Abstract
Transformation to aggressive lymphoma is a critical event in the clinical course of follicular lymphoma (FL) patients. Yet, it is a challenge to reliably predict transformation at the time of diagnosis. Understanding the risk of transformation would be useful for guiding and monitoring patients, as well as for evaluating novel treatment strategies that could potentially prevent transformation. Herein, we review the contribution of clinical, pathological, and genetic risk factors to transformation. Patients with multiple clinical high-risk factors are at elevated risk of transformation but we are currently lacking a prognostic index that would specifically address transformation rather than disease progression or overall survival. From the biological standpoint, multiple studies have correlated individual biomarkers with transformation. However, accurate prediction of this event is currently hampered by our limited knowledge of the evolutionary pathways leading to transformation, as well as the scarcity of comprehensive, large-scale studies that assess both the genomic landscape of alterations within tumor cells and the composition of the microenvironment. Liquid biopsies hold great promise for achieving precision medicine. Indeed, mutations detected within circulating tumor DNA may be a better reflection of the inherent intratumoral heterogeneity than the biopsy of a single site. Last, we will assess whether evidence exists in the literature that transformation might be prevented altogether, based on the choice of therapy for FL.
Medscape Continuing Medical Education online

In support of improving patient care, this activity has been planned and implemented by Medscape, LLC and the American Society of Hematology. Medscape, LLC is jointly accredited by the Accreditation Council for Continuing Medical Education (ACCME), the Accreditation Council for Pharmacy Education (ACPE), and the American Nurses Credentialing Center (ANCC), to provide continuing education for the healthcare team.
Medscape, LLC designates this Journal-based CME activity for a maximum of 1.00 AMA PRA Category 1 Credit(s)™. Physicians should claim only the credit commensurate with the extent of their participation in the activity.
All other clinicians completing this activity will be issued a certificate of participation. To participate in this journal CME activity: (1) review the learning objectives and author disclosures; (2) study the education content; (3) take the post-test with a 75% minimum passing score and complete the evaluation at http://www.medscape.org/journal/blood; and (4) view/print certificate. For CME questions, see page 383.
Disclosures
Editor Nancy Berliner received grants for clinical research from Novartis. Laurie Barclay, freelance writer and reviewer, Medscape, LLC, owns stock, stock options, or bonds from Alnylam, Biogen, and Pfizer. Author Laurie H. Sehn served as an advisor or consultant for AbbVie, Amgen, Roche/Genentech, Celgene, Janssen, Lundbeck, Seattle Genetics, and TG Therapeutics and received grants for clinical research from Roche/Genentech. The remaining authors declare no competing financial interests.
Learning objectives
Upon completion of this activity, participants will be able to:
Identify clinical risk factors predicting transformation from follicular lymphoma to aggressive lymphoma, based on a review.
Identify pathologic and genetic risk factors predicting transformation from follicular lymphoma to aggressive lymphoma.
Evaluate novel strategies for prediction and potential prevention of transformation from follicular lymphoma to aggressive lymphoma.
Release date: July 20, 2017; Expiration date: July 20, 2018
Introduction
Follicular lymphoma (FL) is the most common indolent lymphoma,1,2 with an estimated incidence of 6 new cases per 100 000 persons per year.3,4 With the exception of limited-stage disease, FL is generally considered to be an incurable malignancy, despite the fact that patients typically survive for a prolonged time, with median overall survival times in excess of 18 years reported in the literature.5 Notwithstanding these favorable outlooks, the prudent clinician cannot be universally reassuring when counseling newly diagnosed FL patients, as a subset is at risk of early lymphoma-related mortality. Early progression, most commonly defined as progression within 2 years after starting rituximab-chemotherapy, occurs in ∼20% of patients and has recently been associated with poor outcome.6-9 Similarly, transformation to aggressive lymphoma, occurring in 2% to 3% of patients per year, has repeatedly been linked to adverse prognosis when compared with patients not experiencing transformation.10-15 There is an association between early progression and transformation. Indeed, in the experience from 2 regional cancer centers in Ontario, 27% of early progressors had either documented transformation or were suspected to harbor transformed disease based on clinical grounds.6 Furthermore, in a report from the University of Iowa/Mayo Clinic, transformation within 18 months after diagnosis was correlated with shortened survival when compared with transformation occurring at a later time point.13 Given these considerations, it is becoming increasingly relevant to accurately predict early progression and/or early transformation. This review will focus on prediction of transformation and will explore whether clinical intervention can alter this risk.
The pathogenesis of transformation from underlying FL
FL is composed of cells resembling normal germinal center B cells that thrive within and potentially outside of follicles, whereas aggressive lymphoma is represented by a diffuse proliferation of large cells effacing the follicular architecture.16 The most common histology at the time of transformation is diffuse large B-cell lymphoma, sometimes in the presence of residual low-grade histology (a situation referred to as composite lymphoma), followed by rarer instances of transformation to high-grade B-cell lymphoma (not otherwise specified or with double-hit/triple-hit genetics), or more rarely lymphoblastic lymphoma.16,17 Clinically, transformation is characterized by enlarging disproportionate masses, B symptoms, abnormal laboratory values such as elevated serum lactate dehydrogenase (LDH) or calcium, all of which may appear relatively rapidly (ie, over the course of days to weeks), reflecting an inflection point in the growth properties of the lymphoma. Correlates of an acceleration of proliferation kinetics can be found, for example, in increased expression of Ki67 within the tumor18 or high levels of maximum standardized uptake value (>12-14) on positron emission tomography scanning.19-23
On the molecular level, transformed FL (TFL) differs from preceding indolent FL by higher numbers of single-nucleotide mutations, small insertions and deletions, copy-number changes, and structural rearrangements.24-26 Transformation occurs via the activation of known or putative oncogenes (MYC, CCND3) and inactivation of known or putative tumor suppressor genes (TP53, CDKN2A/B, B2M).26-35 Recent genome-wide studies have contributed to expanding our knowledge on the breadth of genetic alterations that are associated with transformation.25,26,36,37 However, the complete description of the genomic architecture of causative molecular disruptions will require further, comprehensive, integrated, and large-scale studies, ideally complemented by functional genomic analyses. The vast majority of genetic aberrations that are typical of TFL can also be found at lower prevalence in indolent FL, suggesting that a single alteration may not be sufficient to drive transformation and that the additive effect of further biological disruption is largely dependent on the existing contextual molecular landscape. Moreover, it is unclear, at present, whether transformation occurs by determined sequences of successive genetic hits and/or whether certain alterations need to co-occur in order to confer an aggressive phenotype.
Clinical risk factors
Several retrospective studies have assessed the overall incidence of clinical risk factors of transformation (Table 1). The variables most commonly reported to predict transformation include altered performance status, advanced stage, low hemoglobin, elevated LDH, and high Follicular Lymphoma International Prognostic Index (FLIPI). The fact that these same factors are generally prognostic of overall survival in FL begs the conclusion that outcome in FL is either largely driven by transformation or that these factors perform poorly in terms of specifically predicting transformation. One of the most rigorous assessments of features associated with transformation comes from the PRIMA trial in which Sarkozy et al report progression in 463 patients of 1018 after a median follow-up of 6 years.15 Of these 463 cases, 194 patients had histologic confirmation of progression, with TFL and FL found in 40 and 154 instances, respectively. The authors compared the 40 cases with documented transformation to 708 cases with either no progression, or progression but without evidence of transformation. In a multivariate analysis, Eastern Cooperative Oncology Group (ECOG) performance status ≥2 and hemoglobin <12 g/dL were independently associated with transformation. The established prognostic value of clinical risk factors suggests that they will continue to play a role in refining prognosis for FL patients and potentially in predicting transformation, most likely as part of an integrated clinical and biological score.
Clinical risk factors for transformation reported in studies from the last 2 decades
| Reference . | No. of patients . | Incidence TFL . | Median survival after transformation, y . | PS 2-4 . | Stage III-IV . | Hgb low . | LDH high . | FLIPI . | Treatment . | Effect of treatment . |
|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 220 | 31% at 10 y | 0.6 | NS | NS | NR | NS | NR | Chemo | NR |
| 38 | 276 | 15% at 10 y | 1.2 | NR | NR | NR | Sig | Sig | Observation, chemo | No effect |
| 11 | 325 | 28% at 10 y | 1.2 | NR | Sig | NR | NR | Sig | Observation, radiation, chemo | Observation: increased risk |
| 12 | 600 | 30% at 10 y | 1.7 | NS | Sig | NS | NS | NS | BP-VACOP-RT, alkylator and purine analog | • Observation: no effect |
| • Alkylator with purine analog: increased risk | ||||||||||
| 39 | 281 | 15% at 10 y | 2.7 | NS | NS | NS | NS | NS | Observation, chemo, R, R-chemo | • R-chemo: increased risk |
| • R-monotherapy: reduced risk | ||||||||||
| • Chemo, observation: intermediate risk | ||||||||||
| 13 | 631 | 10.7% at 5 y | 4.2 | NS | NS | Sig | Sig | Sig | Observation, chemo, R, R-chemo | • Observation: increased risk |
| • R-monotherapy: reduced risk | ||||||||||
| 17 | 126 | NA | 3.9 | NS | NS | NS | Sig | NS | Observed, R or R-chemo | NR |
| 14 | 2652 | 14.3% at 6.8 y | 5.0 | Sig | NS | NS | Sig | NR | Observed, R, R-chemo, other | • Observation: increased risk |
| • R-CHOP vs R-CVP: no effect | ||||||||||
| 15 | 1018 | NA | 3.8 | Sig | NS | Sig | Sig | Sig | R-chemo ± R-maintenance | • R-maintenance: no effect |
| • R-CHOP vs R-CVP vs R-FCM: no effect (but small numbers) |
| Reference . | No. of patients . | Incidence TFL . | Median survival after transformation, y . | PS 2-4 . | Stage III-IV . | Hgb low . | LDH high . | FLIPI . | Treatment . | Effect of treatment . |
|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 220 | 31% at 10 y | 0.6 | NS | NS | NR | NS | NR | Chemo | NR |
| 38 | 276 | 15% at 10 y | 1.2 | NR | NR | NR | Sig | Sig | Observation, chemo | No effect |
| 11 | 325 | 28% at 10 y | 1.2 | NR | Sig | NR | NR | Sig | Observation, radiation, chemo | Observation: increased risk |
| 12 | 600 | 30% at 10 y | 1.7 | NS | Sig | NS | NS | NS | BP-VACOP-RT, alkylator and purine analog | • Observation: no effect |
| • Alkylator with purine analog: increased risk | ||||||||||
| 39 | 281 | 15% at 10 y | 2.7 | NS | NS | NS | NS | NS | Observation, chemo, R, R-chemo | • R-chemo: increased risk |
| • R-monotherapy: reduced risk | ||||||||||
| • Chemo, observation: intermediate risk | ||||||||||
| 13 | 631 | 10.7% at 5 y | 4.2 | NS | NS | Sig | Sig | Sig | Observation, chemo, R, R-chemo | • Observation: increased risk |
| • R-monotherapy: reduced risk | ||||||||||
| 17 | 126 | NA | 3.9 | NS | NS | NS | Sig | NS | Observed, R or R-chemo | NR |
| 14 | 2652 | 14.3% at 6.8 y | 5.0 | Sig | NS | NS | Sig | NR | Observed, R, R-chemo, other | • Observation: increased risk |
| • R-CHOP vs R-CVP: no effect | ||||||||||
| 15 | 1018 | NA | 3.8 | Sig | NS | Sig | Sig | Sig | R-chemo ± R-maintenance | • R-maintenance: no effect |
| • R-CHOP vs R-CVP vs R-FCM: no effect (but small numbers) |
BP-VACOP-RT, bleomycin, cisplatin, etoposide, doxorubicin, cyclophosphamide, vincristine, prednisone, radiation; Chemo, chemotherapy; Hgb, hemoglobin; NA, not applicable; NR, not reported; NS, not statistically significant; PS, performance status; R, rituximab; R-FCM, rituximab with fludarabine, cyclophosphamide, and mitoxantrone; Sig, statistically significant.
Pathological risk factors including the tumor microenvironment
Biological factors that have been reported to modulate risk of transformation are summarized in Table 2. As studies often contradict each other, with often the same feature associated with favorable risk in 1 study and unfavorable or no risk in another, we will only describe a selection of the most relevant and consistent potential biomarkers.
Biological risk factors for transformation reported in the literature
| Category . | Variable . | Reported risk of transformation . |
|---|---|---|
| Microanatomical structure | Disrupted CD21+ FDC meshworks | Increased40,41 |
| Decreased42 | ||
| Intrafollicular localization of CD14+ FDCs | Increased43 | |
| FL grade | Grade 3A | Increased17,38 |
| No effect39,40 | ||
| IRF4 tumor cell staining by IHC | Increased17 | |
| Tumor microenvironment | Predominantly intrafollicular localization of CD4+ T cells | Increased40 |
| High numbers of PD1+ cells | Decreased44 | |
| Diffuse pattern of PD1+ cells | Increased43 | |
| Higher FOXP3 expression | Increased42 | |
| No effect40 | ||
| Intra- or perifollicular distribution of FOXP3+ cells | Increased45 | |
| No effect40 | ||
| Increased vessel density | Increased46 | |
| Gene expression signatures | Embryonic stem cell–like signature | Increased47 |
| NF-κB target signature scores | Increased48 | |
| Germ line polymorphism | SNP rs6457327 | Increased49,50 |
| Large-scale genetic alterations | eg, Deletions of chromosomes 1p or 6q; gain of chromosomes 2, 3q, or 5 | Increased51-54 |
| Higher numbers of structural rearrangements | Increased24 | |
| Single gene alterations | TP53 mutations, deletions | Inconclusive55 |
| MYC translocations, mutations | Inconclusive | |
| FAS mutations | Increased26 | |
| BCL6 translocations | Increased17,56 | |
| BCL2 mutations | Increased57 | |
| Circulating tumor DNA | Proportion of mutations uniquely found in plasma | Increased58 |
| Category . | Variable . | Reported risk of transformation . |
|---|---|---|
| Microanatomical structure | Disrupted CD21+ FDC meshworks | Increased40,41 |
| Decreased42 | ||
| Intrafollicular localization of CD14+ FDCs | Increased43 | |
| FL grade | Grade 3A | Increased17,38 |
| No effect39,40 | ||
| IRF4 tumor cell staining by IHC | Increased17 | |
| Tumor microenvironment | Predominantly intrafollicular localization of CD4+ T cells | Increased40 |
| High numbers of PD1+ cells | Decreased44 | |
| Diffuse pattern of PD1+ cells | Increased43 | |
| Higher FOXP3 expression | Increased42 | |
| No effect40 | ||
| Intra- or perifollicular distribution of FOXP3+ cells | Increased45 | |
| No effect40 | ||
| Increased vessel density | Increased46 | |
| Gene expression signatures | Embryonic stem cell–like signature | Increased47 |
| NF-κB target signature scores | Increased48 | |
| Germ line polymorphism | SNP rs6457327 | Increased49,50 |
| Large-scale genetic alterations | eg, Deletions of chromosomes 1p or 6q; gain of chromosomes 2, 3q, or 5 | Increased51-54 |
| Higher numbers of structural rearrangements | Increased24 | |
| Single gene alterations | TP53 mutations, deletions | Inconclusive55 |
| MYC translocations, mutations | Inconclusive | |
| FAS mutations | Increased26 | |
| BCL6 translocations | Increased17,56 | |
| BCL2 mutations | Increased57 | |
| Circulating tumor DNA | Proportion of mutations uniquely found in plasma | Increased58 |
FDC, follicular dendritic cell; IHC, immunohistochemistry; SNP, single-nucleotide polymorphism.
With regards to pathological findings, grading of FL has variably been associated with transformation. FL can be designated as grade 1, 2, 3A, and 3B disease, based on the number of centroblasts per high-power field.59 Whereas grade 3B is typically considered to be more closely related to de novo aggressive lymphoma, grades 1 to 3A are considered in keeping with the morphological spectrum of FL. Grade 3A FL is not robustly associated with poorer survival, yet, somewhat paradoxically, has been linked to an increased incidence of transformation in some studies17,38 although not in others.39,40 Grade 3A/3B FL is often associated with other findings such as absent expression of CD10, the presence of a BCL6 translocation, the absence of the prototypical BCL2 translocation, and/or positive expression of IRF4.60,61 Immunohistochemical assessment of IRF4 protein expression deserves further attention. In a study from the BC Cancer Agency in which we accounted for grade, BCL2 and BCL6 translocation status, as well as CD10 expression in a cohort of outcome extremes, only IRF4 expression was significantly predictive of transformation (hazard ratio, 13.3; P < .001).17 IRF4 has also been shown to be correlated with shortened progression-free survival in 2 cohorts from the French study group LYSA and with poor overall survival in 2 phase 2 trials from the Southwest Oncology Group (SWOG).62,63 In a SWOG trial that predated the rituximab era, there was, however, no statistical difference in survival between IRF4+ and IRF4− cases.62 IRF4 requires further validation as a potential predictor of transformation and poor outcome, in part because IRF4 expression is reflective of increased NF-κB activation, suggesting patients whose samples overexpress this protein could potentially be candidates for immunomodulatory drugs that downregulate IRF4.64
The FL tumor microenvironment has received well-deserved attention by numerous studies. Nonneoplastic immune and stromal cells clearly play an important role in the pathogenesis of FL, as evidenced by the influence that recurrent genetic alterations exert on the composition of the tumor microenvironment,24,65,66 and by the association between immune cell–related gene expression changes and overall survival.67 Glas et al performed gene expression and immunohistochemical analyses of samples from 31 patients with rapid transformation and 35 patients without transformation.40 Gene expression changes in early transformers appeared comparable to follicular hyperplasia and there were no differences in total T-cell numbers between cases with early transformation and those without. In terms of the distribution of immune cell subsets, the localization of CD4+ T cells within rather than between follicles was found more commonly in rapidly transforming FL.40 Regarding other specific T-cell populations, high numbers of PD1-expressing (PD1+) cells have been reported to be associated with a reduced risk of transformation.44 This observation may appear intriguing as PD1 is an immune checkpoint whose ligation inhibits T-cell activation and thereby contributes to immune suppression. However, PD1 is typically expressed on follicular T helper cells, high numbers of which may represent nondisrupted lymph node architecture and low-grade FL. In a subsequent study in which a diffuse pattern of PD1+ cells was correlated with shorter time to transformation, PD1 expression outside of follicles was restricted to TIM3+, exhausted T cells.43 Other components of the tumor microenvironment of which the abundance or distribution has inconsistently been reported to be associated with transformation include FOXP3-expressing T regulatory cells40,42,45,68 and lymph node vascularization.46 Further studies are certainly warranted to lend more texture to our understanding of how the microenvironment modulates the risk of transformation. This will become increasingly important, as immune-directed therapies, including immunomodulators and checkpoint inhibitors, represent promising new avenues to improve treatment efficacy for both first-line and relapsed FL.
Gene expression signatures predictive of transformation
Whereas the above-mentioned study by Glas et al uncovered differential gene expression between early and nontransforming FL, the small size of the data set precluded the construction of a robust gene expression predictor.40 A reanalysis of these data showed, however, that a core embryonic stem cell–like signature, including MYC and its target genes, might underlie the risk of histologic transformation.47 A refined model was, indeed, predictive of survival, both in the Glas et al40 and the separate Dave et al67 data sets.47 The average linear predictor score from this model was higher in rapidly vs nontransforming FL, suggesting that it has potential to identify patients at high risk of transformation. These latter findings would need to be corroborated in nonenriched patient cohorts to ensure applicability in populations of patients at average risk of transformation. Brodtkorb et al performed integrated copy-number and gene expression analysis, and found that 6 NF-κB target signature scores, particularly IRAK1 and TRIM37 scores, predicted transformation.48 These scores were, however, not always consistent across serial samples from individual patients, which the authors attributed to divergent clonal evolution. Other scores that predicted transformation were associated with B-cell receptor signaling. Altogether, these findings suggested that an activated B-cell–like phenotype in FL might be predictive of transformation, a finding that is in keeping with the previously mentioned association of IRF4 expression and early transformation.17
Risk of transformation conferred by genetic alterations
Large-scale changes in the structural genome
Certain regional chromosomal imbalances have been linked to poor outcome and an increased risk of transformation. They include, for example, segmental deletions within 1p and 6q, as well as gains involving chromosomes 2, 3q, and 5.51-54 It is not fully elucidated what the precise coding or noncoding targets in these deleted regions are, nor whether their association with poor outcome could merely reflect increased “genomic complexity” as the number of chromosomal alterations by itself has been reported to be correlated with adverse survival.51,53 In our own work, we found that FL cases with subsequent transformation had higher numbers of structural rearrangements as compared with cases experiencing either progression in the absence of transformation, or cases who were event-free for at least 5 years following diagnosis.24 These findings clearly suggest that genome-wide discovery approaches hold great potential for uncovering genomic properties of rapid transformation.
Individual genetic aberrations
Individual somatic gene mutations have been associated with unfavorable outcome in FL. In this regard, TP53 is one of the genes that is most strongly associated with transformation, mutations being rare in diagnostic samples (∼5%) but common in TFL (25%-30%).26-28,55 Whereas TP53 mutations typically occur in the absence of deletion of the other allele in untransformed FL, biallelic hits through deletion, loss of heterozygosity, or further mutation are common in TFL.26,33,69 In a series of 185 diagnostic samples from the Lymphoma/Leukemia Molecular Profiling Project, TP53 mutations were correlated with shortened progression-free and overall survival, yet, somewhat surprisingly, not with transformation.55 In a study of 151 patients from the GLSG2000 trial, TP53 mutations were also associated with poor failure-free and overall survival, with no results for transformation reported.70 Similar to TP53 mutations, MYC translocations are rare at diagnosis but frequently found at transformation.17,26,29,30,71 The prognostic implication of MYC translocation in FL is unclear, despite these cases typically also harboring a BCL2 translocation and de facto being double-hit lymphomas. It is possible that larger patient cohorts would need to be studied to define whether TP53 and MYC alterations predict transformation. FAS mutations are present in about 6% to 7% of FLs.26,72,73 In contrast to TP53 and MYC, they are generally not more common in TFL when compared with preceding FL, yet they appear to be found predominantly in those cases with subsequent transformation.26 This observation, which deserves further validation, suggests that FAS mutations might foster a phenotype that is conducive to transformation. BCL6 translocations have been shown to confer an increased risk of transformation but it is unclear whether BCL6 by itself drives transformation or whether it is the marker of a distinct biological subset of FL with increased propensity to transform.17,56 Lastly, BCL2 mutations have been reported to predict transformation in 1 study,57 without being associated with outcome in other studies,24,70,74 a discrepancy that might be attributable to technical differences between these studies.
As individual genes have been described to increase the likelihood of transformation, the question arises of whether a multigene mutational outcome predictor would be a preferred strategy going forward. In this direction, Pastore et al recently reported on a clinicogenetic risk model that incorporated the mutational status of 7 genes (EZH2, ARID1A, MEF2B, EP300, FOXO1, CREBBP, and CARD11) into a predictive score that also includes the FLIPI and ECOG performance status.70 The new model, named m7-FLIPI, improves risk stratification for failure-free survival when compared with the traditional FLIPI, which is achieved mostly by reclassifying a subset of high-risk FLIPI patients into a low-risk m7-FLIPI risk group. Although of an exploratory nature, it would be relevant to assess whether the m7-FLIPI may help stratifying risk of transformation. Unfortunately, the number of events in the cohorts that formed the basis of this study is currently too small to perform a sufficiently powered analysis for transformation risk (Oliver Weigert, Ludwig Maximilians University Munich, written communication, 9 November 2016). As mutation screening is becoming more and more accessible, we foresee that mutation-based outcome predictors will continue to play a key role in evaluating outcomes for FL patients.
Evolutionary dynamics
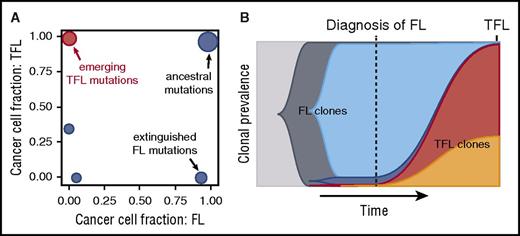
Cancers in general and FL in particular evolve through branching evolution, with genetic lesions that are acquired at an early stage being truncal, that is, found in every descendant cell, with mutations that are acquired at a later time point being confined to 1 branch of the tumor’s phylogenetic tree.25,26,75,76 These latter mutations are referred to as subclonal. Multiple studies have documented that subclonal mutations are common in FL and that transformation typically follows a branching pattern.25,26,66,77 In a set of 15 trios in which DNA from germ line, FL, and TFL samples were subjected to whole-genome sequencing, we deconvoluted the clonal composition and showed that transformation was the result of drastic shifts, with transformation-specific subclones rapidly rising to clonal dominance in the transformed samples (Figure 1).24 These aggressive subclones were typically either undetectable in the diagnostic specimen, or at best detectable at very low levels (<1% of cells), a pattern that was independent of prior treatment or the time to transformation. Our observations suggest that prediction of transformation based on TFL-associated gene mutations may be either impossible or at least technically challenging as deep sampling of tumor alleles may be required for their detection. This might, nonetheless, be a simplistic conclusion because it is conceivable that clonal mutations, present at diagnosis, could increase the likelihood of transformation, for example by providing a fertile soil for the acquisition of additional drivers of transformation. In addition, spatial heterogeneity contributes to diluting out transformation-associated alleles and multisite sampling could prove useful, although challenging to implement in practice.
Temporal analysis of clonal evolution in typical FL and matched TFL samples. (A) Shown are cancer cell fractions of mutation clusters, plotted against time point (FL, TFL). The FL time point is shown on the x-axis, and the TFL time point on the y-axis. The emerging TFL-associated mutations can either not be detected, or are present at very low abundance in the preceding FL (<1%). (B) Time-sweep plot showing evolution over time of FL clones (in blue colors) and the emergence of TFL clones (in red and orange). Time is shown on the x-axis, clonal abundance on the y-axis. The transformed genotype is typically not readily detected in the initial FL biopsy, although it can be present in other anatomical sites, based on the data by Scherer et al.58 Adapted from Kridel et al24 in accordance with the Creative Commons Attribution (CC BY) license.
Temporal analysis of clonal evolution in typical FL and matched TFL samples. (A) Shown are cancer cell fractions of mutation clusters, plotted against time point (FL, TFL). The FL time point is shown on the x-axis, and the TFL time point on the y-axis. The emerging TFL-associated mutations can either not be detected, or are present at very low abundance in the preceding FL (<1%). (B) Time-sweep plot showing evolution over time of FL clones (in blue colors) and the emergence of TFL clones (in red and orange). Time is shown on the x-axis, clonal abundance on the y-axis. The transformed genotype is typically not readily detected in the initial FL biopsy, although it can be present in other anatomical sites, based on the data by Scherer et al.58 Adapted from Kridel et al24 in accordance with the Creative Commons Attribution (CC BY) license.
Circulating tumor DNA
An area of investigation that is becoming increasingly relevant for clinical practice is the detection of circulating tumor DNA (ctDNA) in plasma or serum, allowing for noninvasive monitoring of disease status. It has been known for a long time that cancer patients have quantitatively more ctDNA than healthy individuals78 and that specific cancer-associated mutations can be detected in ctDNA,79,80 but the study of ctDNA certainly gained momentum with the advent of high-throughput sequencing together with methods that suppress sequencing errors. In lymphoma in particular, the promise of ctDNA assessment and monitoring has been illustrated for molecular subtyping, response evaluation, and preclinical detection of relapse.58,81-83 Tumor-specific ctDNA may originate from multiple clones and ctDNA is thus often considered as an integrator of the intrapatient mutational heterogeneity. Recently, Scherer et al comprehensively reported on the application of capture-based targeted sequencing of both tumor and plasma samples.58 Of particular interest for this review is the description of 1 patient, initially diagnosed with FL on the basis of an inguinal lymph node biopsy, who presented with biopsy-proven transformation within a retroperitoneal lymph node after 9 months. Mutational analysis of ctDNA taken at the time of diagnosis revealed mutations that were specific to the transformed clone and that were not found in the initial FL, suggesting that transformed disease was already present at the time of diagnosis and could be detected by ctDNA. The authors developed a predictive model in which the proportion of mutations that are uniquely found in plasma emerged as the main feature, suggesting that clonal divergence is a potential predictive marker. Although evaluation of ctDNA undoubtedly holds great promise, further validation and prospective implementation studies are necessary to move this novel biomarker strategy into clinical practice.
Challenges and conceptual considerations
To date, we do not currently have a tool in hand that would allow us to accurately predict transformation. In part, the above-mentioned biomarkers lack validation. Moreover, it may be challenging to implement them in the routine clinical setting. Immunohistochemistry, for example, suffers from technical variation and suboptimal reproducibility.84,85 Determination of gene mutation status appears to be more robust, yet technical challenges are equally present. This includes determination of the somatic nature of a mutation in the absence of germ line sequence, the difficulty of robustly calling low-abundance variants, or sequencing artifacts.86-88 Beyond these technical challenges, we are facing a knowledge gap with respect to the evolutionary pathways leading to transformation. Different scenarios can be envisioned that have clearly distinct implications for prediction of transformation (Figure 2). It is unclear at present whether transformation occurs after diagnosis of indolent lymphoma, or whether transformed, but quiescent, subclones are already present at the time of initial diagnosis. In our clonal evolution analysis study of 15 patients presenting with transformation, we found evidence for the latter scenario in at least 1 case, as determined by deep amplicon sequencing and digital droplet polymerase chain reaction.24 The article by Scherer et al also reported on 1 patient with preclinical genetic evidence of TFL.58 It is possible that this situation could be proven to be more frequent if one systematically assessed the presence of transformation-associated gene mutations in diagnostic FL samples using sensitive technology, and if one did so in multiple disease sites to account for spatial tumor heterogeneity. If transformed clones are present at low levels well before the onset of clinical transformation, then there is clear potential for predicting transformation.
Different scenarios with varying implications for the predictability of transformation. (A) In this scenario, transformation results from the acquisition of genetic alterations that occur after diagnosis of preceding FL. Transformation cannot be predicted by a test performed at the time of diagnosis. This would be a probabilistic scenario as no feature from the initial FL determines subsequent transformation. (B) In some cases, the transformed clone can be detected when patients present with what is thought to be FL. Detection is typically challenging as it requires experimental strategies such as deep sequencing of DNA obtained from the FL tumor biopsy or of circulating DNA in plasma. This would be either a probabilistic or deterministic scenario as it is unclear whether the presence of a minor aggressive subclone results in increased risk of transformation. (C) FL tumor cells harbor characteristics that increase the likelihood of transformation. These characteristics could be distinct alterations that increase the mutation rate, or even a permissive microenvironment. A biomarker could be particularly useful in this setting. This latter scenario is deterministic. We would like to emphasize that scenarios A-C are simplified conceptualizations of the large number of possible ways that transformation can potentially evolve from underlying low-grade lymphoma and are certainly not mutually exclusive.
Different scenarios with varying implications for the predictability of transformation. (A) In this scenario, transformation results from the acquisition of genetic alterations that occur after diagnosis of preceding FL. Transformation cannot be predicted by a test performed at the time of diagnosis. This would be a probabilistic scenario as no feature from the initial FL determines subsequent transformation. (B) In some cases, the transformed clone can be detected when patients present with what is thought to be FL. Detection is typically challenging as it requires experimental strategies such as deep sequencing of DNA obtained from the FL tumor biopsy or of circulating DNA in plasma. This would be either a probabilistic or deterministic scenario as it is unclear whether the presence of a minor aggressive subclone results in increased risk of transformation. (C) FL tumor cells harbor characteristics that increase the likelihood of transformation. These characteristics could be distinct alterations that increase the mutation rate, or even a permissive microenvironment. A biomarker could be particularly useful in this setting. This latter scenario is deterministic. We would like to emphasize that scenarios A-C are simplified conceptualizations of the large number of possible ways that transformation can potentially evolve from underlying low-grade lymphoma and are certainly not mutually exclusive.
Alternatively, biological transformation could arise entirely after diagnosis, a situation in which detection of the transformed clones at diagnosis would be completely impossible. Within this scenario, one would have to distinguish whether the occurrence of transformation is purely probabilistic or whether the determinants of transformation are already present in the initial indolent clone. The probabilistic scenario would suggest that a certain chance exists per year for a cell to acquire aggressive properties, whereas the deterministic scenario would presuppose that the indolent clones have certain characteristics that predispose them to undergo eventual transformation. The presence of TP53 mutations could be seen as an example of the latter as they can be found in indolent FL but are associated with adverse outcome (although strictly speaking they have not been reported to be predictive of transformation in the literature, to the best of our knowledge). In line with the deterministic scenario is the consideration that, although transformation can occur at any given time and certainly late in the course of FL, most events nonetheless occur within the first 5 to 10 years following diagnosis, at least in some series, suggesting that some patients are predestined to experience transformation.10,12,17 In such a scenario where easily detectable alterations are present at the time of indolent FL, prediction of transformation holds promise although further studies are clearly needed to establish the best biomarker in this setting.
Can histologic transformation be prevented?
The question of whether transformation can be predicted is pertinent because one could potentially modulate therapy, aiming to prevent transformation from occurring. Several studies have explored whether certain treatments are associated with a differential risk of transformation. First, there is some controversy as to whether upfront treatment reduces the incidence of transformation, when compared with observation in asymptomatic, low-tumor burden patients. Retrospective studies are not ideal to answer this question due to inherent selection bias, and report, indeed, conflicting results with some studies suggesting no benefit of upfront treatment,12,39,89 and others actually reporting an increased occurrence of transformation in patients who are initially observed.11,13,14 Stronger evidence can be drawn from prospective studies. In the study by Brice et al, in which 193 low-tumor burden FL patients were randomized to observation, prednimustine, or interferon, the rate of transformation was similar in the 3 arms.90 The study by Ardeshna et al, which randomized 309 asymptomatic, advanced-stage indolent lymphoma patients to either observation or chlorambucil monotherapy, saw no survival difference between treatment arms. The majority of patients in this study (204, or 66%) had been diagnosed with FL. To the best of our knowledge, rates of transformation have not been reported, but given the strong impact that transformation has on survival, we would not expect differences with regards to transformation in the 2 groups.91 A more recent study by Ardeshna et al randomized 379 asymptomatic, advanced-stage FL patients to either watchful waiting or maintenance rituximab. Fewer patients in the rituximab maintenance arm needed treatment at 3 years, but there was no difference in terms of overall survival or incidence of transformation.92 Overall, these prospective studies show relatively convincingly that observation does not negatively impact the rate of transformation. Observation is associated with better outcomes following transformation,93 suggesting that patients that can be observed have more indolent disease, or that the absence of prior therapy prevents selection of resistant subclones.
The next question pertains to whether the type of treatment modulates the risk of transformation in those patients who actually require therapy, such as those with symptoms or with high tumor burden. This question is relevant because it is theoretically conceivable that the more genotoxic chemotherapy is, the greater the risk of acquiring novel gene mutations that contribute to transformation. Or, one could speculate that more efficacious treatments would potentially reduce the likelihood of transformation. Again, the evidence from retrospective studies needs to be considered with caution. Al-Tourah et al reported an increased risk for patients receiving an alkylator-purine analog combination, as opposed to patients treated with bleomycin, cisplatin, etoposide, doxorubicin, cyclophosphamide, vincristine, prednisone and radiation.12 Conconi et al report that a small subset of patients treated with rituximab and chemotherapy were at higher risk of presenting with transformation when compared with chemotherapy without rituximab,39 a finding that is counterintuitive given that improved overall survival was shown when rituximab is added to chemotherapy.94-96 In a report from the National LymphoCare Study on 2652 FL patients of whom 379 or 14.3% experienced lymphoma transformation, the rate of transformation was similar whether patients received rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) or rituximab, cyclophosphamide, vincristine, and prednisone (R-CVP). Lastly, in the study by Sarkozy et al that we mentioned earlier in this review that reports on transformation events in the PRIMA trial, no differences with regard to transformation were seen in the 3 chemotherapy groups (R-CHOP, R-CVP, or rituximab, fludarabine, cyclophosphamide, or mitoxantrone). However, the number of patients in at least some of these groups was small.15
In summary, it appears that transformation is largely independent of treatment received, and that with currently available therapies, it might not be preventable. Such a statement, however, will need to continue to be challenged in the future as newer treatment modalities emerge. Thus, the lack of a relationship between transformation and treatment mirrors our own observation that the typical evolutionary pattern underlying transformation appears to be independent of treatment received, as it was seen both in patients who were observed and in patients who were treated with immunochemotherapy or other approaches.24 In aggregate, these findings point to transformation arising through selective pressures that are yet unidentified.
Conclusions and future directions
Predicting transformation remains a challenge, with no robust biomarker currently available in routine clinical practice. Indeed, several genetic alterations, gene signatures, or immunohistochemical markers have been reported in the literature to be associated with transformation, but have not been sufficiently validated to warrant their assessment outside of the research setting. Future studies are important to address this unmet need. We anticipate that large-scale investigations that comprehensively elucidate the full spectrum of genetic alterations within tumor cells, as well as the composition of the tumor microenvironment, would represent a step forward, provided that they contain detailed and reliable clinical annotation and that therapy is at least relatively uniform. Ideally, such studies would be conducted using the most current treatment to account for the possible interaction between the prognostic value of biomarkers and specific treatments, which may be challenging as standard therapies are under constant flux. It will remain equally important to conduct well-designed tumor evolution studies ascertaining evolutionary patterns pertaining to both progression and transformation to gain further understanding of the genetic underpinnings of transformation. Thus far, no treatment strategy appears to mitigate the risk of transformation. Only a deeper understanding of tumor biology and evolution will allow us to eventually predict transformation, which could open new avenues for the investigation of strategies aiming at circumventing it.
Authorship
Contribution: R.K., L.H.S., and R.D.G. wrote the paper.
Conflict-of-interest disclosure: L.H.S. served as an advisor or consultant for AbbVie, Amgen, Roche/Genentech, Celgene, Janssen, Lundbeck, Seattle Genetics, and TG Therapeutics and received grants for clinical research from Roche/Genentech. The remaining authors declare no competing financial interests.
Correspondence: Robert Kridel, Division of Medical Oncology and Hematology, Princess Margaret Cancer Centre, 610 University Ave, OPG 6th Floor, Suite 6-714, Toronto, ON M5G 2M9, Canada; e-mail: robert.kridel@uhn.ca.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal