Abstract
Inherited risk determinants for follicular lymphoma (FL) have recently been described in the immune gene-rich human leukocyte antigen region on chromosome 6p. The known importance of host immune response to FL survival led us to evaluate these germline factors in FL outcome. We confirm the association of single nucleotide polymorphisms rs10484561 (P = 3.5 × 10−9) and rs6457327 (P = .008) with risk of FL and demonstrate that rs6457327 predicts both time to (P = .02) and risk of (P < .01) FL transformation independently of clinical variables, including the Follicular Lymphoma International Prognostic Index.
Introduction
The first genome-wide association studies of follicular lymphoma (FL) recently identified susceptibility loci, rs10484561 (upstream of HLA-DQB1) and rs6457327 (near C6orf15), in the immune gene–rich human leukocyte antigen (HLA) region on chromosome 6p211,2 (supplemental Figures 1-2, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The role of these single nucleotide polymorphisms (SNPs) and HLA allelotypes3,4 in FL risk and the importance of the immune response to FL outcome5 led us to investigate the clinical significance of rs10484561 and rs6457327 in a series of FL cases from the United Kingdom.
Methods
Patient samples
DNA from fresh frozen uninvolved bone marrow or peripheral blood samples was obtained from 218 patients diagnosed with FL between 1977 and 2005 and managed at Barts and the London NHS Trust. Sample collection followed informed, written consent in accordance with the Declaration of Helsinki, and the study was performed under approval 05/Q0605/140 from East London and the City Health Authority Local Research Ethics Committee. A case-control analysis evaluated FL risk in the United Kingdom for SNPs rs6457327 and rs10484561 by comparing patient genotypes against those of control populations (2691 and 2684 persons for the respective SNPs) established from the Wellcome Trust Case-Control Consortium 2 (1958) birth cohort (www.wtccc.org.uk; Table 1). Clinical outcome was assessed on a subset of cases (n = 130) who presented initially to Barts (supplemental Table 1).
SNPs rs10484561 and rs6457327 association with risk of developing FL in the United Kingdom
| SNP/genotype . | Genotype count . | MAF (cases/control) . | HWE controls . | Variant allele OR (95% CI)‡ . | Allelic OR (95% CI)‡ . | Allelic P . | Trend P . | Genotypic P . | |
|---|---|---|---|---|---|---|---|---|---|
| Cases* . | Controls† . | ||||||||
| rs10484561 | 0.21/0.11 | 0.924 | 2.40 (1.80-3.19) | 2.07 (1.61-2.63) | 3.502 × 10-9 | 2.743 × 10-9 | 6.342 × 10-9 | ||
| GG | 5 | 34 | |||||||
| GT | 82 | 548 | |||||||
| TT | 131 | 2102 | |||||||
| rs6457327 | 0.32/0.39 | 0.808 | 0.77 (0.58-1.01) | 0.75 (0.61-0.93) | .008 | .008 | .0190 | ||
| AA | 19 | 408 | |||||||
| AC | 103 | 1272 | |||||||
| CC | 96 | 1011 | |||||||
| SNP/genotype . | Genotype count . | MAF (cases/control) . | HWE controls . | Variant allele OR (95% CI)‡ . | Allelic OR (95% CI)‡ . | Allelic P . | Trend P . | Genotypic P . | |
|---|---|---|---|---|---|---|---|---|---|
| Cases* . | Controls† . | ||||||||
| rs10484561 | 0.21/0.11 | 0.924 | 2.40 (1.80-3.19) | 2.07 (1.61-2.63) | 3.502 × 10-9 | 2.743 × 10-9 | 6.342 × 10-9 | ||
| GG | 5 | 34 | |||||||
| GT | 82 | 548 | |||||||
| TT | 131 | 2102 | |||||||
| rs6457327 | 0.32/0.39 | 0.808 | 0.77 (0.58-1.01) | 0.75 (0.61-0.93) | .008 | .008 | .0190 | ||
| AA | 19 | 408 | |||||||
| AC | 103 | 1272 | |||||||
| CC | 96 | 1011 | |||||||
MAF indicates minor allele frequency; HWE, Hardy-Weinberg equilibrium; and CI, confidence interval.
FL cases from St Bartholomew's Hospital (n = 218 for both SNPs). In a validation series (10% of FL cases), direct sequencing results for each SNP genotype were fully concordant with the AD-PCR genotype results.
Controls from WTCCC2 1958 birth cohort (after filtering for missing genotype, population outliers, and relatedness: n = 2684 for rs10484561; n = 2691 for rs6457327). Both control populations maintain HWE.
ORs calculated for the variant allele carriers (homozygous or heterozygous) vs homozygous common allele carriers (Variant allele OR) and for the minor vs the major allele (Allelic OR).
Presentation and outcome data
Patient demographics included age, gender, stage, hemoglobin, lactate or hydroxybutyrate dehydrogenase, number of nodal sites involved by disease, Follicular Lymphoma International Prognostic Index risk group, and tumor grade (1, 2, or 3a) at diagnosis. The outcome parameters assessed were initial management, response to first therapy, dates of first relapse and transformation, and date of death or last follow-up (supplemental Table 1). Clinical characteristics are as expected at a tertiary referral center with median age at diagnosis of 52 years and other demographics, including the proportion of cases in Follicular Lymphoma International Prognostic Index risk groups, similar to those previously described.6 In 51 of 130 (39%) cases, initial management was with observation only (“watch and wait”), and 12 of these cases (9%) were never treated. Response to first therapy was available for 113 of 118 treated cases, including 30 (27%) complete responses. Biopsy-proven disease transformation to diffuse large B-cell lymphoma occurred in 45 cases at a median of 2.5 years (range, 0.2-20.1 years), with a rate comparable with recent studies.7,8 Ninety-one cases relapsed at a median of 2.5 years, and estimated median overall survival for all 130 cases was 12 years. The 67 surviving patients were followed up for a median of 9.3 years (range, 1.6-30.5 years) with 94% seen for more than 5 years and 48% for more than 10 years.
Molecular analysis
SNP genotypes were established by allelic-discrimination (AD) PCR using custom TaqMan SNP Genotyping Assays with results interpreted using Sequence Detection Systems software Version 2.3. Direct DNA sequencing of AD-PCR controls, an AD-PCR validation set, and C6orf15 was performed using the Big Dye Terminator kit (Applied Biosystems). The single probe for C6orf15 on current gene expression arrays has shown only limited efficacy in other FL series.5 Therefore, C6orf15 expression was determined by 2-step reverse-transcriptase PCR on 16 FL and 2 transformed-FL tumor samples, 1 benign lymph node, 6 B-cell non-Hodgkin lymphoma, 2 lymphoblastoid (and 2 myeloid) cell lines, and 4 tonsils. (Protocols are available on request.)
Statistical analysis
Single marker association tests were conducted using the one degree of freedom (df) allelic χ2 test, 2 df genotypic test, and 1 df association test under an additive model (Cochran-Armitage trend test) implemented in the PLINK Version 1.07 software (www.pngu.mgh.harvard.edu/∼purcell/plink/). PLINK, Version 1.07 was also used to test deviations from Hardy-Weinberg equilibrium in controls. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated for the minor allele and for the variant allele carriers (homozygous or heterozygous vs homozygous common allele carriers) by median-unbiased estimation using the mid-p method from the epitools R package (www.cran.r-project.org/web/packages/epitools/index.html). Associations with diagnostic parameters were determined using the Wilcoxon-Mann-Whitney, χ2, or Fisher exact tests. Survival analysis was performed using the Kaplan-Meier survival function with log-rank test for equality of survival (censoring at death/last follow-up). Logistic regression models estimated crude OR for transformation risk; Cox regression models estimated crude hazard ratios for time to transformation. Potential confounders were identified by crude regression models and clinical hypotheses (supplemental Table 2). Adjusted models were constructed using forward selection of potential confounders, which gave a more than or equal to 10% change to the estimate of effect. Results were considered statistically significant with P less than .05.
Results and discussion
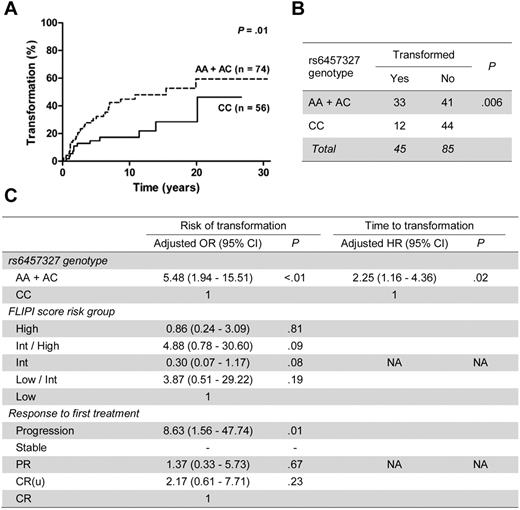
We validated that variant alleles for rs10484561 (allelic P = 3.5 × 10−9) and rs6457327 (allelic P = .008) predict increased and reduced risk of FL, respectively, in our United Kingdom cohort (Table 1). Allele frequencies were similar to those reported in other populations.1,2 Further, we demonstrate, for the first time, that rs6457327 predicts clinical outcome of FL with the variant allele (AA + AC) predicting a shorter time from diagnosis to transformation (P = .01; Figure 1A) and higher risk of transformation occurrence (P = .006; Figure 1B). These effects were independent of rs10484561, which showed no clinical correlation (supplemental Table 1).
SNP rs6457327 predicts time to and risk of transformation of FL. (A) Kaplan-Meier plots of the percentage of FL cases with transformation against time from diagnosis for rs6457327 genotypes. Genotype groups with respective number of cases and the log-rank test statistic P value are indicated on the right. (B) Number of cases that transformed for each rs6457327 genotype group. The χ2P value is indicated. (C) Final adjusted models for risk of transformation and time to transformation. These were generated following multivariate analysis incorporating potential confounders using logistic regression for risk of transformation and Cox regression for time to transformation. No cases with stable disease as response to first treatment transformed, so no data are available. CI indicates confidence interval; HR, hazard ratio; Int, intermediate; PR, partial response; CR(u), complete response unconfirmed; CR, complete response; and NA, not applicable.
SNP rs6457327 predicts time to and risk of transformation of FL. (A) Kaplan-Meier plots of the percentage of FL cases with transformation against time from diagnosis for rs6457327 genotypes. Genotype groups with respective number of cases and the log-rank test statistic P value are indicated on the right. (B) Number of cases that transformed for each rs6457327 genotype group. The χ2P value is indicated. (C) Final adjusted models for risk of transformation and time to transformation. These were generated following multivariate analysis incorporating potential confounders using logistic regression for risk of transformation and Cox regression for time to transformation. No cases with stable disease as response to first treatment transformed, so no data are available. CI indicates confidence interval; HR, hazard ratio; Int, intermediate; PR, partial response; CR(u), complete response unconfirmed; CR, complete response; and NA, not applicable.
Potential confounders for the associations with transformation are detailed in supplemental Table 2. After their incorporation in multivariate analyses, only rs6457327 genotype retained its predictive value for time to transformation (P = .02, hazard ratio = 2.25; 95% CI, 1.16-4.36). For risk of transformation only rs6457327 AA + AC genotype (P < .01, adjusted OR = 5.48; 95% CI, 1.94-15.51) and progression after first therapy (P = .01, adjusted OR = 8.63; 95% CI, 1.56-47.74) remained predictive (Figure 1C).
rs6457327 is in a 26-kb segment of high linkage disequilibrium that includes only one coding locus, C6orf15. This locus is not, however, a promising candidate as we found that C6orf15 expression was restricted to tonsils (3 of 4) and was not detected in other samples, including B-cell non-Hodgkin lymphoma cell lines and primary tumors (n = 24). Furthermore, DNA sequencing of C6orf15 in 50 diagnostic FL tumor samples revealed no mutational events.
Although more than 30 studies have reported SNPs associated with FL risk, only the recent genome-wide association studies were validated in multiple patient cohorts. Our study is the first to provide independent confirmation of the association between SNPs rs10484561 and rs6457327 and FL risk. Moreover, these findings further support the role of rs10484561 as a major susceptibility locus for FL. There are fewer reports regarding SNP associations with FL outcome,9-14 and only one identified an association with transformation.12 Similar to that study, the association of rs6457327 genotype with transformation did not translate into prediction of survival. It seems unlikely that this is the result of limitations of the study cohort, as the transformation frequency (35%) and its association with overall survival (P < .001, supplemental Figure 3) are similar to those of larger FL series.7,8 Nonetheless, it will be important to validate our observations in independent case cohorts, particularly those that accrue prolonged follow-up from the current era of combination immunochemotherapy.
The direct role of C6orf15 in FL remains to be established and, because rs6457327 is also in linkage disequilibrium with HLA-C alleles,15 further studies are needed to determine the functional locus influencing transformation. However, this study represents a step forward in the characterization of inherited predictors of clinical outcome which, together with recently described acquired predictors,16-19 represent a growing pool of molecular outcome markers in FL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr George Wright from the National Cancer Institute, National Institute of Health, Bethesda, MD, for providing guidance regarding C6orf15 in the previous genome-wide expression profiling study of FL.
This work was supported by Cancer Research UK (program grant C1574/A6806; D.W., J.F., J.G.G., and T.A.L.), the National Cancer Institute (grants CA122663 and CA104682), the National Institutes of Health (C.F.S.), the Wellcome Trust (grant 075491/Z/04; J.-B.C.), the Partner Fellowship (2009/01) awarded by the European Hematology Association (C.B.), Olivia Walduck's family (S.M.), and the Mark Ridgwell Family trust (S.M.).
This study makes use of data generated by the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust (award nos. 076113 and 085475).
National Institutes of Health
Wellcome Trust
Authorship
Contribution: D.W. designed the study, performed research, analyzed data, and wrote the paper; P.L., C.F.S., J.-B.C., and L.C. analyzed data; J.M. provided data; S.I., E.C., and C.B. provided samples; M.C. and S.M. collected data; and J.G.G., T.A.L., and J.F. designed the study and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David Wrench, Centre for Haemato-Oncology, Barts Cancer Institute, Barts and the London School of Medicine and Dentistry, Queen Mary University of London, 3rd Fl, John Vane Bldg, Charterhouse Sq, London, EC1M 6BQ United Kingdom; e-mail: d.j.wrench@qmul.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal