Abstract
The secondary genetic events associated with follicular lymphoma (FL) progression are not well defined. We applied genome-wide BAC array comparative genomic hybridization to 106 diagnostic biopsies of FL to characterize regional genomic imbalances. Using an analytical approach that defined regions of copy number change as intersections between visual annotations and a Hidden Markov model–based algorithm, we identified 71 regional alterations that were recurrent in at least 10% of cases. These ranged in size from approximately 200 kb to 44 Mb, affecting chromosomes 1, 5, 6, 7, 8, 10, 12, 17, 18, 19, and 22. We also demonstrated by cluster analysis that 46.2% of the 106 cases could be sub-grouped based on the presence of +1q, +6p/6q−, +7, or +18. Survival analysis showed that 21 of the 71 regions correlated significantly with inferior overall survival (OS). Of these 21 regions, 16 were independent predictors of OS using a multivariate Cox model that included the international prognostic index (IPI) score. Two of these 16 regions (1p36.22-p36.33 and 6q21-q24.3) were also predictors of transformation risk and independent of IPI. These prognostic features may be useful to identify high-risk patients as candidates for risk-adapted therapies.
Introduction
Lymphoid malignancies account for approximately 5% of cases of cancer in the United States and have continued to rise in frequency at 3% to 4% annually.1,2 Of the different types of indolent lymphoma, follicular lymphoma (FL) is most prevalent and has a variable clinical course with a median survival of 10 years.3 While management strategies have changed, advanced-stage FL remains an incurable disease using conventional therapies.4 Approximately 85% of FL is associated with a specific balanced translocation, t(14;18)(q32;q21), that leads to overexpression of the antiapoptotic gene BCL2 due to its relocation in proximity to an IgH enhancer element.5-8 This genetic abnormality alone, however, is unlikely to produce clinical FL, as BCL2-overexpressing transgenic mice do not develop lymphoma9,10 and t(14;18)-bearing lymphocytes have been frequently demonstrated in healthy individuals.11,12
If the pathogenesis of FL results from a sequential accumulation of genetic alterations,13 the analysis of early neoplastic lesions may define the critical events associated with the initial development and further progression. To the best of our knowledge, there have been 12 large studies reported in the Western Hemisphere in the last decade that have investigated chromosomal imbalances in FL using a combination of techniques including conventional karyotyping, comparative genomic hybridization (CGH) and single nucleotide polymorphism (SNP) technology. The reported recurrent copy number alterations have consistently included losses of 1p32-36, 6q, 10q, and 17p, and gains of 1q, 2p, 7, 9p, 12, 17q, 18q, and X.14-25 The analysis by Hoglund et al utilized computational analysis of a large number of published G-banded FL karyotypes to define early from late accruing genetic imbalances and demonstrated 4 putative pathways of clonal evolution in FL.26 This karyotype-based study was hampered by the inherent inaccuracies of G-banding analysis and excluded from consideration all marker chromosomes and unbalanced chromosomal additions that are common features of FL karyotypes. Further examination of such complex karyotypes by multicolor karyotyping may improve the definition of these recurrent aberrations.27 However, the metaphases typically obtained from short-term lymph node cultures allow only for the detection of DNA imbalances that exceed 5 to 10 Mb in size and may represent only a fraction of the sideline diversity present in FL genomes. No studies to date have utilized a combination of high-resolution genomic analysis and a large FL cohort composed exclusively of diagnostic biopsies.
The advent of array comparative genomic hybridization (array CGH) technologies now provides the capability to detect sub-cytogenetic DNA copy number gains and losses. These techniques have led to improvements in the characterization of both acquired and inherited genetic abnormalities.28 In this study we have applied whole-genome tiling-path BAC-array CGH, with a resolution of at least 200 kb for detection of copy number alterations in clinical specimens and a reported tolerance of up to 70% contamination by nontumor cells,29 to a cohort of 106 FL diagnostic specimens with complete clinical information. We have generated a comprehensive profile of regional copy number imbalances with which to identify significant prognostic correlates in relation to both survival and transformation risk.
Methods
Patient materials
The 106 FL cases were selected from the Lymphoid Cancer Research Database of the British Columbia Cancer Agency (BCCA) in Vancouver, British Columbia, identified between 1987 and 1996 based on the availability of sufficient frozen diagnostic tumor material and information on clinical outcome. Importantly, these cases were enriched in part for cases where 2 or more sequential specimens were available from the indolent phase or when transformation had occurred. Transformation was defined as either histologically proven (biopsy demonstrating large B-cell lymphoma), or clinically proven (one or more of the following: sudden rise in LDH to > twice the normal level, rapid discordant localized nodal enlargement, and new unusual extranodal involvement of organs such as brain, lung and bone). The time to transformation was defined as the time from diagnosis to clinical or pathological end point described above. The International Prognostic Index (IPI) Score was used to risk-stratify these patients because information on the hemoglobin level and number of nodal sites were not available to generate a FLIPI score.30 All cases were classified as FL based on the criteria defined by the World Health Organization classification of tumors of hematopoietic and lymphoid tissues.31 Of the 106 cases, 20 have been included in previously reported studies.24,26 This study was approved by the University of British Columbia IRB, per certificate #H05-60103 and was performed in accordance with the Declaration of Helsinki.
Cytogenetic analysis
Cytogenetic analysis of lymph node specimens was performed as previously described.24 Fluorescence in situ hybridization (FISH) was performed using the LSI IGH/BCL2 probe according to the manufacturer's protocol (Vysis, Downers Grove, IL) to detect the presence of IGH/BCL2 genomic fusion. For validation of deletion of the 1p36.32 locus, the RP13-493G06 or RP11-756P03 BAC clones were selected from the array CGH profile and prepared for use as FISH probes as previously described, while BAC RP11-229M05 at 1q32.3 was used for copy number control.32 For validation of the 6q23.3 locus deletion, the RP11-703G08 BAC was used, while RP11-516E15 at 6p12.3 served as copy number control. All BAC clones had previously been identity-verified by BAC-end sequencing and hybridized to normal metaphases to confirm the expected site of chromosomal localization. The frequency of false deletion for each BAC FISH probe was established by hybridization to normal lymphocyte cell suspensions and ranged from 0.5% to 3.0%. For the purpose of this study the cutoff value for true deletion was set at more than 5%.
DNA extraction
Genomic DNA extraction was performed according to standard procedures using proteinase K digestion and fresh frozen tissue or cells stored at −80°C. The DNA was further purified using the Gentra Puregene Tissue Kit (QIAGEN, Mississauga, ON).
Whole-genome tiling-path BAC-array CGH
The sub-megabase resolution tiling array contains 26 819 BAC clones spotted in duplicate and covers more than 95% of the human genome.33 Array CGH was performed as previously described.34 The array slide was scanned using a charged-couple device camera system to capture the cyanine-3 and cyanine-5 channels (Applied Precision, Issiquah, WA). The images were then analyzed by SoftWoRx microarray analysis software (Applied Precision), followed by a stepwise normalization procedure.35 Microarray data were deposited at Gene Expression Omnibus (GEO), the accession number assigned is: GSE12393. Data were filtered based on both replicate standard deviation (data points with > 0.1 standard deviation removed) and signal to noise ratio (data points with a signal to noise ratio < 3 removed). Copy number alterations were visualized using the “SeeGH” software available at http://www.flintbox.ca/technology.asp?tech = FB312FB.36
Computational analysis
Intersection analysis.
Scoring of array CGH data was performed separately by 2 methods: visual analysis by a cytogeneticist (D.E.H.), using a criterion for an aberration defined as an apparent log-ratio shift away from baseline in a minimum of 3 adjacent BACs (∼200 kb or larger), and computational analysis by determining probability of aberration (loss, neutral, or gain) for each clone using the program CNA-HMMer version 0.1 (available at http://www.cs.ubc.ca/∼sshah/acgh/), which is based on a Hidden Markov Model (HMM).37 Only those alterations identified by both HMM and visual interpretation were accepted as true. We modified the emission model of the HMM described in Shah et al to be a mixture of Student t distributions, achieving the equivalent robustness to outliers while producing output that was more interpretable to the investigator.37 In addition, this modification required fewer hyperparameters to be set, which were selected automatically using an ‘empirical Bayes’–type approach.38 Concordance between the visual calls and the HMM predictions was assessed by calculating the area under the receiver operator characteristic (ROC) curve. ROC curves are a plot of the true positive rate (TPR—proportion of clones called as an aberration that were also predicted by the HMM) against the false positive rate (FPR—proportion of clones predicted as aberrant by the HMM that were not called visually). The area under the ROC curve (AUC) is a single measurement that represents the tradeoff between TPR and FPR. AUC was calculated for each sample. The average AUC in this study was 0.93. A perfect AUC would be 1. All analyses were run using default settings.
Cluster analysis.
Clustering of the 106 cases was performed using the K-medoids (also called partitioning around medoids) algorithm. The input data X(i,j) represented the copy number of clone j in case i. Only clones that showed a 10% rate of recurrent loss or gain determined by intersection analysis were used for clustering. A Hamming distance function of a case to a medoid was used and the algorithm was run 1000 times using random initializations of the medoids. The run producing the lowest total distance of cases to their assigned medoids was reported. The number of clusters was chosen to be 5 based on the previous work by Hoglund et al26
Clinical correlations
The log-rank test using the Kaplan-Meier method was performed for univariate analysis assessing the prognostic significance of each of the 71 regional aberrations on survival and the risk of transformation. Each case was dichotomized as positive or negative for each of the 71 regions, where positive was defined as having at least one alteration in the region. The Cox proportional-hazards model was used to identify only those regions reaching significance independant of the IPI score. All clinical statistical data were computed using the SPSS version 11 software.
Results
Clinical data
The clinical, morphologic, and cytogenetic information on the cohort is presented in Table 1. In brief, 56% were male and 44% were female with a median age of 53 years. Forty-two percent of the patients died, with a median overall survival time of 10.83 years after diagnosis. Overall, 50% of patients had developed transformed lymphoma over a median follow-up time of 7.33 years. The median time to transformation was 6.61 years. The majority of patients who developed transformed lymphoma had biopsy proven transformation (64%). These patients had a similar clinical outcome as those whose transformed lymphoma was diagnosed on clinical grounds. Transformed lymphoma was the cause of death in 64% of patients, supporting the observation that transformation is an important cause of mortality in these patients and may be a confounding factor in assessing the risk of genetic alterations affecting survival in patients who develop transformed lymphoma.
Patient characteristics of 106 FL specimens acquired at diagnosis
| Clinical characteristics . | N = 106 (%) . | Log-rank P value for survival . | Log-rank P value for transformation . |
|---|---|---|---|
| Median age, y | 53 | ||
| Male sex | 59 (56) | .4 | .7 |
| Age > 60 y | 33 (31) | .2 | .5 |
| PS > 1 | 13 (13) | .003 | .3 |
| LDH > normal | 24 (25) | .008 | .1 |
| Extranodal sites > 1 | 13 (12) | .4 | .6 |
| Stage III/IV | 74 (70) | .01 | .1 |
| IPI score | .003 | .02 | |
| 0-1 | 66 (62) | ||
| 2-3 | 34 (32) | ||
| 4-5 | 6 (6) | ||
| Diagnostic pathology | |||
| FOLL1 | 63 (60) | ||
| FOLL2 | 33 (31) | ||
| FOLL3A | 9 (9) | ||
| Primary therapy | |||
| Observation | 26 (24) | ||
| Rad alone | 15 (14) | ||
| Single agent chemo | 12 (11) | ||
| Multi-agent chemo +/− rad | 41 (39) | ||
| Multi-agent chemo + rituximab | 12 (11) | .7 | .7 |
| Outcome | |||
| Transformation | 53 (50) | .02 | |
| Biopsy proven | 34 (64) | .6 | |
| Clinical | 19 (36) | ||
| Death | 45 (42) | ||
| Unrelated | 3 (7) | ||
| From transformation | 29 (64) | ||
| From progressive indolent FL | 13 (29) | ||
| Median follow-up alive = 7.33 y | |||
| Median overall survival = 10.83 y | |||
| Median time to transformation = 6.61 y |
| Clinical characteristics . | N = 106 (%) . | Log-rank P value for survival . | Log-rank P value for transformation . |
|---|---|---|---|
| Median age, y | 53 | ||
| Male sex | 59 (56) | .4 | .7 |
| Age > 60 y | 33 (31) | .2 | .5 |
| PS > 1 | 13 (13) | .003 | .3 |
| LDH > normal | 24 (25) | .008 | .1 |
| Extranodal sites > 1 | 13 (12) | .4 | .6 |
| Stage III/IV | 74 (70) | .01 | .1 |
| IPI score | .003 | .02 | |
| 0-1 | 66 (62) | ||
| 2-3 | 34 (32) | ||
| 4-5 | 6 (6) | ||
| Diagnostic pathology | |||
| FOLL1 | 63 (60) | ||
| FOLL2 | 33 (31) | ||
| FOLL3A | 9 (9) | ||
| Primary therapy | |||
| Observation | 26 (24) | ||
| Rad alone | 15 (14) | ||
| Single agent chemo | 12 (11) | ||
| Multi-agent chemo +/− rad | 41 (39) | ||
| Multi-agent chemo + rituximab | 12 (11) | .7 | .7 |
| Outcome | |||
| Transformation | 53 (50) | .02 | |
| Biopsy proven | 34 (64) | .6 | |
| Clinical | 19 (36) | ||
| Death | 45 (42) | ||
| Unrelated | 3 (7) | ||
| From transformation | 29 (64) | ||
| From progressive indolent FL | 13 (29) | ||
| Median follow-up alive = 7.33 y | |||
| Median overall survival = 10.83 y | |||
| Median time to transformation = 6.61 y |
PS indicates ECOG performance status; LDH, lactate dehydrogenase; IPI, International Prognostic Index; FOLL1, follicular lymphoma grade 1; FOLL2, grade 2; FOLL3A, grade 3A; rad, radiation; chemo, chemotherapy; and FL, follicular lymphoma.
Treatment of these patients varied due to changes in era-specific approaches to management (Table 1). The effect of the addition of rituximab to standard chemotherapy could not be assessed reliably because of small numbers of patients (n = 12; log-rank test on overall survival and transformation, P = .7), recent incorporation of rituximab into primary treatment (after 2004) varying times of introduction (diagnosis, first progression, relapse, multiple relapses), and variable combination with standard agents (single agent or combination in multiple drug regimens).
Cytogenetic data
Ninety-three of the 106 cases had been studied by karyotype analysis and/or FISH using the IGH/BCL2 fusion probe. The t(14;18) or variant was present in 75 of these 93 cases (81%) but was absent in 18. Thirteen cases were not investigated by these techniques but had the standard morphologic features of FL.
Profile of copy number alterations in FL
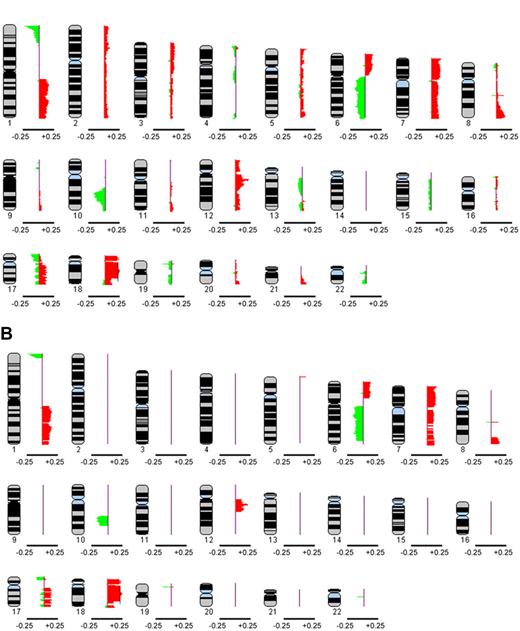
Each array CGH profile was annotated individually by visual inspection and by computation without knowledge of the associated karyotype. The individual profiles were combined to generate a genome-wide copy number profile of the 106 diagnostic FL specimens. Figure 1 A represents a global composite profile ideogram of all aberrations affecting the 22 autosomes as determined by the intersection analysis. Figure 1B shows an ideogram of only those regions that were affected in at least 10% of cases. This 10% cutoff produced 71 altered regions ranging in size from 200 kb to 44 Mb. Overall, 97 of 106 cases (91.5%) had aberrations detected by array CGH with a median of 16.1% and a range from 0% to 32.2%. The most frequently altered region was band 1p36.22-p36.33 (∼11 Mb in size), showing 25.5% frequency of deletion. Table 2 provides details on the 71 regions of alteration. Figure S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article) provides a representative array CGH profile of an individual. Array CGH raw data of the 106 patients are included in the supplemental data.
Composite frequency ideogram plot of genome-wide copy number alterations in 106 diagnostic FL cases based on intersection analysis. (A) The frequencies of aberrations, represented by green signals for losses and red signals for gains, in the autosomes were derived from intersection analysis, where the union was taken between calls made visually by a cytogenetic pathologist and those determined by CNA-HMMer version 0.1. (B) Composite frequency ideogram plot showing only those aberrations affecting at least 10% of cases. The data were visualized using the SeeGH software. Genetic losses or gains are represented by green and red signals, respectively. The horizontal bar below each ideogram represents gain and loss frequencies of +0.25 and −0.25, respectively.
Composite frequency ideogram plot of genome-wide copy number alterations in 106 diagnostic FL cases based on intersection analysis. (A) The frequencies of aberrations, represented by green signals for losses and red signals for gains, in the autosomes were derived from intersection analysis, where the union was taken between calls made visually by a cytogenetic pathologist and those determined by CNA-HMMer version 0.1. (B) Composite frequency ideogram plot showing only those aberrations affecting at least 10% of cases. The data were visualized using the SeeGH software. Genetic losses or gains are represented by green and red signals, respectively. The horizontal bar below each ideogram represents gain and loss frequencies of +0.25 and −0.25, respectively.
Detailed information on the 71 regional aberrations affecting at least 10% of FL cases in intersection analysis (data based on NCBI build 36.1)
| A . | B . | C . | D . | E . | F . | G . | H . | I . | J . | M . | N . | O . | P . | Q . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr . | ID no. . | Start (bp) . | End (bp) . | Chr band . | Upper flanking clone . | Lower flanking clone . | No. clones . | Size (bp) . | Frequency of aberration . | Univariate survival (P value) . | Cox survival with IPI (P value) . | Univariate transformation (P value) . | Cox transformation with IPI (P value) . | Genes of interest . |
| 1p− | 1 | 1 120 588 | 12 511 370 | p36.22-p36.33 | RP11-158F02 | RP11-333P22 | 95 | 11 390 782 | 0.25 | .012 | .023 | .004 | .006 | |
| 1q+ | 2 | 144 203 231 | 149 149 179 | q21.1-q25.1 | RP11-026E04 | RP11-724H12 | 46 | 4 945 948 | 0.13 | NS | NS | |||
| 3 | 149 346 998 | 171 525 137 | q21.13-q25.1 | RP11-068I18 | RP11-723L18 | 206 | 22 178 139 | 0.16 | NS | NS | ||||
| 4 | 173 318 349 | 186 631 007 | q25.1-q31.1 | RP11-288P06 | RP11-076I10 | 111 | 13 312 658 | 0.14 | .039 | NS | NS | |||
| 5 | 192 104 220 | 215 373 858 | q31.2-q41 | RP11-167B22 | RP11-260A12 | 239 | 23 269 638 | 0.17 | NS | NS | ||||
| 6 | 216 564 705 | 223 861 673 | q41-q42.12 | RP11-680C18 | RP11-797E11 | 61 | 7 296 968 | 0.12 | .049 | .041 | NS | |||
| 7 | 225 671 030 | 233 337 746 | q42.13-q42.3 | CTD-2063G05 | RP13-539L11 | 60 | 7 666 716 | 0.12 | .049 | .041 | NS | |||
| 8 | 236 546 601 | 244 497 402 | q43-q44 | RP11-102F13 | RP11-744G19 | 86 | 7 950 801 | 0.11 | .014 | .018 | NS | |||
| 9 | 246 412 897 | 246 741 718 | q44 | RP11-462C05 | RP11-059M10 | 2 | 328 821 | 0.10 | NS | NS | ||||
| 5p+ | 10 | 568 397 | 2 059 719 | p15.33 | RP11-773M18 | RP11-369A06 | 13 | 1 491 322 | 0.10 | .003 | .002 | NS | ||
| 6p+ | 11 | 101 435 | 8 166 300 | p24.3-p25.3 | RP11-812K10 | CTD-2145K22 | 73 | 8 064 865 | 0.12 | NS | .010 | .023 | ||
| 12 | 9 910 293 | 15 045 693 | p23-p24.3 | RP11-240G16 | RP11-698I24 | 52 | 5 135 400 | 0.11 | NS | .002 | .009 | |||
| 13 | 15 322 212 | 17 396 937 | p22.3-p23 | RP11-277N23 | RP11-772A24 | 22 | 2 074 725 | 0.11 | NS | .002 | .009 | |||
| 14 | 18 182 851 | 19 088 319 | p22.3 | CTD-2300B02 | RP11-641M20 | 10 | 905 468 | 0.10 | NS | .000 | .001 | |||
| 15 | 19 741 746 | 23 462 851 | p22.3 | RP11-702J22 | CTD-2325C20 | 36 | 3 721 105 | 0.10 | NS | .000 | .001 | |||
| 16 | 23 588 018 | 37 906 515 | p21.2-p22.2 | RP11-063M15 | RP11-644C02 | 151 | 14 318 497 | 0.14 | NS | .006 | .009 | |||
| 17 | 39 096 919 | 42 527 171 | p21.1-p21.2 | RP11-685F06 | CTD-2125F20 | 38 | 3 430 252 | 0.11 | NS | .028 | .046 | |||
| 18 | 42 763 847 | 44 815 780 | p21.1 | RP11-711C14 | RP11-079F13 | 18 | 2 051 933 | 0.10 | .035 | NS | .008 | .007 | ||
| 6q− | 19 | 67 856 203 | 102 485 267 | q12-q16.3 | RP11-616K23 | RP11-543M18 | 329 | 34 629 064 | 0.17 | .003 | .007 | NS | ||
| 20 | 104 959 688 | 145 932 243 | q21-q24.3 | RP11-639C21 | RP11-724O10 | 406 | 40 972 555 | 0.17 | .016 | .038 | .002 | .001 | ||
| 21 | 146 006 611 | 158 842 383 | q24.3-q25.3 | RP11-272E07 | CTD-2245D13 | 132 | 12 835 772 | 0.12 | NS | NS | ||||
| 7p+ | 22 | 76 475 | 9 330 205 | p21.3-p22.3 | RP11-379K15 | RP11-646H18 | 79 | 9 253 730 | 0.16 | .007 | NS | NS | ||
| 23 | 11 733 679 | 12 411 228 | p21.3 | RP11-720O24 | CTD-2028D24 | 12 | 677 549 | 0.11 | .021 | NS | NS | |||
| 24 | 13 834 918 | 57 826 849 | p11.1-p21.2 | RP11-708O01 | RP11-415F22 | 408 | 43 991 931 | 0.15 | .049 | NS | NS | |||
| 7q+ | 25 | 65 183 346 | 75 792 082 | q11.21-q11.23 | RP11-763P10 | RP11-818A14 | 91 | 10 608 736 | 0.11 | NS | NS | |||
| 26 | 77 519 343 | 79 287 368 | q21.11 | RP11-157F12 | RP11-327P19 | 17 | 1 768 025 | 0.11 | NS | NS | ||||
| 27 | 79 508 881 | 81 760 593 | q21.11 | RP11-176M05 | RP11-459E23 | 17 | 2 251 712 | 0.11 | NS | NS | ||||
| 28 | 82 203 999 | 82 396 484 | q21.11 | RP11-056M07 | RP11-562O03 | 2 | 192 485 | 0.11 | NS | NS | ||||
| 29 | 84 076 954 | 88 755 109 | q21.11-q21.13 | RP11-750F10 | RP11-693A17 | 44 | 4 678 155 | 0.11 | NS | NS | ||||
| 30 | 89 067 084 | 95 807 510 | q21.13-q21.3 | RP11-734C11 | RP11-814L20 | 57 | 6 740 426 | 0.11 | NS | NS | CYP51 | |||
| 31 | 97 314 220 | 99 254 745 | q21.3-q22.1 | RP11-380G21 | RP11-757A13 | 15 | 1 940 525 | 0.10 | NS | NS | ||||
| 32 | 101 780 109 | 102 036 700 | q22.1 | RP11-342G18 | RP11-676J21 | 2 | 256 591 | 0.10 | NS | NS | ||||
| 33 | 107 062 787 | 108 434 145 | q22.3-q31.1 | RP11-467F09 | RP11-372G06 | 12 | 1 371 358 | 0.10 | NS | NS | ||||
| 34 | 114 602 302 | 115 391 679 | q31.2 | RP11-103A01 | RP11-022K23 | 6 | 789 377 | 0.10 | NS | NS | ||||
| 35 | 115 770 526 | 117 527 587 | q31.2-q31.31 | RP11-691L23 | RP11-099N14 | 15 | 1 757 061 | 0.10 | NS | NS | ||||
| 36 | 119 932 154 | 124 328 532 | q31.31-q31.33 | RP11-367A17 | RP11-661D08 | 39 | 4 396 378 | 0.10 | NS | NS | ||||
| 37 | 124 779 872 | 128 206 833 | q31.33-q32.1 | RP11-818I12 | RP11-526K05 | 30 | 3 426 961 | 0.11 | NS | NS | ||||
| 38 | 131 217 474 | 135 972 389 | q32.3-q33 | RP11-193I17 | RP11-811K22 | 50 | 4 754 915 | 0.11 | NS | .034 | NS | |||
| 39 | 136 860 444 | 140 073 345 | q33-q34 | RP11-019L13 | RP11-788O06 | 27 | 3 212 901 | 0.11 | NS | NS | ||||
| 40 | 140 508 716 | 144 516 051 | q34-q35 | CTD-2253O07 | RP11-409M07 | 41 | 4 007 335 | 0.12 | NS | NS | ||||
| 41 | 147 152 251 | 151 714 055 | q35-q36.1 | RP11-564O04 | RP11-208G20 | 43 | 4 561 804 | 0.12 | NS | NS | ||||
| 42 | 151 938 143 | 158 777 885 | q36.1-q36.3 | RP11-452J20 | RP11-083D03 | 57 | 6 839 742 | 0.12 | NS | NS | ||||
| 42 | 151 938 143 | 158 777 885 | q36.1-q36.3 | RP11-452J20 | RP11-083D03 | 57 | 6 839 742 | 0.12 | NS | NS | ||||
| 8q+ | 43 | 86 699 540 | 86 900 037 | q21.2 | RP11-198E24 | RP11-048I22 | 5 | 207 338 | 0.12 | .046 | .020 | NS | ||
| 44 | 127 567 638 | 144 615 332 | q24.13-q24.3 | RP11-435M05 | RP13-618I02 | 140 | 17 047 694 | 0.16 | NS | NS | KCNK9, NIBP, PTK2/FAK, PTP4A3 | |||
| 10q− | 45 | 83 519 320 | 109 743 759 | q23.1-q25.1 | RP11-535H04 | RP11-799H24 | 238 | 26 224 439 | 0.20 | NS | NS | PTEN | ||
| 12q+ | 46 | 39 241 592 | 40 479 848 | q12 | RP11-425I22 | RP11-810RP11-4 | 13 | 1 238 256 | 0.10 | NS | NS | |||
| 47 | 41 164 595 | 41 849 783 | q12 | RP11-702F02 | RP11-118A03 | 6 | 685 188 | 0.10 | NS | NS | ||||
| 48 | 42 875 295 | 43 431 071 | q12 | RP11-462E15 | RP11-374RP11-9 | 4 | 555 776 | 0.10 | NS | NS | ||||
| 49 | 45 222 971 | 47 260 459 | q13.11 | RP11-755G20 | RP11-810G14 | 21 | 2 037 488 | 0.12 | NS | NS | ||||
| 50 | 47 315 713 | 48 862 941 | q13.11-q13.13 | RP11-270J09 | RP11-411RP11-4 | 15 | 1 547 228 | 0.11 | NS | NS | ||||
| 51 | 49 766 128 | 53 769 742 | q13.13-q13.2 | RP11-573G09 | RP11-321D04 | 39 | 4 003 614 | 0.21 | NS | .0380 | NS | |||
| 52 | 54 124 295 | 71 253 094 | q13.2-q21.1 | RP11-222A15 | RP11-349G03 | 170 | 17 128 799 | 0.17 | NS | NS | MDM2 | |||
| 17p− | 53 | 433 730 | 5 025 418 | p13.2-p13.3 | RP11-411G07 | CTD-2103L01 | 42 | 4 591 688 | 0.13 | NS | NS | |||
| 54 | 5 418 209 | 6 976 115 | p13.1-p13.2 | RP11-328G09 | RP11-417F20 | 9 | 1 557 906 | 0.11 | .021 | .021 | NS | |||
| 55 | 7 297 479 | 7 963 869 | p13.1 | RP13-626G05 | CTD-2205J22 | 6 | 666 390 | 0.10 | .019 | .010 | NS | p53 | ||
| 56 | 7 873 762 | 8 172 436 | p13.1 | RP11-613J07 | RP11-452D01 | 2 | 298 674 | 0.11 | NS | NS | ||||
| 57 | 8 633 350 | 9 331 199 | p13.1 | RP11-476A23 | RP11-165P17 | 7 | 697 849 | 0.11 | NS | NS | ||||
| 17q+ | 58 | 28 335 370 | 34 341 092 | q11.2-q12 | RP11-626C08 | RP11-154N09 | 49 | 6 005 722 | 0.12 | .003 | .017 | NS | ||
| 59 | 35 115 643 | 37 009 923 | q12-q21.2 | RP11-610O22 | RP11-619I14 | 16 | 1 894 280 | 0.12 | .003 | .017 | NS | |||
| 60 | 40 291 911 | 49 011 357 | q21.31-q22 | RP11-419E16 | RP11-577I13 | 89 | 8 719 446 | 0.13 | .012 | .037 | NS | |||
| 61 | 50 468 106 | 53 853 636 | q22 | RP11-767L04 | RP11-620A23 | 33 | 3 385 530 | 0.12 | NS | NS | ZNF161 | |||
| 62 | 58 325 527 | 62 428 590 | q23.2-q24.2 | RP11-579J02 | RP11-622E22 | 48 | 4 103 063 | 0.13 | .031 | .019 | NS | |||
| 63 | 64 191 239 | 69 931 286 | q24.2-q25.1 | RP11-618E04 | RP11-478P05 | 50 | 5 740 047 | 0.12 | .007 | .007 | NS | |||
| 64 | 73 418 360 | 77 524 868 | q25.3 | RP11-102I17 | RP11-712H22 | 34 | 4 106 508 | 0.11 | .021 | .017 | NS | |||
| 18p+ | 65 | 35 421 | 15 060 997 | p11.21-p11.32 | RP11-683L23 | RP11-692N17 | 112 | 15 025 576 | 0.23 | NS | NS | |||
| 18q+ | 66 | 16 793 577 | 59 123 838 | q11.1-q21.33 | RP11-063P19 | RP11-796C21 | 370 | 42 330 261 | 0.25 | NS | NS | BCL2 | ||
| 67 | 60 284 518 | 61 729 287 | q22.1 | RP11-252H14 | RP11-284D06 | 11 | 1 444 769 | 0.10 | NS | NS | ||||
| 68 | 63 894 258 | 64 156 108 | q22.1 | RP11-255L11 | RP11-261M15 | 2 | 261 850 | 0.10 | NS | NS | ||||
| 69 | 70 236 233 | 76 098 439 | q22.3-q23 | RP11-277E17 | RP11-565D23 | 50 | 5 862 206 | 0.12 | NS | NS | ||||
| 19p− | 70 | 8 566 514 | 8 784 792 | p13.2 | RP11-092E05 | RP11-014H08 | 6 | 218 278 | 0.13 | NS | NS | |||
| 22q− | 71 | 21 112 875 | 21 296 725 | q11.22 | RP11-757F24 | RP11-757F24 | 1 | 183 850 | 0.10 | NS | NS |
| A . | B . | C . | D . | E . | F . | G . | H . | I . | J . | M . | N . | O . | P . | Q . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr . | ID no. . | Start (bp) . | End (bp) . | Chr band . | Upper flanking clone . | Lower flanking clone . | No. clones . | Size (bp) . | Frequency of aberration . | Univariate survival (P value) . | Cox survival with IPI (P value) . | Univariate transformation (P value) . | Cox transformation with IPI (P value) . | Genes of interest . |
| 1p− | 1 | 1 120 588 | 12 511 370 | p36.22-p36.33 | RP11-158F02 | RP11-333P22 | 95 | 11 390 782 | 0.25 | .012 | .023 | .004 | .006 | |
| 1q+ | 2 | 144 203 231 | 149 149 179 | q21.1-q25.1 | RP11-026E04 | RP11-724H12 | 46 | 4 945 948 | 0.13 | NS | NS | |||
| 3 | 149 346 998 | 171 525 137 | q21.13-q25.1 | RP11-068I18 | RP11-723L18 | 206 | 22 178 139 | 0.16 | NS | NS | ||||
| 4 | 173 318 349 | 186 631 007 | q25.1-q31.1 | RP11-288P06 | RP11-076I10 | 111 | 13 312 658 | 0.14 | .039 | NS | NS | |||
| 5 | 192 104 220 | 215 373 858 | q31.2-q41 | RP11-167B22 | RP11-260A12 | 239 | 23 269 638 | 0.17 | NS | NS | ||||
| 6 | 216 564 705 | 223 861 673 | q41-q42.12 | RP11-680C18 | RP11-797E11 | 61 | 7 296 968 | 0.12 | .049 | .041 | NS | |||
| 7 | 225 671 030 | 233 337 746 | q42.13-q42.3 | CTD-2063G05 | RP13-539L11 | 60 | 7 666 716 | 0.12 | .049 | .041 | NS | |||
| 8 | 236 546 601 | 244 497 402 | q43-q44 | RP11-102F13 | RP11-744G19 | 86 | 7 950 801 | 0.11 | .014 | .018 | NS | |||
| 9 | 246 412 897 | 246 741 718 | q44 | RP11-462C05 | RP11-059M10 | 2 | 328 821 | 0.10 | NS | NS | ||||
| 5p+ | 10 | 568 397 | 2 059 719 | p15.33 | RP11-773M18 | RP11-369A06 | 13 | 1 491 322 | 0.10 | .003 | .002 | NS | ||
| 6p+ | 11 | 101 435 | 8 166 300 | p24.3-p25.3 | RP11-812K10 | CTD-2145K22 | 73 | 8 064 865 | 0.12 | NS | .010 | .023 | ||
| 12 | 9 910 293 | 15 045 693 | p23-p24.3 | RP11-240G16 | RP11-698I24 | 52 | 5 135 400 | 0.11 | NS | .002 | .009 | |||
| 13 | 15 322 212 | 17 396 937 | p22.3-p23 | RP11-277N23 | RP11-772A24 | 22 | 2 074 725 | 0.11 | NS | .002 | .009 | |||
| 14 | 18 182 851 | 19 088 319 | p22.3 | CTD-2300B02 | RP11-641M20 | 10 | 905 468 | 0.10 | NS | .000 | .001 | |||
| 15 | 19 741 746 | 23 462 851 | p22.3 | RP11-702J22 | CTD-2325C20 | 36 | 3 721 105 | 0.10 | NS | .000 | .001 | |||
| 16 | 23 588 018 | 37 906 515 | p21.2-p22.2 | RP11-063M15 | RP11-644C02 | 151 | 14 318 497 | 0.14 | NS | .006 | .009 | |||
| 17 | 39 096 919 | 42 527 171 | p21.1-p21.2 | RP11-685F06 | CTD-2125F20 | 38 | 3 430 252 | 0.11 | NS | .028 | .046 | |||
| 18 | 42 763 847 | 44 815 780 | p21.1 | RP11-711C14 | RP11-079F13 | 18 | 2 051 933 | 0.10 | .035 | NS | .008 | .007 | ||
| 6q− | 19 | 67 856 203 | 102 485 267 | q12-q16.3 | RP11-616K23 | RP11-543M18 | 329 | 34 629 064 | 0.17 | .003 | .007 | NS | ||
| 20 | 104 959 688 | 145 932 243 | q21-q24.3 | RP11-639C21 | RP11-724O10 | 406 | 40 972 555 | 0.17 | .016 | .038 | .002 | .001 | ||
| 21 | 146 006 611 | 158 842 383 | q24.3-q25.3 | RP11-272E07 | CTD-2245D13 | 132 | 12 835 772 | 0.12 | NS | NS | ||||
| 7p+ | 22 | 76 475 | 9 330 205 | p21.3-p22.3 | RP11-379K15 | RP11-646H18 | 79 | 9 253 730 | 0.16 | .007 | NS | NS | ||
| 23 | 11 733 679 | 12 411 228 | p21.3 | RP11-720O24 | CTD-2028D24 | 12 | 677 549 | 0.11 | .021 | NS | NS | |||
| 24 | 13 834 918 | 57 826 849 | p11.1-p21.2 | RP11-708O01 | RP11-415F22 | 408 | 43 991 931 | 0.15 | .049 | NS | NS | |||
| 7q+ | 25 | 65 183 346 | 75 792 082 | q11.21-q11.23 | RP11-763P10 | RP11-818A14 | 91 | 10 608 736 | 0.11 | NS | NS | |||
| 26 | 77 519 343 | 79 287 368 | q21.11 | RP11-157F12 | RP11-327P19 | 17 | 1 768 025 | 0.11 | NS | NS | ||||
| 27 | 79 508 881 | 81 760 593 | q21.11 | RP11-176M05 | RP11-459E23 | 17 | 2 251 712 | 0.11 | NS | NS | ||||
| 28 | 82 203 999 | 82 396 484 | q21.11 | RP11-056M07 | RP11-562O03 | 2 | 192 485 | 0.11 | NS | NS | ||||
| 29 | 84 076 954 | 88 755 109 | q21.11-q21.13 | RP11-750F10 | RP11-693A17 | 44 | 4 678 155 | 0.11 | NS | NS | ||||
| 30 | 89 067 084 | 95 807 510 | q21.13-q21.3 | RP11-734C11 | RP11-814L20 | 57 | 6 740 426 | 0.11 | NS | NS | CYP51 | |||
| 31 | 97 314 220 | 99 254 745 | q21.3-q22.1 | RP11-380G21 | RP11-757A13 | 15 | 1 940 525 | 0.10 | NS | NS | ||||
| 32 | 101 780 109 | 102 036 700 | q22.1 | RP11-342G18 | RP11-676J21 | 2 | 256 591 | 0.10 | NS | NS | ||||
| 33 | 107 062 787 | 108 434 145 | q22.3-q31.1 | RP11-467F09 | RP11-372G06 | 12 | 1 371 358 | 0.10 | NS | NS | ||||
| 34 | 114 602 302 | 115 391 679 | q31.2 | RP11-103A01 | RP11-022K23 | 6 | 789 377 | 0.10 | NS | NS | ||||
| 35 | 115 770 526 | 117 527 587 | q31.2-q31.31 | RP11-691L23 | RP11-099N14 | 15 | 1 757 061 | 0.10 | NS | NS | ||||
| 36 | 119 932 154 | 124 328 532 | q31.31-q31.33 | RP11-367A17 | RP11-661D08 | 39 | 4 396 378 | 0.10 | NS | NS | ||||
| 37 | 124 779 872 | 128 206 833 | q31.33-q32.1 | RP11-818I12 | RP11-526K05 | 30 | 3 426 961 | 0.11 | NS | NS | ||||
| 38 | 131 217 474 | 135 972 389 | q32.3-q33 | RP11-193I17 | RP11-811K22 | 50 | 4 754 915 | 0.11 | NS | .034 | NS | |||
| 39 | 136 860 444 | 140 073 345 | q33-q34 | RP11-019L13 | RP11-788O06 | 27 | 3 212 901 | 0.11 | NS | NS | ||||
| 40 | 140 508 716 | 144 516 051 | q34-q35 | CTD-2253O07 | RP11-409M07 | 41 | 4 007 335 | 0.12 | NS | NS | ||||
| 41 | 147 152 251 | 151 714 055 | q35-q36.1 | RP11-564O04 | RP11-208G20 | 43 | 4 561 804 | 0.12 | NS | NS | ||||
| 42 | 151 938 143 | 158 777 885 | q36.1-q36.3 | RP11-452J20 | RP11-083D03 | 57 | 6 839 742 | 0.12 | NS | NS | ||||
| 42 | 151 938 143 | 158 777 885 | q36.1-q36.3 | RP11-452J20 | RP11-083D03 | 57 | 6 839 742 | 0.12 | NS | NS | ||||
| 8q+ | 43 | 86 699 540 | 86 900 037 | q21.2 | RP11-198E24 | RP11-048I22 | 5 | 207 338 | 0.12 | .046 | .020 | NS | ||
| 44 | 127 567 638 | 144 615 332 | q24.13-q24.3 | RP11-435M05 | RP13-618I02 | 140 | 17 047 694 | 0.16 | NS | NS | KCNK9, NIBP, PTK2/FAK, PTP4A3 | |||
| 10q− | 45 | 83 519 320 | 109 743 759 | q23.1-q25.1 | RP11-535H04 | RP11-799H24 | 238 | 26 224 439 | 0.20 | NS | NS | PTEN | ||
| 12q+ | 46 | 39 241 592 | 40 479 848 | q12 | RP11-425I22 | RP11-810RP11-4 | 13 | 1 238 256 | 0.10 | NS | NS | |||
| 47 | 41 164 595 | 41 849 783 | q12 | RP11-702F02 | RP11-118A03 | 6 | 685 188 | 0.10 | NS | NS | ||||
| 48 | 42 875 295 | 43 431 071 | q12 | RP11-462E15 | RP11-374RP11-9 | 4 | 555 776 | 0.10 | NS | NS | ||||
| 49 | 45 222 971 | 47 260 459 | q13.11 | RP11-755G20 | RP11-810G14 | 21 | 2 037 488 | 0.12 | NS | NS | ||||
| 50 | 47 315 713 | 48 862 941 | q13.11-q13.13 | RP11-270J09 | RP11-411RP11-4 | 15 | 1 547 228 | 0.11 | NS | NS | ||||
| 51 | 49 766 128 | 53 769 742 | q13.13-q13.2 | RP11-573G09 | RP11-321D04 | 39 | 4 003 614 | 0.21 | NS | .0380 | NS | |||
| 52 | 54 124 295 | 71 253 094 | q13.2-q21.1 | RP11-222A15 | RP11-349G03 | 170 | 17 128 799 | 0.17 | NS | NS | MDM2 | |||
| 17p− | 53 | 433 730 | 5 025 418 | p13.2-p13.3 | RP11-411G07 | CTD-2103L01 | 42 | 4 591 688 | 0.13 | NS | NS | |||
| 54 | 5 418 209 | 6 976 115 | p13.1-p13.2 | RP11-328G09 | RP11-417F20 | 9 | 1 557 906 | 0.11 | .021 | .021 | NS | |||
| 55 | 7 297 479 | 7 963 869 | p13.1 | RP13-626G05 | CTD-2205J22 | 6 | 666 390 | 0.10 | .019 | .010 | NS | p53 | ||
| 56 | 7 873 762 | 8 172 436 | p13.1 | RP11-613J07 | RP11-452D01 | 2 | 298 674 | 0.11 | NS | NS | ||||
| 57 | 8 633 350 | 9 331 199 | p13.1 | RP11-476A23 | RP11-165P17 | 7 | 697 849 | 0.11 | NS | NS | ||||
| 17q+ | 58 | 28 335 370 | 34 341 092 | q11.2-q12 | RP11-626C08 | RP11-154N09 | 49 | 6 005 722 | 0.12 | .003 | .017 | NS | ||
| 59 | 35 115 643 | 37 009 923 | q12-q21.2 | RP11-610O22 | RP11-619I14 | 16 | 1 894 280 | 0.12 | .003 | .017 | NS | |||
| 60 | 40 291 911 | 49 011 357 | q21.31-q22 | RP11-419E16 | RP11-577I13 | 89 | 8 719 446 | 0.13 | .012 | .037 | NS | |||
| 61 | 50 468 106 | 53 853 636 | q22 | RP11-767L04 | RP11-620A23 | 33 | 3 385 530 | 0.12 | NS | NS | ZNF161 | |||
| 62 | 58 325 527 | 62 428 590 | q23.2-q24.2 | RP11-579J02 | RP11-622E22 | 48 | 4 103 063 | 0.13 | .031 | .019 | NS | |||
| 63 | 64 191 239 | 69 931 286 | q24.2-q25.1 | RP11-618E04 | RP11-478P05 | 50 | 5 740 047 | 0.12 | .007 | .007 | NS | |||
| 64 | 73 418 360 | 77 524 868 | q25.3 | RP11-102I17 | RP11-712H22 | 34 | 4 106 508 | 0.11 | .021 | .017 | NS | |||
| 18p+ | 65 | 35 421 | 15 060 997 | p11.21-p11.32 | RP11-683L23 | RP11-692N17 | 112 | 15 025 576 | 0.23 | NS | NS | |||
| 18q+ | 66 | 16 793 577 | 59 123 838 | q11.1-q21.33 | RP11-063P19 | RP11-796C21 | 370 | 42 330 261 | 0.25 | NS | NS | BCL2 | ||
| 67 | 60 284 518 | 61 729 287 | q22.1 | RP11-252H14 | RP11-284D06 | 11 | 1 444 769 | 0.10 | NS | NS | ||||
| 68 | 63 894 258 | 64 156 108 | q22.1 | RP11-255L11 | RP11-261M15 | 2 | 261 850 | 0.10 | NS | NS | ||||
| 69 | 70 236 233 | 76 098 439 | q22.3-q23 | RP11-277E17 | RP11-565D23 | 50 | 5 862 206 | 0.12 | NS | NS | ||||
| 19p− | 70 | 8 566 514 | 8 784 792 | p13.2 | RP11-092E05 | RP11-014H08 | 6 | 218 278 | 0.13 | NS | NS | |||
| 22q− | 71 | 21 112 875 | 21 296 725 | q11.22 | RP11-757F24 | RP11-757F24 | 1 | 183 850 | 0.10 | NS | NS |
NS indicates not significant.
Association between copy number alterations and clinical parameters
When using the relative number of alterations per case to predict clinical outcome, defined as the number of altered BAC clones determined by intersection analysis divided by the total BAC clones (26 819) expressed as a percentage, we showed that there was no significant correlation between cases with at least 10% alterations and those with no more than 10% in terms of overall survival and transformation risk (log-rank test, P = .7 and P = .06, respectively). However, significant correlation with overall survival and transformation risk was observed if cases with at least 5% alterations were compared with those containing no more than 5% (log-rank test, P = .02 and P = .03, respectively).
Univariate analysis indicated that 21 regions correlated significantly with poor survival (column M of Table 2), as did performance status, LDH, stage, and the IPI group (Table 1). Of these, 16 regions were independent of IPI in multivariate analysis (column N of Table 2). Univariate analysis showed that 12 regions correlated significantly with risk of transformation (column O of Table 2). The IPI group, but not the individual factors, was also predictive of transformation (Table 1). Ten of the 12 regions were identified as IPI-independent predictors of risk of transformation in multivariate analysis (column P of Table 2). Del(1)(p36.22-p36.33) and del(6)(q21-q24.3) (identified by ID numbers 1 and 20, respectively) were not only associated with transformation and inferior outcome (Figure 2A,B), but were also IPI-independent predictors for both clinical variables (shown in bold in Table 2) and thus were selected as candidate regions for validation.
Correlation analysis of the 71 regional aberrations with clinical data. (A) Kaplan-Meier survival of ID numbers 1 [del(1)(p36.22-p36.33)] and 20 [del(6)(q21-q24.3)], and (B) transformation graphs of ID numbers 1 [del(1)(p36.22-p36.33)] and 20 [del(6)(q21-q24.3)]. Log-rank test was performed to assess significance (P ≤ .05).
Correlation analysis of the 71 regional aberrations with clinical data. (A) Kaplan-Meier survival of ID numbers 1 [del(1)(p36.22-p36.33)] and 20 [del(6)(q21-q24.3)], and (B) transformation graphs of ID numbers 1 [del(1)(p36.22-p36.33)] and 20 [del(6)(q21-q24.3)]. Log-rank test was performed to assess significance (P ≤ .05).
Validation of array CGH data
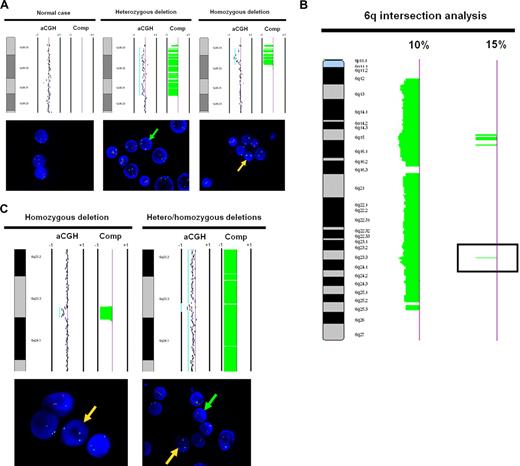
Two BAC clones, RP13-493G06 and RP11-756P03, that mapped to 1p36.32 and spaced approximately 200 kb apart, were used for validation of the array CGH-detected 1p36 deletion. We performed FISH using these BAC clones on 10 selected cases. The RP11-229M05 probe at 1q32.3 was used as a control. Two of the 10 cases were determined by CGH intersection analysis to have no log ratio shift (no alteration), while 8 cases had evident deletions at 1p36.3 of variable size. The concordance rate between FISH and intersection analysis was 10 of 10 cases. Figure 3A shows the array CGH ideogram and FISH results for 3 representative cases, 1 without a deletion, 1 with a heterozygous deletion, and 1 with a homozygous deletion.
Array CGH and FISH correlation of 1p36.3 and 6q23.3. (A) FISH validation of the 1p36.22-p36.33 region (ID no. 1), which presented significant correlation with clinical outcome. Array CGH (top panel) and FISH (bottom panel) demonstrating a case without deletion at 1p36.3 (normal), a case with heterozygous deletion at 1p36.3, and a case with homozygous deletion at 1p36.3. The 1p36.32 probe was labeled red, while the control probe at 1q32 was labeled green. (B) A composite-array CGH ideogram profile of 6q alterations affecting at least 10% and at least 15% of FL cases was generated by intersection analysis. The alteration indicated by the black box in the 15% ideogram corresponds to the 6q23.3 region targeted for FISH validation. (C) FISH validation of the 6q23.3 region (ID no. 20), which presented significant correlation with clinical outcome. Array CGH (top panel) and FISH (bottom panel) demonstrating a case with homozygous deletion and a case with homozygous deletion at 6q23.3 and proximal and distal heterozygous deletion. The 1p36.3 and 6q23.3 probes were labeled red, while the control probes at 1q32.3 and 6p12.3 were labeled green. Green arrows in FISH indicate the presence of heterozygous deletion, while yellow arrows indicate the presence of homozygous deletion. For array CGH, each dot represents a BAC clone and the light blue lines represent visual calling of aberrations. Loss is indicated by a shift to the left of center and gain by a shift to the right of center. Vertical lines are −1 and + 1 scale bars of log2 ratios. “Comp” refers to the HMM computational analysis of aberration frequency.
Array CGH and FISH correlation of 1p36.3 and 6q23.3. (A) FISH validation of the 1p36.22-p36.33 region (ID no. 1), which presented significant correlation with clinical outcome. Array CGH (top panel) and FISH (bottom panel) demonstrating a case without deletion at 1p36.3 (normal), a case with heterozygous deletion at 1p36.3, and a case with homozygous deletion at 1p36.3. The 1p36.32 probe was labeled red, while the control probe at 1q32 was labeled green. (B) A composite-array CGH ideogram profile of 6q alterations affecting at least 10% and at least 15% of FL cases was generated by intersection analysis. The alteration indicated by the black box in the 15% ideogram corresponds to the 6q23.3 region targeted for FISH validation. (C) FISH validation of the 6q23.3 region (ID no. 20), which presented significant correlation with clinical outcome. Array CGH (top panel) and FISH (bottom panel) demonstrating a case with homozygous deletion and a case with homozygous deletion at 6q23.3 and proximal and distal heterozygous deletion. The 1p36.3 and 6q23.3 probes were labeled red, while the control probes at 1q32.3 and 6p12.3 were labeled green. Green arrows in FISH indicate the presence of heterozygous deletion, while yellow arrows indicate the presence of homozygous deletion. For array CGH, each dot represents a BAC clone and the light blue lines represent visual calling of aberrations. Loss is indicated by a shift to the left of center and gain by a shift to the right of center. Vertical lines are −1 and + 1 scale bars of log2 ratios. “Comp” refers to the HMM computational analysis of aberration frequency.
As the region 6q21-q24.3 was too large (> 40 Mb) for case-specific FISH validation, we further narrowed this region by seeking areas of overlapping deletions affecting more than 15% of cases. Figure 3B shows the refinement of a broadly deleted region of the 6q arm (at the 10% cutoff level) to 4 small discrete regions of deletion (at the 15% cutoff level). The area that correlated significantly with survival and transformation risk corresponded to the single peak in band 6q23.3. The size of this peak was less than 200 kb and spanned from 138 148 563 to 138 307 923 bp (NCBI build 36.1). We used BAC clone RP11-703G08 for this region and RP11-516E15 from 6p12.3 as control for FISH analysis of 10 selected cases. Two of 10 cases were determined by intersection analysis to have no alteration, while 8 cases had deletions by array CGH (as small as 810 kb) overlapping at 6q23.3. The concordance rate between FISH and intersection analysis was 10 of 10 cases. Figure 3C shows the array CGH ideogram data and FISH results for 2 representative cases, one with homozygous deletion and the other showing homozygous deletion at 6q23.3 with proximal and distal heterozygous deletion.
Correlation of array CGH findings with cytogenetic data
To illustrate the sensitivity of the array CGH platform compared with karyotype analysis, we examined the extent of correlation between array CGH and cytogenetic data in the 1p and 6q regions. Of the 27 cases with deletion of 1p36 detected by array CGH, 17 had karyotype data and, of these, only 7 (41.2%) showed an evident deletion or unbalanced translocation. Similarly, of the 22 cases with deletion of 6q detected by array CGH, 14 had karyotype data and 9 (64.3%) showed either whole chromosome loss, iso(6p) or deletion of 6q, whereas 5 cases showed normal 6q morphology.
Identification of high-level amplicons
In an attempt to identify high-level amplification in our cohort, we performed a simple computational thresholding approach where an amplicon was defined as (1) one that consisted of 3 or more contiguous BACs, (2) the log ratio of a BAC in an amplicon was at least 4 standard deviations above the mean log ratio of the sample, and (3) the frequency with which an amplicon occurred was at least 5% in order to minimize random aberrations in an individual case due to genomic instability, we found a high-level amplicon in 18q12.2 recurrent in 6.6% of cases (Table 3; Figure 2 of the supplemental data illustrates the distribution of amplicons identified by our method on chromosome 18). By visual annotation using more than 1 log2 ratio shift as the definition of high-level amplification, 11 instances were found in 4 cases (Table 3).
Detailed information on high-level amplicons in the 106 FL cohort (data based on NCBI build 36.1)
| Region . | DNA coordinates . | Genes of interest . | No. of incidents . |
|---|---|---|---|
| 18q12.2 | 33 040 900-33 600 413 | BRUNOL4 | 7 |
| 1p11.2-p12 | 119 415 209-120 497 765 | NOTCH2 | 2 |
| 1q21.1-q22 | 143 222 462-153 482 569 | PIAS3, BCL9, MCL1, IL6R, ADAM15, TNFAIP8L2, mir-554, mir-190b, mir-92b | 1 |
| 1q23.2-q23.3 | 158 419 998-161 795 584 | PEX19, DDR2, UHMK1, mir-556 | 1 |
| 1q23.3 | 159 989 344-161 290 912 | DDR2, UHMK1, mir-556 | 1 |
| 12p11.21-p12.3 | 18 053 500-31 175 376 | RERGL, PIK3C2G, KRAS, RASSF8, SSPN, mir-920 | 1 |
| 12q13.3-q21.1 | 56 316 396-72 147 648 | OS9, TSPAN31, CDK4, RASSF3, TBK1, WIF1, DYRK2, IL22, MDM2, RAP1B, YEATS4, FRS2, RAB21, mir-548c | 1 |
| 18q21.1 | 42 698 818-42 831 630 | 1 | |
| Xp11.4 | 37 696 318-40 787 875 | 1 | |
| Xp11.3 | 42 402 004-42 902 505 | 1 | |
| Xp11.1 | 57 834 572-58 333 582 | 1 |
| Region . | DNA coordinates . | Genes of interest . | No. of incidents . |
|---|---|---|---|
| 18q12.2 | 33 040 900-33 600 413 | BRUNOL4 | 7 |
| 1p11.2-p12 | 119 415 209-120 497 765 | NOTCH2 | 2 |
| 1q21.1-q22 | 143 222 462-153 482 569 | PIAS3, BCL9, MCL1, IL6R, ADAM15, TNFAIP8L2, mir-554, mir-190b, mir-92b | 1 |
| 1q23.2-q23.3 | 158 419 998-161 795 584 | PEX19, DDR2, UHMK1, mir-556 | 1 |
| 1q23.3 | 159 989 344-161 290 912 | DDR2, UHMK1, mir-556 | 1 |
| 12p11.21-p12.3 | 18 053 500-31 175 376 | RERGL, PIK3C2G, KRAS, RASSF8, SSPN, mir-920 | 1 |
| 12q13.3-q21.1 | 56 316 396-72 147 648 | OS9, TSPAN31, CDK4, RASSF3, TBK1, WIF1, DYRK2, IL22, MDM2, RAP1B, YEATS4, FRS2, RAB21, mir-548c | 1 |
| 18q21.1 | 42 698 818-42 831 630 | 1 | |
| Xp11.4 | 37 696 318-40 787 875 | 1 | |
| Xp11.3 | 42 402 004-42 902 505 | 1 | |
| Xp11.1 | 57 834 572-58 333 582 | 1 |
Identification of secondary pathways
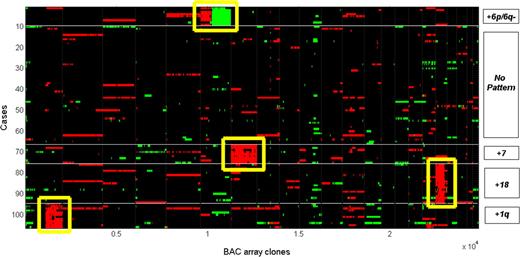
Based on a computational analysis of karyotype data, it was reported that dup(1q), del(6q), dup7 and der18 may constitute 4 distinct events arising secondary to t(14;18) in the early development of FL.26 Using a clone-based approach (high-resolution array CGH) and the application of a robust computational analysis, we attempted to determine whether similar pathway definitions could be obtained in our cohort. We first extracted the 4912 BAC clones from the 71 regions of alterations and applied the k-medoids algorithm to the 106 cases to find clusters based on Hamming distance. Figure 4 presents a heat map where green signals indicate losses and red signals indicate gains. Of the 106 cases, 12 (11.3%) were clustered with dup(1q), 9 (8.5%) with dup(6p)/del(6q), 9 (8.5%) with dup7 and 19 (17.9%) with dup18. The remaining cases clustered into a group that exhibited no obvious pattern of alterations.
Cluster analysis of BAC array clones from the 71 regional aberrations in 106 cases. The K-medoids algorithm (where K = 5) was applied to cluster both 106 cases (x axis) and 4912 BAC clones derived from 71 regions of aberrations. Four distinct clusters, +1q, +6p and 6q−, +7, and +18, were identified. The rest exhibited no obvious pattern of aberrations.
Cluster analysis of BAC array clones from the 71 regional aberrations in 106 cases. The K-medoids algorithm (where K = 5) was applied to cluster both 106 cases (x axis) and 4912 BAC clones derived from 71 regions of aberrations. Four distinct clusters, +1q, +6p and 6q−, +7, and +18, were identified. The rest exhibited no obvious pattern of aberrations.
Discussion
The study reported here describes tiling path array CGH data for 106 cases of FL based exclusively on diagnostic biopsies. Seventy-one regions of alteration were identified to be recurrent in 10% or more cases. Of the 71 regions, 21 were shown to correlate significantly with inferior survival; however, only 16 were considered independent predictors from the IPI in a Cox multivariate model. Some of these regions, including deletion at 1p36.22-p36.33 (ID no. 1), deletion at 6q21-q24.3 (ID no. 20), gains at 17q11.2-q12 (ID no. 55), 17q12-q21.2 (ID no. 56), 17q21.31-q22 (ID no. 57), and 17q24.2-q25.1 (ID no. 60), provide a refinement of regions that were previously shown to be prognostic factors in overall survival.6,39 Our study also demonstrates that 12 of 71 regions were predictors of transformation risk in univariate analysis including: deletion of 1p36.22-p36.33 (ID no. 1), gains of nearly the entire p arm of chromosome 6 (from p21.1 to p25.3, ID no. 11-18), deletion of 6q21-q24.3 (ID no. 20), gain of 7q32.3-q33 (ID no. 38), and gain of 12q13.13-q13.2 (ID no. 51). Other groups have reported CGH gains on both 6p and 7p to correlate with transformation from FL to DLBCL;17,18 however, the 1p36 region has not been previously correlated with transformation.
While our investigation shows that 2 regions, deletions at 1p36.22-p36.33 and 6q21-q24.3, correlated with both inferior survival and higher transformation risk and were independent IPI prognostic predictors, many aberrant regions showed no positive correlation between these 2 clinical parameters. For instance, gains of 6p were associated with higher transformation risk but had no effect on survival rate; likewise, gains of 5p and 17q that were associated with poor survival did not correlate with higher rate of transformation. Three possible explanations could be offered for these findings: (1) transformation and overall survival were not tightly linked in all cases, (2) some cases of FL behaved very aggressively but did not show histological or clinical features used to define transformation in this study, or (3) the median follow-up was too short to appreciate an obvious relationship between transformation and overall survival in this cohort. This might explain why we did not observe a higher percentage of patients (42%) who had died during the observation period.
The most frequently altered region identified in this study was deletion of chromosome band 1p36.22-p36.33 (∼11 Mb in size) that occurred in 25.5% of cases. From the perspective of karyotype analysis, the faint subbands in 1p34 through 1p36 render cytogenetic analysis of 1p36 difficult and even relatively large deletions of this region may be overlooked, resulting in underreporting of deletions that could negatively affect previous correlations with prognosis. Of interest, a recent study by Ross et al found that 50% of 58 low-grade FL showed copy-neutral loss of heterozygosity (LOH; also called acquired uniparental disomy [aUPD]) at 1p36, demonstrating that other mechanisms of gene inactivation may be involved at this and other sites.22 Both deletions and copy-neutral LOH at the terminal portion of 1p have been implicated in other cancer types, including neuroblastoma, melanoma, germ cell tumors, lung cancer, and epithelial ovarian cancers, suggesting the presence of a tumor suppressor in the region.40 A number of candidate genes reside in this region, including the cell cycle protein CDC2L1, the tumor necrosis factor (TNF)–related receptor proteins such as TNFRSF9/14/18/25, the zinc finger transcription factor PRDM16, and the apoptotic factor DFFB. In related studies based on karyotype data we have observed that deletion of 1p36 occurs more frequently in transformed FL than in diagnostic cases (40% vs 24%, 1-tailed P = .028) and that deletion of this region is seen in 50% of high-grade transformations with associated MYC translocations (manuscript in preparation). This latter association may in part explain the strong correlation of 1p36 with inferior survival and transformation observed in this study, as our cases may be enriched for patients that had experienced these events. It suggests that deletion of 1p36 may predispose patients to subsequent transformations with high proliferation rate, as described by Davies et al and Lossos et al, rather than lower proliferative transformations.41,42
Deletion of 6q is detected frequently in acute lymphoblastic leukemia, chronic lymphocytic leukemia, multiple myeloma, DLBCL, and FL.32,43 Different studies have shown various regions to be involved; however, only 1 of these has focused specifically on FL in which a 2.3-Mb region of deletion was identified at 6q16.3 in approximately 15% FL cases.32 Our data show that nearly the entire 6q arm was involved in the majority of cases, though at the 15% cutoff level, only 4 very small regions of loss were defined. None of these regions overlap with that of the Henderson et al study.32 The approximately 500-kb deletion in 6q15 contains the CASP8- associated protein 2 involved in Fas-mediated apoptosis. Of particular interest is deletion of the 6q23.3 band that has been reported in 30% to 38% of ocular MALT lymphomas and FL.22,43,44 Our array CGH data indicate that the approximately 150-kb deletion at 6q23.3 affecting more than 15% of FL cases coincided with the TNFAIP3 (TNF-α induced protein 3) gene. Deletion of this region was validated by FISH, suggesting that TNFAIP3 may be critical in FL development and/or progression. Furthermore, TNFAIP3 was implicated in a recent study based on correlation between genomic loss and gene expression, while it was unclear in another study whether TNFAIP3 or PERP (TP53 apoptosis effector) was implicated since sequencing analysis failed to uncover any mutations.22 Deletion of TNFAIP3 can constitutively activate the NF-kB–signaling pathway as it is a zinc finger protein inhibitor of NF-kB.45 However, since deregulation of NF-kB appears to be uncommon in FL,46 other functions of TNFAIP3 may be responsible, or PERP may be involved since it lies only approximately 200 kb from TNFAIP3. Current evidence indicates that PERP induces TP53-mediated apoptosis, and its deletion could lead to the promotion of tumor growth.47
Although our cohort was partly enriched for diagnostic cases where a specimen was also available from a later transformation event, this selection did not significantly alter the expected median overall survival time (8-10 years vs our observed 10.83 years after diagnosis) and the expected median time to transformation (7 years vs our reported 6.61 years) for this disease. Based on the 3% annual transformation rate observed in FL,2 we would expect 22% of cases to have transformed, whereas 50% of the study patients had transformed to DLBCL. This enrichment allowed us to detect genetic changes that are associated with transformation that would have been missed if too few patients in our study did not have that event. Given the high correlation between transformation and death, confirmation of the full clinical impact of cytogenetic alterations detected in this study should be validated on an unselected series of FL patients.
To exclude the possibility of random aberrant events, we also validated the copy number profile generated from the 106 FL diagnostic cases against an independent cohort of 37 FL (with similar age, grade, and stage characteristics), 30 cases of mantle-cell lymphoma (MCL), and 30 normal specimens. Our results indicated that 40 of the 71 (56.4%) aberrant regions, such as 1p36.22-p36.33 (ID no. 1), 1q42.13-q42.3 (ID no. 7), and 18p11.21-p11.32 (ID no. 65), were unique to FL (Kruskal-Wallis test, P < .05; data not shown), while others, such as 6p23-p24.3 (ID no. 12) and 6q21-q24.3 (ID no. 20), were shared by both FL and MCL. Overlapping regions between the normal controls, FL and MCL were also evident in regions such as 8q21.2 (ID no. 43) and 5p15.33 (ID no. 10), which represented copy number polymorphisms identified by this platform.
A number of CNVs as small as approximately 80 to 200 kb can be detected by the array CGH platform48 and may be evident in the global profile as discrete regions showing both duplication and deletion. Some CNVs may not exhibit this pattern and have been shown to be as large as a few megabases in size.49 Since many CNV breakpoints cannot be precisely defined, and most importantly, that 58% of CNVs overlap with known RefSeq genes,49 we have elected not to filter these CNVs using any stringent criteria. A full understanding of the significance of the CNVs will require additional information on population-based frequencies, observed frequencies in specific types of lymphoma and the possible functional consequences exerted through associated genes, SNPs, or other mechanisms.
Using only the percentage of alterations as a predictive measure of clinical outcome, we found that significant correlation with overall survival and transformation risk was present only if a criterion of 5% or more alterations (rather than 10%) was applied to dichotomize the cases. An explanation for this observation may be that as the criterion of percentage becomes too extreme (as in the example of 10% or more alterations), fewer and fewer cases will constitute one of the groups, thereby significantly affecting statistical comparisons. Nevertheless, our findings were in general agreement with the commonly held notion that with increasing number of aberrations in the genome, prognosis is negatively affected.19
This study has also shed further light on other recurrently altered genomic regions in FL. Potential genes that have been implicated in lymphomas are presented in column Q of Table 2. For instance, the cytochrome gene CYP51 on 7q has been found overexpressed in lymphomas.16 In 8q, 4 candidate genes have been suggested: a potassium channel protein KCNK9, NF-KB–activating protein NIBP, the protein tyrosine kinase PTK2/FAK, and the protein tyrosine phosphatise PTP4A3.22 The PTEN phosphatase tumor suppressor at 10q23.1-q25.1 has been of constant interest.50 MDM2 at 12q13.2-q21.1 has been reported to have altered expression that may negatively affect the stability of p53.51 Correlation between gene expression and genomic changes provided evidence that the interleukin-3 zinc finger transcription factor ZNF161 at 17q23.2 may be involved.39 BCL2 may be overexpressed as a result of extra copies of chromosome 18, especially in DLBCL52 ; however, the bands 18q11-18q21.33 proximal to the BCL2 locus are also consistently overrepresented in FL, implicating a gene proximal to the t(14;18) breakpoint in this amplification.22,24
Our search for high-level amplicons that occurred in at least 5% of cases led to the localization of a region in 18q22, which contains the BRUNOL4 gene. This gene belongs to a family of RNA-binding proteins involved in multiple aspects of RNA processing.53 Its function in hematopoietic cells, however, has not been studied. Eleven other amplicons were found that occurred in less than 5% of cases. Viardot et al showed that among 8 amplicons found in their 124 patients, their regions in 1q23-q25 and 12q13 overlapped with ours,19 as did the bands 8q24 and 12q13-q14 in the Bentz et al study.15
In an attempt to dissect the sequence of cytogenetic events occurring in the clonal evolution of FL, Hoglund et al conducted one of the first studies using published karyotype data to reconstruct the common pathways of clonal evolution secondary to the t(14;18).26 Using principal component analysis to reduce data complexity for multivariate correlations, 4 major events consisting of dup(1q), dup7 del(6q), and der18 were identified to arise independently after the t(14;18). We attempted to use a clone-based approach to cluster both the 106 diagnostic patients and 4912 BAC clones extracted from the 71 regions of alteration. Using different methods and a mostly nonoverlapping FL cohort, we have replicated the findings of the previous karyotype-based study. These data, in conjunction with previous findings, suggest that the early events of clonal progression in FL may evolve along a number of distinct pathways. These events may represent alternative critical steps following the primary event of BCL2 deregulation that are essential to the promotion of early clonal expansion, leading eventually to clinical manifestation of disease and transformation to more aggressive histologies. It is interesting to note that altogether 46.2% of our cases were represented in one of the 4 clustered groups, dup(1q), dup(6p)/del(6q), dup7, and dup(18q), while the rest showed alterations that could not be explained by this approach. These may be cases where other types of biological mechanisms, such as copy-neutral LOH and/or methylation of genes of critical importance may be operative.
In conclusion, our data have confirmed and refined regions of aberrations found in previous findings and provided further insight into the distinct molecular pathways related to FL development using the clone-based cluster analysis. Most importantly, our study has identified deletion of 1p36 and 6q23 as significant prognostic indicators of clinical outcome. These correlations have been strengthened by the ability of high resolution analysis to detect submicroscopic deletions not previously detectable using other methods. The clinical relevance of these genetic alterations and their impact on disease progression will require additional studies of large patient cohorts, ideally managed with uniform therapy and lacking a selection bias.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank B. Chi and R. De Leeuw for computational assistance in the seeGH software.
This work was supported by an NCIC Terry Fox Foundation New Frontiers Program Project Grant (016003/grant type 230/project title: Biology of Cancer: Follicular Lymphoma as a Model of Cancer Progression), Genome Canada/Genome BC Grant Competition III (project title: High Resolution Analysis of Follicular Lymphoma Genomes; J.M.C., R.D.G., and D.E.H), the Deutsche Forschungsgemeinschaft (STE-1706/1-1; C.S.), and the CIHR fellowship award (STP-53912; N.J.).
Authorship
Contribution: K.-J.J.C. contributed to the design and analysis of this study and the writing of the manuscript; S.P.S. designed and implemented all computational methods and engaged in data interpretation; C.S. provided scientific input, performed FISH validation, and assisted in data interpretation of clinical information; N.J. managed the follicular lymphoma database, provided advice of clinical relevance, and contributed to the statistical analysis of clinical variables; T.R. contributed to array CGH experimental work; A.T. and B.L. performed experiments and maintained the array CGH database; R.T.N. and K.P.M. provided scientific advice in experimental design related to computation; W.L. provided array CGH technology; A.J.A.-T. assembled clinical information related to transformation; J.M.C., R.D.G., and D.E.H. were directly or indirectly involved in the selection and procurement of clinical specimens, conceived the study, and involved in the writing of the manuscript; and all authors agreed on the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Doug Horsman, Center for Lymphoid Cancer, British Columbia Cancer Agency, 600 West 10th Avenue, Vancouver, BC V5Z 4E6; e-mail: dhorsman@bccancer.bc.ca.
References
Author notes
*K.-J.J.C. and S.P.S. contributed equally to this study.

![Figure 2. Correlation analysis of the 71 regional aberrations with clinical data. (A) Kaplan-Meier survival of ID numbers 1 [del(1)(p36.22-p36.33)] and 20 [del(6)(q21-q24.3)], and (B) transformation graphs of ID numbers 1 [del(1)(p36.22-p36.33)] and 20 [del(6)(q21-q24.3)]. Log-rank test was performed to assess significance (P ≤ .05).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/1/10.1182_blood-2008-02-140616/4/m_zh80200826210002.jpeg?Expires=1767741480&Signature=FT1bd4ee~pOZggq14SO49Ob-hXc65VQ6aVDN8av1dnMNNR23JFWs~cZM5AMAmZL8tC5ohG0N~6QSk9DEaVHkqJSRS85ksDCo4lnwq4Efx2mpKrb9TaeHLdIxQqPnMTWzuWl7FbUoVnfhgkl8YAugfIQndvF6wGtEyUPjE7UyKtkN1K5uMrOYzB0l~VR2I9Z2parbS5QxsTy~dnAFQJ8xPZ8PsopILcVs-5YJtMxPe3LjlItjpgVULr7TpjIEKzyByAlzXMaxqwq60ozTIyc6Ue-RdyY~erTa1TPrUdlxzeh~CzIYOWCI-OAd6nIoKxTiRRWpDegxNzli-UAGBhj0hw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal