Key Points
Whole-genome integrative analyses in FL reveal that genes strongly influenced by copy number are highly enriched for NF-kB pathway regulators.
Subsignatures of the NF-kB targets predict transformation in FL.
Transformation of follicular lymphoma (FL) to a more aggressive disease is associated with rapid progression and death. Existing molecular markers for transformation are few and their clinical impact is limited. Here, we report on a whole-genome study of DNA copy numbers and gene expression profiles in serial FL biopsies. We identified 698 genes with high correlation between gene expression and copy number, and the molecular network most enriched for these cis-associated genes. This network includes 14 cis-associated genes directly related to the nuclear factor κB (NF-κB) pathway. For each of these 14 genes, the correlated NF-κB target genes were identified and corresponding expression scores were defined. The scores for 6 of the cis-associated NFκB pathway genes (BTK, IGBP1, IRAK1, ROCK1, TMED7-TICAM2, and TRIM37) were significantly associated with transformation. The results suggest that genes regulating B-cell survival and activation are involved in transformation of FL.
Introduction
Transformation of follicular lymphoma (FL) to a more aggressive histology is commonly followed by rapid progression, treatment resistance and death. Finding clinical and molecular markers for transformation risk has been challenging. Existing markers have limited clinical impact.
Several recurring DNA copy number alterations in FL have been associated with transformation and survival.1,,-4 Most alterations span large genomic regions involving hundreds of genes, thus offering limited cues for identification of genes driving the development of FL. Elevated expression of c-MYC and other genes promoting proliferation, inactivation of TP53 and CDKN2A, dysregulation of p38-mitogen-activated protein kinase (MAPK), and mutations of BCL6 have been implicated in subsets of cases with transformation.5,,-8 The immunologic microenvironment of FL may influence transformation risk9 and a combination of 2 gene expression signatures reflecting the presence of T-cells and macrophages was shown to predict FL survival.10 Furthermore, an expression signature reflecting pluripotency was shown to predict transformation and survival in FL.11
Methods
The material comprises 100 biopsies from 44 patients diagnosed with FL and is subsequently referred to as follicular lymphoma serial biopsies (FLSB) (supplemental Table 1, available on the Blood Web site). Copy number profiles were obtained with a custom BAC/PAC array1 (resolution ∼1 Mb) and were segmented with PCF.16 Expression profiles were obtained from 81 of the biopsies using Affymetrix HG U133 Plus 2.0 Gene Chip. The data are accessible through GEO Series Accession number GSE53820 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE53820). Of these, 51 were from 25 patients with subsequent transformation (45 with histological diagnosis FL and 6 with diffuse large B-cell lymphoma (DLBCL)) and 30 from 16 patients without transformation. Median observation time was 84 and 97 months in patients with and without transformation, respectively. In addition to the FLSB data, previously published expression profiles from 191 FL-samples10 with corresponding copy number profiles for 131 cases17 (referred to as the Lymphoma/Leukemia Molecular Profiling Project [LLMPP]) were used for validation of the unsupervised analysis. Cis-genes were defined as genes with Pearson correlation between copy number and expression exceeding 0.4 and differential expression (P < .05) in samples with genomic gain (or loss) vs samples with no change. Ingenuity Systems Pathway Analysis and the Ingenuity Knowledge Database identified the molecular network most significantly enriched for cis-genes. For each associated cis-gene, the significantly correlated (P < .01) downstream target genes were identified and a score defined by averaging the expression of the target genes. Associations between scores and transformation were assessed by Student’s t test and logistic regression. For significant scores (P < .05), cis-gene messenger RNA was quantified by quantitative polymerase chain reaction (qPCR) in 80 biopsies. Details about copy number and expression profiling, qPCR and statistics are provided in the supplemental Material and Methods. The study was approved by the regional committee for research ethics (protocol number S-05209) and was conducted in accordance with the Declaration of Helsinki.
Results and discussion
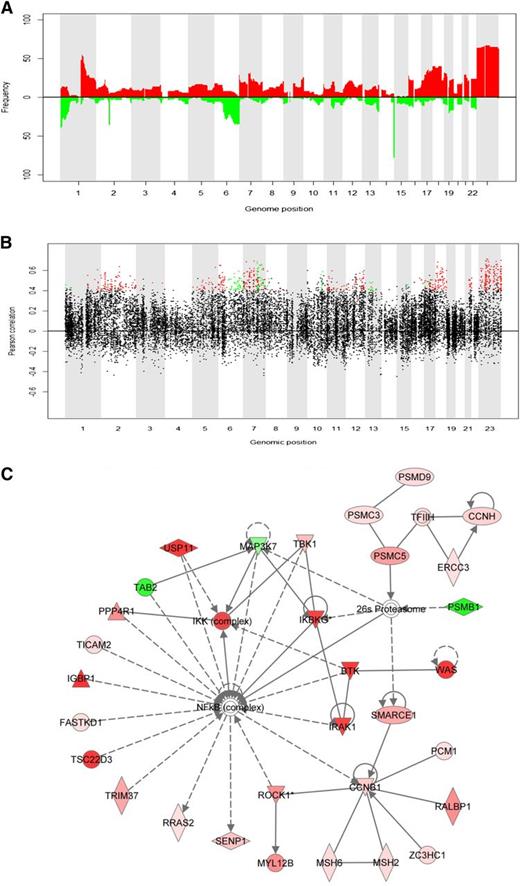
Correlations between copy number and gene expression were positively skewed in commonly aberrant genomic regions, indicating influence of copy number on gene expression (Figure 1A-B). Recurring aberrations with concurrent significant transcriptional changes comprised 698 cis-genes with 23% residing on chromosome X (supplemental Table 2). Gain of X is associated with inferior outcome in FL.1,,-4 Analysis of 131 FL-samples in the LLMPP data set10,17 confirmed these results (supplemental Figures 2-5). Using Ingenuity Systems Pathway Analysis, the molecular network most enriched for cis-genes was identified (Figure 1C). Among 35 genes in the network, 14 cis-genes (43%) encode regulators and signaling molecules upstream of the nuclear factor κB (NFκB) transcription factor and are henceforth referred to as the NFκB-linked cis-genes. Two genes (MAP3K7 and TAB2) were downregulated in regions of loss and the remaining upregulated in regions of gain (supplemental Figure 6). Cis-regulation was confirmed for the 14 genes in an independent set of 131 FL-samples10,17 (supplemental Table 3). For each NFκB-linked cis-gene, a permutation test (n = 25 000) identified correlated NFκB targets (P < .01), resulting in 14 signatures of associated NFκB target genes (supplemental Table 4). The overall correlation pattern between NFκB-linked cis-genes and NFκB targets was independently confirmed in 191 FL-samples10 (supplemental Figure 7). Correlations in the 2 data sets were positively and significantly associated for all 14 signatures. Scores found by averaging gene expressions in each signature followed a very similar distribution in the 2 data sets (supplemental Figure 8).
In cis association between copy number and gene expression. (A) Frequency of gain and loss as a function of genomic position, calculated across all the serial FL samples from which DNA copy number were obtained (see supplemental Material and Methods). Colors indicate type of aberration, with red representing gains and green representing losses. (B) Pearson’s correlation coefficient as a function of genomic position (total base pairs) for all 21 461 genes. The genes marked in green have high cis-correlation (Pearson's r > 0.4) and differential gene expression in samples with loss vs samples with normal copy number (P < .05). The genes marked in red have high cis-correlation (Pearson's r > 0.4) and differential gene expression in samples with gain vs samples with normal copy number (P < .05). Further details about the selection of cis-genes are provided in the supplemental Material and Methods and in supplemental Figure 1. Correlations between copy number and gene expression were positively skewed in commonly aberrant genomic regions, indicating an influence of copy number on gene expression. (C) The molecular network most highly enriched with cis-genes. The network (denoted Network 1) was identified by using Ingenuity Systems Pathway Analysis with the Ingenuity Pathway Knowledge Database and specifying small network size. The NFκB transcription factor plays the role as a hub in this network, and 14 genes in the network encode regulators and signaling molecules upstream of the NFκB transcription factor: BTK, FASTKD1, IGBP1, IKBKG, IRAK1, MAP3K7, PPP4R1, ROCK1, TAB2, TBK1, TMED7-TICAM2, TRIM37, TSCD22, and USP11. The genes/molecules colored red and green in Network 1 represent cis-genes located in regions of gain and loss, respectively. The color intensity increases with increasing frequency of the respective genomic aberration (as shown in supplemental Figure 6).
In cis association between copy number and gene expression. (A) Frequency of gain and loss as a function of genomic position, calculated across all the serial FL samples from which DNA copy number were obtained (see supplemental Material and Methods). Colors indicate type of aberration, with red representing gains and green representing losses. (B) Pearson’s correlation coefficient as a function of genomic position (total base pairs) for all 21 461 genes. The genes marked in green have high cis-correlation (Pearson's r > 0.4) and differential gene expression in samples with loss vs samples with normal copy number (P < .05). The genes marked in red have high cis-correlation (Pearson's r > 0.4) and differential gene expression in samples with gain vs samples with normal copy number (P < .05). Further details about the selection of cis-genes are provided in the supplemental Material and Methods and in supplemental Figure 1. Correlations between copy number and gene expression were positively skewed in commonly aberrant genomic regions, indicating an influence of copy number on gene expression. (C) The molecular network most highly enriched with cis-genes. The network (denoted Network 1) was identified by using Ingenuity Systems Pathway Analysis with the Ingenuity Pathway Knowledge Database and specifying small network size. The NFκB transcription factor plays the role as a hub in this network, and 14 genes in the network encode regulators and signaling molecules upstream of the NFκB transcription factor: BTK, FASTKD1, IGBP1, IKBKG, IRAK1, MAP3K7, PPP4R1, ROCK1, TAB2, TBK1, TMED7-TICAM2, TRIM37, TSCD22, and USP11. The genes/molecules colored red and green in Network 1 represent cis-genes located in regions of gain and loss, respectively. The color intensity increases with increasing frequency of the respective genomic aberration (as shown in supplemental Figure 6).
Six NFκB target signature scores corresponding to the NFκB-linked cis-genes BTK, IGBP1, IRAK1, ROCK1, TMED7-TICAM2, and TRIM37 were associated with transformation (P < .05 with Student’s t test and logistic regression) (supplemental Table 5 and supplemental Figures 9 and 10). Of interest, the average expression of all NFκB target genes was not associated with transformation. Subsequent qPCR quantification of the gene expression of the 6 NFκB-linked cis-genes showed positive correlations (median, 0.40; range, 0.10-0.62) to array expression and weak positive correlations (median, 0.21; range, 0.01-0.32) to copy number.
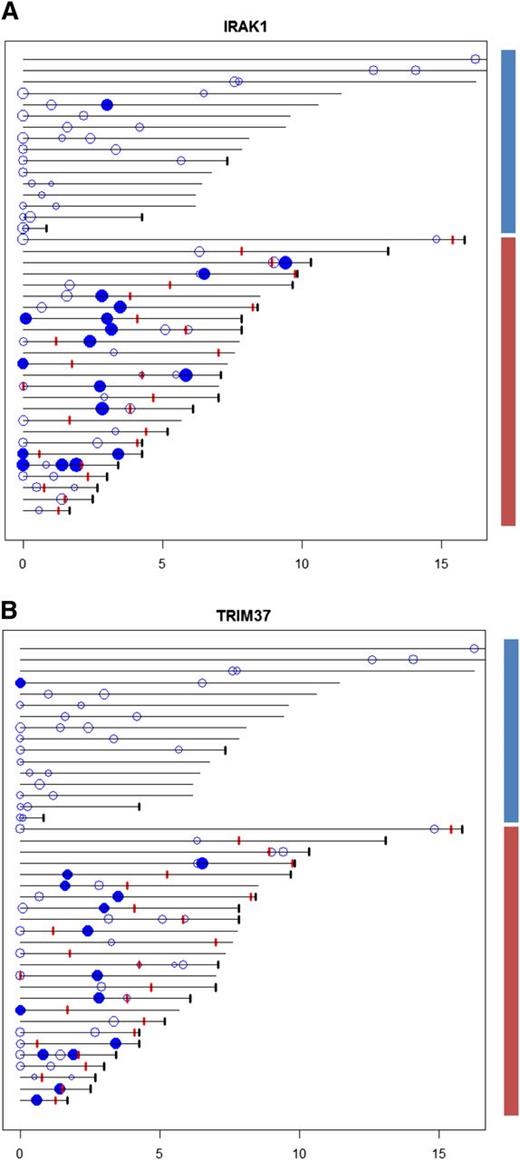
The most significant predictors for transformation were the IRAK1- and TRIM37-associated scores, high scores being associated with transformation (Figure 2A-B). A combination of the IRAK1 and TRIM37 scores resulted in improved discrimination of cases with and without transformation (supplemental Figure 11). Most patients with high-scoring FL biopsies (with respect to IRAK1 and TRIM37) also had FL biopsies with low scores, suggesting variable activation of transcriptional programs in different FL clones from the same patient. This is consistent with divergent clonal evolution in FL and may explain the limited success in predicting transformation at diagnosis from genetic changes in a single biopsy.
Time diagram of clinical observations, FL biopsies, TRIM37, and IRAK1 scores. Each horizontal line represents the time line for a particular patient, the length indicating total observation time since primary diagnosis. Patients are ordered according to total observation time, with FL biopsies from patients without transformation (blue bar; 30 FL biopsies from 16 patients) and FL biopsies from patients with transformation (red bar; 45 FL biopsies from 25 patients) shown separately. The patients are ordered as shown in supplemental Table 1 with patient number 1 at the bottom of the diagram. Black tick marks indicate time of death. Red tick marks represent time of transformation. Circles indicate FL samples with copy number and gene expression data available, the size reflecting the magnitude of the TRIM37- and IRAK1-associated scores (left and right panel, respectively). Filled circles represent scores exceeding a threshold chosen for TRIM37- and IRAK1-associated scores separately to maximize the discrimination between patients with and without transformation (indicated with red and blue bars, respectively, at the right side of the figure). Most patients with samples showing high scores have additional samples showing low scores. Of note, in most cases the samples showing high scores precede transformation. A combination of the TRIM37- and IRAK1-associated scores resulted in improved discrimination between the cases with and without transformation (supplemental Figure 11).
Time diagram of clinical observations, FL biopsies, TRIM37, and IRAK1 scores. Each horizontal line represents the time line for a particular patient, the length indicating total observation time since primary diagnosis. Patients are ordered according to total observation time, with FL biopsies from patients without transformation (blue bar; 30 FL biopsies from 16 patients) and FL biopsies from patients with transformation (red bar; 45 FL biopsies from 25 patients) shown separately. The patients are ordered as shown in supplemental Table 1 with patient number 1 at the bottom of the diagram. Black tick marks indicate time of death. Red tick marks represent time of transformation. Circles indicate FL samples with copy number and gene expression data available, the size reflecting the magnitude of the TRIM37- and IRAK1-associated scores (left and right panel, respectively). Filled circles represent scores exceeding a threshold chosen for TRIM37- and IRAK1-associated scores separately to maximize the discrimination between patients with and without transformation (indicated with red and blue bars, respectively, at the right side of the figure). Most patients with samples showing high scores have additional samples showing low scores. Of note, in most cases the samples showing high scores precede transformation. A combination of the TRIM37- and IRAK1-associated scores resulted in improved discrimination between the cases with and without transformation (supplemental Figure 11).
It is poorly understood how the signaling pathways that regulate B-cell survival, proliferation, and differentiation are involved in FL pathogenesis. Most cases of transformed FL are classified as GCB subtype of DLBCL by gene expression profile or immunophenotype.18 Using a high-resolution SNP array, copy number profiles of FL, and transformed FL showed similarities with the GCB, as well as the ABC subtype of DLBCL,3,19 and genes involved in NFκB signaling were targeted by recurrent copy number aberrations. The predictive power of 6 NFκB target gene scores suggests that attainment of a more ABC-like phenotype predisposes FL for transformation. Then again, the lack of association between the whole NFκB target gene signature and transformation, combined with the correlation of 6 NFκB-linked cis-genes only to subsets of the target genes, suggest that a trans-effect of these genes on transcription is mediated by alternative transcription factors.
The protein encoded by IRAK1 mediates Toll-like receptor (TLR) signaling and promotes downstream NFκB activation. Approximately 30% of ABC-DLBCLs harbor mutations in MyD88 and are dependent on MyD88, IRAK1, and IRAK4 expression for survival.20 The ability of the IRAK1-associated target gene score to predict transformation suggests a role for TLR-signaling in FL progression. TRIM37 encodes an ubiquitin ligase in the TRIM family which comprises several proto-oncogenes.21 A role of TRIM37 in lymphoma, to our knowledge, has not been investigated.
The predictive roles of the BTK- and IGBP1-associated scores suggest involvement of B-cell receptor (BCR) signaling in FL transformation. In ABC-DLBCL, chronic BCR signaling is a mechanism for constitutive NFκB activation.22 In FL, BCR-induced phosphorylation of SYK was more potent in FL B-cells than in nonmalignant B-cells. Then again, intratumoral heterogeneity is important in FL, as a lymphoma subset with abnormal BCR signaling was identified, and the prevalence of the subset was associated with inferior outcome.23 Clinical studies have demonstrated a positive effect of the BTK-inhibitor ibrutinib in FL patients.24 These findings encourage further testing of therapeutic approaches targeting BCR signaling in FL.
In summary, the expression level of several genes involved in NFκB signaling is affected by copy number in FL. Associated expression scores of downstream target genes predict transformation.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE53820).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Lise Forfang for assisting in the laboratory and Rolf Skotheim and Anita Sveen at the Department of Cancer Prevention, Institute for Cancer Research, Oslo University Hospital, for assisting in the analysis of raw gene expression data.
This work was supported by grants from The Norwegian Cancer Society (ES 33260) and The Faculty of Medicine, The University of Oslo; and the gene expression data from 191 lymphoma samples with corresponding conventional comparative genomic hybridization data for 131 samples were obtained from published data from the Lymphoma/Leukemia Molecular Profiling Project, headed by L. M. Staudt, National Cancer Institute, Rockville, MD.
Authorship
Contribution: M.B., K.H., M.H., V.I.H., and G.T. performed experiments; M.B. and O.C.L. performed analyses and made figures; E.L. and A.R. performed experiments and raw data analyses of the validation data set; E.B.S., J.D., M.B., O.C.L., J.H.M., and H.H. designed the research; M.B., O.C.L., E.B.S., H.H., and J.D. wrote the paper; and all authors have approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marianne Brodtkorb, Department of Oncology, Division of Cancer Medicine, Surgery and Transplantation, Oslo University Hospital, The Norwegian Radium Hospital, PO Box 4950, Nydalen, Oslo, N-0424 Norway; e-mail: marianne.brodtkorb@ous-hf.no.