A (1 µM) and nicotinamide (5 mM) for the last 4 hours of culture to enrich for acetylated proteins. Protein expression was determined by western blot and normalized to GAPDH (input samples). Acetylation (Ac) of p53 was detected using an antiacetylated lysine antibody and normalized to total p53 (IP samples) and to cells transduced with the NT or to DMSO-treated cells, respectively. Asterisk indicates an unspecific band. Ac-BSA, acetylated bovine serum albumin, positive control; vorinostat, 1 μM; entinostat, 500 nM; mocetinostat, 500 nM.
HDAC inhibition decreases p53 levels similarly to TRRAP silencing. (A) Impact of different small-molecule inhibitors on p53 levels. Lymphoma cell lines were treated with 0.1% DMSO or the indicated inhibitors. 24 hours after treatment, cells were subjected to p53 flow cytometry. wtp53 cell lines were additionally treated with 25 µM etoposide for 5 hours before harvest to stabilize p53. The p53 mutation status is specified for each cell line (green, wtp53; red, mutp53). Values in heatmap denote log2 fold changes of p53 MFI normalized to DMSO-treated or etoposide-treated cells, respectively. Values in fluorescence-activated cell sorter (FACS) plots denote the ratio of the p53 MFI between vorinostat-treated (red) and DMSO-treated cells (gray). For cell lines with a bimodal p53 level, gates and the corresponding percentages for the p53-low and p53-high population are indicated (10 µM Nutlin-3a, MDM2 inhibitor; 5 µM 17-AAG, Hsp90 inhibitor; 5 µM MG-132, proteasome inhibitor; 10 µM chloroquine, autophagy inhibitor; 5 µM vorinostat, HDAC inhibitor). (B) Effect of vorinostat on mutp53 expression and evaluation of MDM2-dependency. Namalwa cells were treated with DMSO (0.1%, gray), vorinostat (5 µM, red), or with a combination of vorinostat and Nutlin-3a (10 µM, blue) for 24 hours before they were harvested. mRNA expression values (mean + SD, n = 2) determined by qRT-PCR and normalized to GAPDH and to DMSO-treated cells (left). p53 flow cytometry (right). Values denote p53 MFI normalized to DMSO-treated cells. (C) p53 flow cytometry of Namalwa cells after treatment with 0.1% DMSO (gray) or different HDAC inhibitors (red). The target proteins of the different inhibitors are indicated. Values denote p53 MFI normalized to DMSO-treated cells (7.5 µM tubacin; 7.5 µM tubastatin A; 10 µM PCI-34051; 500 nM entinostat; 500 nM mocetinostat). (D) Correlation of the impact of TRRAP silencing (8 days after transduction) and Vorinostat treatment (5 µM, 24 hours) on mutp53 levels determined by flow cytometry. Values denote p53 MFIs normalized to either untransduced or DMSO-treated cells, respectively. FACS plots in parts reproduced from panel A. The Pearson correlation coefficient (r) and the corresponding P value are indicated. (E) Gene set enrichment analysis of differentially expressed genes after TRRAP knockdown (shRNA #233) in mutp53 Namalwa, wtp53 Seraphina, and p53KO Seraphina cells (compared with nontargeting control). Cells were selected with puromycin for 48 hours and harvested 6 to 8 days after transduction. Analysis was performed using PAGE with the MSigDB gene set collection “c2” filtered for the term “HDAC.” Only gene sets that were significantly altered in at least 1 cell line are shown (adjusted P < .05), and nonsignificant enrichments are shown in gray. HDAC1/2/3-related pathways are highlighted.50 (F) Impact of HDAC inhibition and TRRAP silencing on mutp53 acetylation. Namalwa cells were treated with DMSO (0.05%) or different HDAC inhibitors before they were subjected to p53 or control (IgG) immunoprecipitation (IP). Silencing experiments were performed using Namalwa cells with IPTG-inducible shRNAs against TRRAP (shRNA #233) or a nontargeting control (NT). Cells were harvested 18 hours after drug exposure or 4 days after shRNA induction and were treated with the deacetylase inhibitors trichostatin A (1 µM) and nicotinamide (5 mM) for the last 4 hours of culture to enrich for acetylated proteins. Protein expression was determined by western blot and normalized to GAPDH (input samples). Acetylation (Ac) of p53 was detected using an antiacetylated lysine antibody and normalized to total p53 (IP samples) and to cells transduced with the NT or to DMSO-treated cells, respectively. Asterisk indicates an unspecific band. Ac-BSA, acetylated bovine serum albumin, positive control; vorinostat, 1 μM; entinostat, 500 nM; mocetinostat, 500 nM.
To address whether the vorinostat-mediated effect on mutp53 was dependent on MDM2, we treated cells with either vorinostat alone or in combination with Nutlin-3a, and quantified p53 mRNA and protein expression (Figure 6B). TP53 mRNA expression was strongly reduced after Vorinostat treatment48,49 and not rescued with Nutlin-3a treatment, suggesting a MDM2-independent effect on TP53 mRNA transcription or stability. In contrast, vorinostat-mediated depletion of mutp53 protein was MDM2-dependent, since Nutlin-3a treatment augmented mutp53 protein abundance following vorinostat treatment.46,47 This indicates that although HDAC inhibition regulates mutp53 both transcriptionally and posttranscriptionally, its inhibitory effect on mutp53 protein levels is achieved primarily via posttranslational mechanisms. Notably, HDAC inhibition did not decrease mutp53 levels by downregulating TRRAP (Figure 6B).
Because vorinostat is a broad range HDAC inhibitor, we tested subgroup-specific HDAC inhibitors for their effect on mutp53 levels (Figure 6C). While HDAC6 inhibition (tubacin, tubastatin A) and HDAC8 inhibition (PCI-34051) reduced mutp53 only mildly (53% to 91% of the basal level), HDAC1/2/3 inhibition (entinostat and mocetinostat) strongly depleted mutp53 (12% to 42%), indicating that these HDACs selectively control mutp53 stability. In primary lymphoma samples, we observed a heterogeneous effect of HDAC1/2/3 inhibition on mutp53 levels (supplemental Figure 9C).
When we compared the mutp53 phenotypes after HDAC inhibition and TRRAP silencing (Figure 6D), we observed a striking similarity of response (r = 0.80, P = .018); ie, cell lines responded either strongly (eg, BL-41 or Raji) or weakly (eg, CA-46) to both perturbations, which suggests that both converged on similar mechanisms. To evaluate whether TRRAP silencing and HDAC inhibition could target similar genes transcriptionally, we performed gene expression profiling after TRRAP knockdown in mutp53, wtp53, and p53KO BL cells, specifically focusing on gene sets regulated by HDACs (Figure 6E). Indeed, several HDAC-related gene sets were significantly altered upon TRRAP silencing across cell lines, indicating that this function of TRRAP is independent of p53 status. TRRAP knockdown deregulated 4 gene sets composed of genes altered upon HDAC1/2/3 knockdown50 in mutp53 cells, which supports our finding that HDAC1/2/3 inhibition had the strongest impact on mutp53 levels.
Based on these observations, we investigated whether TRRAP silencing or HDAC inhibition altered mutp53 acetylation. Because p53 can be acetylated on multiple lysine residues,51 we immunoprecipitated mutp53 and quantified acetylation by western blot using an antiacetylated lysine antibody (Figure 6F). Basal acetylation of mutp53 was reduced to ∼70% upon TRRAP silencing, and HDAC inhibition reduced mutp53 acetylation to a similar extent (∼40% to 60%). This suggests that mutp53 acetylation may contribute to its stability and that TRRAP silencing and HDAC1/2/3 inhibitors putatively regulate mutp53 levels by functional convergence.
Altogether, our results indicate that acetylation-modifying complexes, including TRRAP-containing HAT complexes and HDAC1/2/3, participate in regulation of p53 protein levels.
Discussion
Excessive mutp53 protein in tumors is at the root of mutp53 GOF properties and contributes to tumorigenesis. Targeting mutp53 accumulation may represent a tumor-specific strategy for therapy. However, our understanding of the mechanisms through which mutp53 is stabilized in cancer cells is presently still limited. We conducted an RNAi screen to identify regulators of mutp53 protein levels, which revealed a central role of the HAT complex and PIKK family member TRRAP in mediating mutp53 stabilization, by protecting mutp53 from the natural p53 degradation machinery. Therefore, our data support the idea that MDM2 retains the capacity to degrade mutp53.14,15 The specific role of MDM2 in mutp53 degradation is, however, still under debate; it is not limited to ubiquitination,52 but may also include other effects, such as facilitating access to the proteasome.53
mutp53 is intrinsically unstable and requires additional oncogenic events, such as chronic DNA damage, to be stabilized.8-12 Because TRRAP silencing also impaired wtp53 accumulation upon genotoxic stress, TRRAP may be a physiological positive regulator of p53 stability in stressed conditions. This may explain our finding that TRRAP regulates mutp53 accumulation independently of p53 mutation sequence and cancer entity. Overall, our results support the notion that mutp53 accumulation in tumors is not achieved by proteins that specifically gain a mutp53-stabilizing function but rather by hijacking the physiological machinery of stress-induced wtp53 activation.
Several lines of evidence support a specific link between TRRAP and p53 regulation and function. First, other members of the PIKK family (ATM,54,55 ATR,56,57 DNA-PKcs,58,59 SMG1,60,61 and mTOR62,63 ) are either direct regulators of p53 or are under the control of p53. Secondly, TRRAP has been reported to bind to wtp53 and to be essential for wtp53-dependent MDM2 expression.64 Thirdly, 3 of the 4 human TRRAP-containing HAT complexes (PCAF,65,66 TIP60,67,68 and STAGA69 ) have been reported to regulate p53, although TRRAP’s specific function within these complexes remains elusive.33
We identified a ∼100-amino-acid stretch mapping to the N-clasp and ring in TRRAP’s N-terminal HEAT repeat region, which was essential for cell proliferation and mutp53 stabilization. The clasps are important for closing the ring structure of TRRAP,37 suggesting that targeting of this region may have a detrimental impact on TRRAP’s overall structure. In yeast, this region is among the ones mediating binding to the HAT complexes SAGA (human: STAGA) and NuA4 (human: TIP60).70 Notably, this region is also in proximity to the hotspot GOF S722F TRRAP mutation in melanoma.71
Therapeutic targeting of TRRAP in cancers harboring TP53 mutations is appealing. Given the fact that we found no evidence either for preferential toxicity of TRRAP silencing in mutp53 cells or for a specific domain mediating mutp53 regulation, direct targeting may be challenging. However, our finding that HDAC inhibition may be used to target mutp53 levels is of importance, because we found this drug class to resemble TRRAP silencing in terms of p53 regulation. Notably, 2 studies found evidence for preferential toxicity of pan-HDAC inhibition in mutp53 cancer cells, as compared with wtp53 cancer cells.46,47 In line with our results, HDAC inhibition has been shown to downregulate mutp53 levels by diverse mechanisms: while HDAC6 facilitates mutp53 stabilization via activation of Hsp90,13 HDAC1, 2, and 8 are crucial for maintaining TP53 transcription.48,49
The surprising finding that silencing of the HAT complex member TRRAP and HDAC inhibition both converge on decreasing mutp53 acetylation argues for an indirect mode of action. In general, our data are consistent with studies that have linked p53 deacetylation to its destabilization.72 Because the impact of acetylation on mutp53 stabilization remains incompletely understood,16 this warrants further studies. In particular, it remains unclear whether deacetylation is the signal which initiates the degradation cascade.
In summary, our study provides a strong link between acetylation-modifying complexes and regulation of p53 protein levels, which may be exploitable in cancer therapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the High Throughput Sequencing unit and the Microarray unit of the German Cancer Research Center Genomics & Proteomics Core Facility, especially Sabine Schmidt, for technical support and expertise. The authors also thank Ina Oehme and Olaf Witt for providing HDAC inhibitors, Shihui Wang for preparation of cell lysates, and Georg Stoecklin for help with revision of the manuscript. The lentiviral shRNA library was kindly provided and developed by Cellecta based on National Institutes of Health–funded research (grants 44RR024095 [National Center for Research Resources] and 44HG003355 [National Human Genome Research Institute]).
This work was supported by the German-Israeli Foundation for Scientific Research and Development (grant 1206-257.13/2012), the German-Israeli Helmholtz Research School in Cancer Biology, and the European Union (Horizon 2020 project SOUND). T.Z. was supported by the Deutsche Krebshilfe (Mildred-Scheel Professorship) and the Monique Dornonville de la Cour Foundation.
Authorship
Contribution: A.J. designed and performed research, analyzed and interpreted data, and wrote the manuscript; M.S. designed and performed research and interpreted data; J.H., M.J., V.D., S.R., L.W., T.W., and H.B. performed research; W.K. performed and analyzed tissue microarray; M.R. performed mass spectrometry experiments; J.L. contributed to data analysis; A.H.S. and F.S. analyzed mass spectrometry data; M.M.S. supervised mass spectrometry experiments; W.H. contributed to data analysis and interpretation; Y.A. designed research and contributed to data interpretation; M.O. and T.Z. designed research, interpreted data, supervised the work, and wrote the manuscript; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of the members of the MMML Network Project appears in “Appendix.”
Correspondence: Thorsten Zenz, Department of Hematology, University Hospital and University of Zürich, Rämistr 100, 8091 Zürich, Switzerland; e-mail: thorsten.zenz@usz.ch.
Appendix: MMML Network Project members
The members of the Molecular Mechanisms in Malignant Lymphomas (MMML) Network Project of the Deutsche Krebshilfe are (in alphabetical order within groups): Thomas F. E. Barth (Institute of Pathology, University Hospital of Ulm, Germany), Heinz-Wolfram Bernd (Institute of Pathology, University Hospital Schleswig-Holstein Campus Lübeck, Germany), Sergio B. Cogliatti (Institute of Pathology, Kantonsspital St. Gallen, Switzerland), Alfred C. Feller (Institute of Pathology, University Hospital Schleswig-Holstein Campus Lübeck, Germany), Martin L. Hansmann (Institute of Pathology, University Hospital of Frankfurt, Germany), Michael Hummel (Institute of Pathology, Campus Benjamin Franklin, Charité Universitätsmedizin Berlin, Germany), Wolfram Klapper (Department of Pathology, Hematopathology Section, University Hospital Schleswig-Holstein Campus Kiel/Christian-Albrechts University Kiel, Germany), Dido Lenze (Institute of Pathology, Campus Benjamin Franklin, Charité Universitätsmedizin Berlin, Germany), Peter Möller (Institute of Pathology, University Hospital of Ulm, Germany), Hans-Konrad Müller-Hermelink (Institute of Pathology, University of Würzburg, Germany), Ilske Oschlies (Department of Pathology, Hematopathology Section, University Hospital Schleswig-Holstein Campus Kiel/Christian-Albrechts University Kiel, Germany), German Ott (Institute of Clinical Pathology, Robert Bosch Krankenhaus, Stuttgart, Germany), Andreas Rosenwald (Institute of Pathology, University of Würzburg, Germany), Harald Stein (Institute of Pathology, Campus Benjamin Franklin, Charité Universitätsmedizin Berlin, Germany), and Monika Szczepanowski (Department of Pathology, Hematopathology Section, University Hospital Schleswig-Holstein Campus Kiel/Christian-Albrechts University Kiel, Germany) (pathology group); Thomas F. E. Barth (Institute of Pathology, University Hospital of Ulm, Germany), Petra Behrmann (University Medical Center Hamburg-Eppendorf, Hamburg, Germany), Peter Daniel (Department of Hematology, Oncology and Tumor Immunology, University Medical Center Charité, Germany), Judith Dierlamm (University Medical Center Hamburg-Eppendorf, Hamburg, Germany), Stefan Gesk (Institute of Human Genetics, University Hospital Schleswig-Holstein Campus Kiel/Christian-Albrechts University Kiel, Germany), Eugenia Haralambieva (Institute of Pathology, University of Würzburg, Germany), Lana Harder (Institute of Human Genetics, University Hospital Schleswig-Holstein Campus Kiel/Christian-Albrechts University Kiel, Germany), Paul-Martin Holterhus (Division of Pediatric Endocrinology and Diabetes, Department of Pediatrics, University Hospital Schleswig-Holstein Campus Kiel/Christian-Albrechts University Kiel, Germany), Ralf Küppers (Institute for Cell Biology [Tumor Research], University of Duisburg-Essen, Germany), Dieter Kube (Department of Hematology and Oncology, Georg-August University of Göttingen, Germany), Peter Lichter (German Cancer Research Center, Heidelberg, Germany), Jose I. Martín-Subero (Institute of Human Genetics, University Hospital Schleswig-Holstein Campus Kiel/Christian-Albrechts University Kiel, Germany), Peter Möller (Institute of Pathology, University Hospital of Ulm, Germany), Eva M. Murga-Peñas (University Medical Center Hamburg-Eppendorf, Hamburg, Germany), German Ott (Institute of Clinical Pathology, Robert Bosch Krankenhaus, Stuttgart, Germany), Shoji Pellissery (Institute of Human Genetics, University Hospital Schleswig-Holstein Campus Kiel/Christian-Albrechts University Kiel, Germany), Claudia Philipp (Institute for Cell Biology [Tumor Research], University of Duisburg-Essen, Germany), Christiane Pott (Second Medical Department, University Hospital Schleswig-Holstein Campus Kiel/Christian-Albrechts University Kiel, Germany), Armin Pscherer (German Cancer Research Center, Heidelberg, Germany), Julia Richter (Institute of Human Genetics, University Hospital Schleswig-Holstein Campus Kiel/Christian-Albrechts University Kiel, Germany), Andreas Rosenwald (Institute of Pathology, University of Würzburg, Germany), Itziar Salaverria (Institute of Human Genetics, University Hospital Schleswig-Holstein Campus Kiel/Christian-Albrechts University Kiel, Germany), Carsten Schwaenen (Cytogenetic and Molecular Diagnostics, Internal Medicine III, University Hospital of Ulm, Germany), Reiner Siebert (Institute of Human Genetics, University Hospital Schleswig-Holstein Campus Kiel/Christian-Albrechts University Kiel, Germany), Heiko Trautmann (Second Medical Department, University Hospital Schleswig-Holstein Campus Kiel/Christian-Albrechts University Kiel, Germany), Martina Vockerodt (Department of Pediatrics I, Georg-August University of Göttingen, Germany), and Swen Wessendorf (Cytogenetic and Molecular Diagnostics, Internal Medicine III, University Hospital of Ulm, Germany) (genetics group); Stefan Bentink (Institute of Functional Genomics, University of Regensburg, Germany), Hilmar Berger (Institute for Medical Informatics, Statistics and Epidemiology, University of Leipzig, Germany), Christian W. Kohler (Institute of Functional Genomics, University of Regensburg, Germany), Dirk Hasenclever (Institute for Medical Informatics, Statistics and Epidemiology, University of Leipzig, Germany), Markus Kreuz (Institute for Medical Informatics, Statistics and Epidemiology, University of Leipzig, Germany), Markus Loeffler (Institute for Medical Informatics, Statistics and Epidemiology, University of Leipzig, Germany), Maciej Rosolowski (Institute for Medical Informatics, Statistics and Epidemiology, University of Leipzig, Germany), and Rainer Spang (Institute of Functional Genomics, University of Regensburg, Germany) (bioinformatics group); Birgit Burkhardt (NHL-BFM Study Center Department of Pediatric Hematology and Oncology, Justus-Liebig University, Giessen, Germany), Alfred Reiter (NHL-BFM Study Center Department of Pediatric Hematology and Oncology, Justus-Liebig University, Giessen, Germany), and Willhelm Woessmann (NHL-BFM Study Center Department of Pediatric Hematology and Oncology, Justus-Liebig University, Giessen, Germany) (clinical group for pediatric lymphoma); Benjamin Stürzenhofecker (Department of Hematology and Oncology, Georg-August University of Göttingen, Germany), Lorenz Trümper (Department of Hematology and Oncology, Georg-August University of Göttingen, Germany), and Maren Wehner (Department of Hematology and Oncology, Georg-August University of Göttingen, Germany) (project coordination); and Markus Loeffler (Institute for Medical Informatics, Statistics and Epidemiology, University of Leipzig, Germany), Reiner Siebert (Institute of Human Genetics, University Hospital Schleswig-Holstein Campus Kiel/Christian-Albrechts University Kiel, Germany), Harald Stein (Institute of Pathology, Campus Benjamin Franklin, Charité Universitätsmedizin Berlin, Germany), and Lorenz Trümper (Department of Hematology and Oncology, Georg-August University of Göttingen, Germany) (steering committee).

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal