Abstract
Mycosis fungoides (MF), the most common cutaneous T-cell lymphoma, is a malignancy of mature, skin-homing T cells. Sézary syndrome (Sz) is often considered to represent a leukemic phase of MF. In this study, the pattern of numerical chromosomal alterations in MF tumor samples was defined using array-based comparative genomic hybridization (CGH); simultaneously, gene expression was analyzed using microarrays. Highly recurrent chromosomal alterations in MF include gain of 7q36, 7q21-7q22 and loss of 5q13 and 9p21. The pattern characteristic of MF differs markedly from chromosomal alterations observed in Sz. Integration of data from array-based CGH and gene-expression analysis yielded several candidate genes with potential relevance in the pathogenesis of MF. We confirmed that the FASTK and SKAP1 genes, residing in loci with recurrent gain, demonstrated increased expression. The RB1 and DLEU1 tumor suppressor genes showed diminished expression associated with loss. In addition, it was found that the presence of chromosomal alterations on 9p21, 8q24, and 1q21-1q22 was associated with poor prognosis in patients with MF. This study provides novel insight into genetic alterations underlying MF. Furthermore, our analysis uncovered genomic differences between MF and Sz, which suggest that the molecular pathogenesis and therefore therapeutic requirements of these cutaneous T-cell lymphomas may be distinct.
Introduction
Mycosis fungoides (MF), the most common type of primary cutaneous T-cell lymphoma (CTCL), is a malignancy of mature, skin-homing T cells. MF commonly presents with erythematous patches and plaques and generally behaves as a low-grade lymphoma with an indolent disease course.1,2 A subset of patients with MF experiences disease progression, which is characterized by the formation of skin tumors, the appearance of blast-like cells in the tumoral infiltrate, and extracutaneous dissemination of malignant T cells. Progressive MF is often refractory to treatment and has an unfavorable prognosis.3 In recent years, progress has been made in defining cytogenetic alterations in MF using conventional comparative genomic hybridization (CGH) and fluorescence in situ hybridization methods.4,5 In addition, mutations affecting the CDKN2A, FAS, and JUNB genes and alterations of Janus kinase (JAK)/signal transducer and activator of transcription (STAT) and death receptor signaling have been identified in subgroups of patients with MF.6-11 However, the molecular genetic alterations underlying this T-cell lymphoma remain poorly understood.12,13
A CTCL that is closely related to MF is Sézary syndrome (Sz). Sz is characterized by the triad of erythroderma, generalized lymphadenopathy, and presence of malignant T cells in peripheral blood. Patients with Sz have a considerable leukemic T-cell burden and a dismal prognosis with an estimated 5-year survival rate of 24%.14 Recently, we identified several highly recurrent copy number alterations (CNAs) in Sz, including gain of loci on chromosome 17q24 and 8q24 and loss of regions on 17p13 and 10q25, occurring in up to 85% of patients.15 Additional evaluation of candidate oncogenes and tumor suppressor genes residing in loci with chromosomal alteration pointed to dysregulation of the MYC oncogene, several of its regulators, and IL-2 receptor signaling pathway components in Sz.
MF and Sz are both clonal proliferations of T cells with cerebriform nuclei and a CD4+, CD45RO+, CLA+ immunophenotype.13 Despite differences in clinical presentation and disease behavior of these 2 disease entities, Sz is often designated as a leukemic phase or variant of MF, and it has been suggested that differences between both conditions are a matter of stage of disease.16-18 Therefore, these 2 CTCLs, sometimes collectively termed MF/Sz, share the same classification and staging system and are managed using similar treatment regimens.19,20 Although previously classified as a variant of MF, the current World Health Organization-European Organization of Research and Treatment of Cancer classification lists Sz as a separate disease entity based on its distinctive clinical features and disease behavior.16 Because of the existence of both shared and dissimilar immunophenotypic and genetic properties, controversy has remained as to whether MF and Sz should be regarded as distinct disorders with a different pathogenesis and therapeutic requirements or whether differences reflect distinct stages of a similar disease process.
In this study, numerical chromosomal alterations in malignant T cells from tumor-stage MF samples were mapped using array-based CGH. The first purpose was to define the pattern of recurrent chromosomal alterations characteristic of MF. We then evaluated whether this pattern corresponds to the highly recurrent gains and losses observed in Sz. The second purpose was to identify candidate oncogenes and tumor suppressor genes residing in chromosomal regions with recurrent copy number alteration in MF. To this end, chromosomal alteration and gene-expression patterns of MF tumor samples were integrated to determine which genes located in minimal common regions (MCRs) with CNA demonstrated dysregulated expression associated with chromosomal alteration. A third line of enquiry we pursued was aimed at finding chromosomal alterations with prognostic significance.
Methods
Selection of patients
Lesional skin tumor biopsy samples containing at least 70% malignant T cells from 22 patients with tumor-stage MF (TNM stage T3N0M0B0 in 21 patients and T3N3M0B0 in 1 patient) were included in this study. They included 18 male and 4 female patients with a mean age at the time of biopsy of 66 years. All biopsy samples were obtained before treatment, except in patients diagnosed with plaque-stage disease (T1N0M0B0 or T2 N0M0B0) previously who had been treated with local corticosteroids or phototherapy. The malignant phenotype of T cells in tumoral infiltrates was assessed on the basis of cytonuclear atypia and immunophenotypical characteristics by an expert panel of pathologists. Lymphoid cells were CD4+ and CD8− in all cases. Histopathologically, all included tumor samples showed large cell transformation, indicated by the presence of at least 25% large cells in the tumoral infiltrate. After a mean follow-up period of 23 months, 12 patients had died because of MF, 3 had died of other causes, and 7 patients were alive. Results of array-based CGH analysis were compared with those previously obtained from peripheral blood mononuclear cells of 20 patients with Sz using identical methods.15 In that study, Sz was defined according to criteria of the World Health Organization–European Organization of Research and Treatment of Cancer classification. Clinical characteristics of MF and Sz patients are summarized in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Approval was received from the Leiden University Medical Center institutional review board for these studies. Informed consent was obtained in accordance with the Declaration of Helsinki.
Extraction of DNA and RNA
DNA and RNA were isolated from the same tumor biopsy for array-based CGH and gene-expression analysis, using oligonucleotide arrays and quantitative real-time polymerase chain reaction (qPCR), respectively, of all 22 included patients. DNA was isolated from 25 × 20 μM frozen sections using the Genomic-tips 20/G Kit (QIAGEN, Hilden, Germany), yielding 10 to 60 μg genomic DNA. RNA was extracted from 25 × 50 μM frozen sections using the RNeasy kit (QIAGEN), yielding 25 to 60 μg total RNA. RNA used for gene-expression analysis and for confirmatory qPCR analysis was isolated from the same tumor biopsy sample.
Array-based CGH analysis
Genome-wide analysis of CNAs was performed using array-based CGH containing approximately 3500 bacterial artificial chromosomes (BACs) produced at the Leiden University Medical Center. The particular BAC set used to produce the arrays was distributed by the Wellcome Trust Sanger Institute (Hinxton, United Kingdom) and contains large insert clones spaced at approximately 1 Mb density over the full genome, a set of subtelomeric sequences for each chromosome arm, and a few hundred probes selected for their involvement in oncogenesis. Fabrication and validation of the array, hybridization methods, and analytical procedures have been described in detail elsewhere, whereas the clone content is available in the Cytoview window of the Sanger Center mapping database site Ensembl (http://www.ensembl.org).21 Data were analyzed using CAPWeb and visualized using vesicle-associated membrane protein (VAMP).22 Log2 ratios were classified as copy number gain (> 0.25) or genomic loss (< −0.25). Identified CNA of regions with copy number variations described in the Database of Genomic Variants (http://projects.tcag.ca/variation) were excluded from analysis.
Gene-expression profiling
Samples and microarrays (Human Genome U133plus2.0 array; Affymetrix, Santa Clara, CA), interrogating more than 47 000 human transcripts and variants, were processed according to the manufacturer's protocol as described previously.23 The array images were quantified using the Genechip operating system (version 1.2 software; Affymetrix). The average fluorescence intensity was determined for each microarray, and then the output of each experiment was globally scaled to a target value of 200. Further normalization and variance stabilization were performed using variance-stabilizing normalization in the R statistical software package.24 All microarray data have been deposited with Gene Expression Omnibus under accession number GSE12902.25
Data analysis
BAC clone and oligonucleotide probe positions were established based on Ensembl release 44 (April 2007). Recurrent MCRs with CNA affecting at least 35% of analyzed samples were computed in CAPWeb using the algorithm proposed by Rouveirol et al.26 Only CNAs characterized by gain or loss of at least 2 clones were taken into consideration. The nearby borders of adjacent clones were chosen to delineate MCRs. Copy number was divided into the categories gain, normal, and loss. To determine whether MCRs with recurrent CNA contained a statistically significantly higher number of genes showing increased expression in case of gain, or diminished expression in case of loss the sign test was performed. The normalized expression levels of genes residing in these MCRs as measured by oligonucleotide microarray analysis were then compared between tumor samples with and without the particular CNA. Independent-samples t tests were performed (equality of variances not assumed) using the SPSS 14.0 statistical software package. Genes demonstrating a statistically significant increased expression in MF samples with gain or decreased expression in case of loss were considered of primary interest (P < .05). From this collection of genes with CNA-associated expression, candidate genes with pathobiologic relevance were selected by focusing on genes listed as oncogene or tumor suppressor gene in the European Bioinformatics Institute cancer gene prediction database (http://cgg.ebi.ac.uk/services/cgp) with a probability exceeding 30%. Disease-specific actuarial survival rates of patients were calculated from the date of tumor biopsy for array-based CGH analysis and compared using the log-rank test.
Quantitative real-time PCR
cDNA synthesis was performed on 1 μg of total RNA, after treatment with RQ1 DNase I (Promega, Madison, WI), using IScript reverse transcriptase (Bio-Rad, Hemel Hempstead, United Kingdom), oligo(dT)12-18 and random hexamer priming (Bio-Rad) in a final volume of 20 μL. Real-time PCR was performed with the MyIQ instrument and the SYBR Green Supermix (Bio-Rad). The cycle parameters for transcripts of interest and for the reference genes U1A and RPS11 used for normalization were as follows: denaturing for 15 seconds at 97°C; annealing and extension for 20 seconds at 60°C, for 40 cycles. Primer sequences (Invitrogen, Carlsbad, CA) for selected transcripts are given in Table S2. Data were evaluated using MyIQ software (Bio-Rad) and the second derivative maximum algorithm, whereas confirmation of the specificity of the PCR product and standard curves were performed as previously described.27
Immunohistochemistry
Immunostaining on formalin-fixed, paraffin-embedded skin sections with antibodies against RB1 (dilution 1:400; phosphorylation-nonspecific, 14001A; BD PharMingen, San Diego, CA) and SKAP1 (dilution 1:400; HPA002969; Sigma-Aldrich, St Louis, MO) was performed using a standard 3-step streptavidin-biotin-peroxidase–based technique after antigen retrieval with microwave heating as described previously.28
Results
Pattern of copy number alterations of MF
Clinical characteristics and follow-up data of the 22 patients with tumor-stage MF included in the study are presented in Table S1. Array-based CGH methodology was used to catalog CNAs in the genomes of malignant T cells present in skin tumor biopsy samples. All MF tumor samples showed extensive losses and gains of both large and smaller chromosomal regions. Copy number gains were more frequent than losses. The frequency and cumulative pattern of gains and losses in the tumor samples are depicted in Figure 1A. As a first step toward determining biologically significant patterns of genomic alterations in MF, we computed MCRs with CNA. MCRs represent the smallest recurrent chromosomal region with altered probes common to the set of CGH profiles and are considered to harbor genes with biologic relevance in tumor progression.26,29 We identified 24 MCRs present in at least 35% of the 22 MF patients, ranging in size from 1.2 to 41 Mb. These MCRs are presented in Table 1 and are indicated by vertical lines in a visual representation of averaged CGH data in Figure 1B. Fifteen of these recurrent MCRs with CNA represent gains of chromosomal regions and 9 correspond to losses. Among the most frequently observed alterations were gain of regions on the long arm of chromosome 7 with a MCR on 7q36, observed in 59% of samples, and gains of several other regions on 7q32-7q35, 7q21-7q22, and 7q11.2. The chromosomal regions second most frequently affected with gain were 7p13-7p14, 7p21-7p22, 1q31-1q32, and 1p36.2, occurring in 45% of the patients. Losses were most frequently observed on 5q13, 9p21, and 13q14-13q31.
Visualization of the array-based CGH results by VAMP. (A) Overall frequency of CNAs in MF patient tumor biopsies, calculated using the FrAGL (frequency of amplifications, gains, and losses) option of VAMP. Losses are represented on the negative scale as green bars; gains are presented on the positive scale as red bars. (B) Averaged CGH pattern of all 22 MF tumor samples. MCRs with loss occurring in at least 35% of patients are indicated as green vertical lines, and MCRs with gain as red vertical lines. All data are presented ordered by chromosomal map position of the clones, excluding sex chromosomes.
Visualization of the array-based CGH results by VAMP. (A) Overall frequency of CNAs in MF patient tumor biopsies, calculated using the FrAGL (frequency of amplifications, gains, and losses) option of VAMP. Losses are represented on the negative scale as green bars; gains are presented on the positive scale as red bars. (B) Averaged CGH pattern of all 22 MF tumor samples. MCRs with loss occurring in at least 35% of patients are indicated as green vertical lines, and MCRs with gain as red vertical lines. All data are presented ordered by chromosomal map position of the clones, excluding sex chromosomes.
Minimal common regions with copy number alteration in MF
| Cytogenetic band . | Copy number alteration . | Adjacent clones . | Clone position, Mb . | Affected patients, % . | ||
|---|---|---|---|---|---|---|
| Start . | Stop . | Start . | Stop . | |||
| 1p36.2 | Gain | RP4-539L13 | RP11-196P5 | 11098993 | 12351219 | 45 |
| 1q21-1q22 | Gain | RP4-790G17 | RP11-172I6 | 146342686 | 156056126 | 41 |
| 1q31-1q32 | Gain | RP11-572A16 | RP11-534L20 | 198714422 | 205087972 | 45 |
| 5q13 | Loss | RP11-551B22 | RP11-497H16 | 67677068 | 70179512 | 45 |
| 7p22-7p21 | Gain | RP11-510K8 | RP4-733B9 | 1081263 | 7947777 | 45 |
| 7p15-7p14 | Gain | RP11-99O17 | RP11-302L6 | 24659178 | 37825117 | 41 |
| 7p14-7p13 | Gain | RP11-36H20 | RP11-52M17 | 43272694 | 45048103 | 45 |
| 7q11.2 | Gain | RP11-313P13 | RP11-107L23 | 71274704 | 76190020 | 50 |
| 7q21-7q22 | Gain | RP4-550A13 | RP11-333G13 | 97314794 | 102514284 | 55 |
| 7q32-7q35 | Gain | RP11-329I5 | RP11-298A10 | 130270796 | 143852574 | 55 |
| 7q36 | Gain | RP11-24N19 | RP4-548D19 | 148089302 | 151558264 | 59 |
| 8q24.2 | Gain | RP11-71N3 | RP11-343P9 | 132799581 | 137773461 | 32 |
| 8q24.3 | Gain | RP5-1118A7 | RP5-1056B24 | 142790550 | Telomere | 36 |
| 9p21 | Loss | RP11-113D19 | RP11-149I2 | 20351121 | 22479496 | 41 |
| 9p21 | Loss | RP11-495L19 | RP11-33K8 | 22579721 | 24877888 | 32 |
| 9p13-9p11.1 | Loss | RP11-211N8 | RP11-475I24 | 39990599 | 42614658 | 32 |
| 9q21 | Loss | RP11-490H9 | RP11-336N8 | 78213759 | 80495074 | 32 |
| 9q21 | Loss | RP11-174K23 | RP11-432M2 | 79930787 | 84622895 | 32 |
| 9q21 | Loss | RP11-439A18 | RP1-292F10 | 84783002 | 86180561 | 32 |
| 9q22-9q31 | Loss | RP11-463M14 | RP11-75J9 | 101410218 | 105214273 | 32 |
| 13q14-13q31 | Loss | RP11-168P13 | RP11-464I4 | 42301191 | 83766576 | 36 |
| 17q21 | Gain | RP5-905N1 | RP11-361M10 | 39091531 | 44639847 | 41 |
| 17q22-17q23 | Gain | RP11-312B18 | RP11-156L14 | 48664511 | 59626448 | 32 |
| 17q25 | Gain | RP11-478P5 | GS-362-K4 | 69639765 | Telomere | 36 |
| Cytogenetic band . | Copy number alteration . | Adjacent clones . | Clone position, Mb . | Affected patients, % . | ||
|---|---|---|---|---|---|---|
| Start . | Stop . | Start . | Stop . | |||
| 1p36.2 | Gain | RP4-539L13 | RP11-196P5 | 11098993 | 12351219 | 45 |
| 1q21-1q22 | Gain | RP4-790G17 | RP11-172I6 | 146342686 | 156056126 | 41 |
| 1q31-1q32 | Gain | RP11-572A16 | RP11-534L20 | 198714422 | 205087972 | 45 |
| 5q13 | Loss | RP11-551B22 | RP11-497H16 | 67677068 | 70179512 | 45 |
| 7p22-7p21 | Gain | RP11-510K8 | RP4-733B9 | 1081263 | 7947777 | 45 |
| 7p15-7p14 | Gain | RP11-99O17 | RP11-302L6 | 24659178 | 37825117 | 41 |
| 7p14-7p13 | Gain | RP11-36H20 | RP11-52M17 | 43272694 | 45048103 | 45 |
| 7q11.2 | Gain | RP11-313P13 | RP11-107L23 | 71274704 | 76190020 | 50 |
| 7q21-7q22 | Gain | RP4-550A13 | RP11-333G13 | 97314794 | 102514284 | 55 |
| 7q32-7q35 | Gain | RP11-329I5 | RP11-298A10 | 130270796 | 143852574 | 55 |
| 7q36 | Gain | RP11-24N19 | RP4-548D19 | 148089302 | 151558264 | 59 |
| 8q24.2 | Gain | RP11-71N3 | RP11-343P9 | 132799581 | 137773461 | 32 |
| 8q24.3 | Gain | RP5-1118A7 | RP5-1056B24 | 142790550 | Telomere | 36 |
| 9p21 | Loss | RP11-113D19 | RP11-149I2 | 20351121 | 22479496 | 41 |
| 9p21 | Loss | RP11-495L19 | RP11-33K8 | 22579721 | 24877888 | 32 |
| 9p13-9p11.1 | Loss | RP11-211N8 | RP11-475I24 | 39990599 | 42614658 | 32 |
| 9q21 | Loss | RP11-490H9 | RP11-336N8 | 78213759 | 80495074 | 32 |
| 9q21 | Loss | RP11-174K23 | RP11-432M2 | 79930787 | 84622895 | 32 |
| 9q21 | Loss | RP11-439A18 | RP1-292F10 | 84783002 | 86180561 | 32 |
| 9q22-9q31 | Loss | RP11-463M14 | RP11-75J9 | 101410218 | 105214273 | 32 |
| 13q14-13q31 | Loss | RP11-168P13 | RP11-464I4 | 42301191 | 83766576 | 36 |
| 17q21 | Gain | RP5-905N1 | RP11-361M10 | 39091531 | 44639847 | 41 |
| 17q22-17q23 | Gain | RP11-312B18 | RP11-156L14 | 48664511 | 59626448 | 32 |
| 17q25 | Gain | RP11-478P5 | GS-362-K4 | 69639765 | Telomere | 36 |
Comparison of genomic profiles of MF and Sz
We then evaluated the similarity of chromosomal alterations observed in MF with those present in Sz. Recently, we have studied chromosomal alterations in malignant T cells from peripheral blood of 20 Sz patients using an identical array-based CGH platform and bioinformatic analysis.15 Malignant T cells from patients with Sz are characterized by several highly recurrent alterations, including gain of 17q23-25 (in 80% of patients), gain of 8q24 (in 75%, harboring the MYC oncogene), and loss of 17p13 (in 75%, harboring the TP53 tumor suppressor gene). These specific chromosomal alterations are present much less frequently in MF tumor samples, with frequencies of 32%, 23%, and 9%, respectively. Conversely, many highly recurrent alterations in MF, including gain on 7q36, only rarely occur in Sz. Whereas the overall pattern of chromosomal alterations of MF is characterized by gains on chromosome 1 and 7 and losses on chromosome 9, Sz demonstrates gains of regions on chromosome 8 and 17 and loss on chromosome 10. More detailed comparison of MCRs with recurrent CNAs in MF and Sz revealed clear differences, including many gains and losses that were present at a high frequency in MF but not in Sz. In Table 2, the 10 most frequent MCRs with CNA in MF and Sz are highlighted, and frequencies in both entities are indicated. These findings argue against the notion that differences between these CTCLs are a matter of stage and strongly suggest that the molecular pathogenesis of MF and Sz follows distinct pathways.
Comparison of most highly recurrent CNAs in MF and Sz
| Cytogenetic band . | CNA . | Affected MF patients, % . | Affected Sz patients, % . |
|---|---|---|---|
| Mycosis fungoides | |||
| 7q36 | Gain | 59 | 15 |
| 7q21-7q22 | Gain | 55 | 20 |
| 7q32-7q35 | Gain | 55 | 10 |
| 7q11.2 | Gain | 50 | 15 |
| 1p36.2 | Gain | 45 | 15 |
| 1q31-1q32 | Gain | 45 | 0 |
| 5q13 | Loss | 45 | 40 |
| 7p22-7p21 | Gain | 45 | 20 |
| 7p14-7p13 | Gain | 45 | 15 |
| 1q21-1q22 | Gain | 41 | 5 |
| Sézary syndrome | |||
| 17q23 | Gain | 85 | 32 |
| 17q22-17q23 | Gain | 80 | 32 |
| 17q24-17q25 | Gain | 80 | 27 |
| 8q24.1-8q24.2 | Gain | 75 | 23 |
| 8q24.2-8q24.3 | Gain | 75 | 27 |
| 8q22-8q23 | Gain | 70 | 18 |
| 17p13 | Loss | 70 | 9 |
| 17q25 | Gain | 70 | 32 |
| 8q12-8q21.1 | Gain | 65 | 18 |
| 8q11.2-8q12 | Gain | 60 | 18 |
| Cytogenetic band . | CNA . | Affected MF patients, % . | Affected Sz patients, % . |
|---|---|---|---|
| Mycosis fungoides | |||
| 7q36 | Gain | 59 | 15 |
| 7q21-7q22 | Gain | 55 | 20 |
| 7q32-7q35 | Gain | 55 | 10 |
| 7q11.2 | Gain | 50 | 15 |
| 1p36.2 | Gain | 45 | 15 |
| 1q31-1q32 | Gain | 45 | 0 |
| 5q13 | Loss | 45 | 40 |
| 7p22-7p21 | Gain | 45 | 20 |
| 7p14-7p13 | Gain | 45 | 15 |
| 1q21-1q22 | Gain | 41 | 5 |
| Sézary syndrome | |||
| 17q23 | Gain | 85 | 32 |
| 17q22-17q23 | Gain | 80 | 32 |
| 17q24-17q25 | Gain | 80 | 27 |
| 8q24.1-8q24.2 | Gain | 75 | 23 |
| 8q24.2-8q24.3 | Gain | 75 | 27 |
| 8q22-8q23 | Gain | 70 | 18 |
| 17p13 | Loss | 70 | 9 |
| 17q25 | Gain | 70 | 32 |
| 8q12-8q21.1 | Gain | 65 | 18 |
| 8q11.2-8q12 | Gain | 60 | 18 |
Identification of genes relevant in the pathobiology of MF through integrated genomic analysis
Chromosomal gains and losses can contribute to the development and progression of lymphoma by altering the expression levels of genes residing in loci with CNA. We sought to identify such biologically relevant genes in MF by evaluating the expression levels of genes located in MCRs with recurrent CNA. A schematic representation of the strategy used for identifying these genes is depicted in Figure 2. First, we asked which genes, residing in any of the 24 MCRs affecting at least 35% of patients, showed increased expression associated with gain or decreased expression associated with loss. A total of 1504 annotated genes interrogated by the Affymetrix oligonucleotide arrays are located in the 24 MCRs. In tumor samples affected by gain of any of the 15 identified highly recurrent MCRs, significantly more genes residing in these chromosomal regions showed increased expression. In addition, in 8 of the 9 MCRs with loss, there was a significant excess of genes displaying decreased expression in the tumor samples affected by loss of these MCRs according to the sign test (Table S3). To examine the effect of gene dosage on mRNA abundance, we tested whether gene expression correlated with CNA for each individual gene residing within these MCRs by comparing the gene-expression levels in samples harboring chromosomal gain or loss to the samples not affected by CNA using the Student t test. A total of 223 annotated genes showed increased expression associated with gain and 30 genes decreased expression associated with loss (P < .05). Genes demonstrating such CNA-associated expression pattern, ie, significantly increased expression in samples with gain or decreased expression in samples with loss of a certain chromosomal region, are listed for each of the 24 MCRs in order of frequency of occurrence in Table 3. For each chromosomal region, we then prioritized these genes for potential biologic relevance by triangulating with genes listed as cancer-related in the European Bioinformatics Institute cancer gene database, indicated in bold in Table 3. The resulting list of candidate oncogenes and tumor suppressor genes includes MDMX, MCL1, and RB1. In addition, the CDKN2A gene with an established role in MF progression is among this refined list of candidate genes.6 The 2 CDKN2A probe sets emerging from integrated genomic analysis indicated in Figure 2B both target a region common to the p16 and p14 transcripts. The chromosomal region most frequently affected by gain is 7q36, amplified in 59% of MF patients. Only 3 of the 56 genes residing at the 7q36 locus demonstrate increased expression in the tumor samples with gain (FASTK, NUB1, and LOC791120). The FASTK gene encodes FAS-activated serine/threonine kinase, an antiapoptotic protein expressed in T cells.30,31
Integration of array-based CGH and gene-expression data. (A). Stepwise approach to identification of genes potentially relevant in the development or progression of MF residing in loci with frequent CNA. Transcripts corresponding to genes localized in MCRs (Figure 1B) were identified and extracted using the Ensembl genome browser and cross-referenced with microarray probes. Gene dose effects on expression levels were then statistically evaluated. Genes demonstrating significantly higher expression associated with gain or lower expression associated with loss are summarized in Table 3. (B) Visual illustration of the integration method applied for aCGH and gene-expression data. For 2 exemplary MCRs, with gain on 7q36 and loss on 9p21, heat maps of resident gene-expression patterns were generated. Genes with a CNA-associated expression pattern are indicated with ◀.
Integration of array-based CGH and gene-expression data. (A). Stepwise approach to identification of genes potentially relevant in the development or progression of MF residing in loci with frequent CNA. Transcripts corresponding to genes localized in MCRs (Figure 1B) were identified and extracted using the Ensembl genome browser and cross-referenced with microarray probes. Gene dose effects on expression levels were then statistically evaluated. Genes demonstrating significantly higher expression associated with gain or lower expression associated with loss are summarized in Table 3. (B) Visual illustration of the integration method applied for aCGH and gene-expression data. For 2 exemplary MCRs, with gain on 7q36 and loss on 9p21, heat maps of resident gene-expression patterns were generated. Genes with a CNA-associated expression pattern are indicated with ◀.
Results from integration of expression and aCGH results: candidate oncogenes and tumor suppressor in MF
| Cytogenetic band . | Copy number alteration . | Clone position . | Affected patients, % . | Candidate genes . | |
|---|---|---|---|---|---|
| Start . | Stop . | ||||
| 7q36 | Gain | 148089302 | 151558264 | 59 | FASTK, NUB1, LOC791120 |
| 7q21-7q22 | Gain | 97314794 | 102514284 | 55 | AP1S1, SMURF1, ZKSCAN1, C7orf38, CLDN15, ZNF789, RASA4, ZNF498, ZNF789, ARMC10, POLR2J2, ZNHIT1, ZCWPW1, MGC40499 |
| 7q32-7q35 | Gain | 130270796 | 143852574 | 55 | TRIM24, CNOT4, PTN, C7orf49, KIAA0738, LUC7L2 |
| 7q11.2 | Gain | 71274704 | 76190020 | 50 | GTF2IRD1, ABHD11, NSUN5, NSUN5B, NSUN5C, ELN, WBSCR22, TRIM73 |
| 1p36.2 | Gain | 11098993 | 12351219 | 45 | MFN2 |
| 1q31-1q32 | Gain | 198714422 | 205087972 | 45 | MDM4, NAV1, RBBP5, IPO9, CSRP1, KIF21B, PPP1R15B, NUCKS1, TIMM17A, SNRPE, KIF14 |
| 5q13 | Loss | 67677068 | 70179512 | 45 | TAF9, SERF1A, SERF1B, SMN1, TAF9, GUSBP1 |
| 7p14-7p13 | Gain | 43272694 | 45048103 | 45 | CAMK2B, POLR2J4 |
| 7p22-7p21 | Gain | 1081263 | 7947777 | 45 | WIPI2, LOC222967, FTSJ2, MICALL2 |
| 1q21-1q22 | Gain | 146342686 | 156056126 | 41 | MCL1, CLK2, PRCC, ARHGEF11, HDGF, GPATCH4, JTB, MSTO1, FLAD1, CRTC2, SMG5, ADAR, MRPL24, KRTCAP2, SETDB1, C1orf2, SF3B4, PRPF3, SEMA4A, MTX1, ISG20L2, SNAPAP, ENSA, PLEKHO1, ISG20L2, DAP3, GON4L, C1orf85, APOA1BP, C1orf43, RUSC1, UBAP2L, CDC42SE1, MAPBPIP, SCAMP3, C1orf77, PYGO2, PSMD4, GATAD2B, PEAR1, FDPS, VPS72, MRPL9, IQGAP3, DENND4B, TNFAIP8L2, UBQLN4, SLC39A1, TPM3, PRUNE |
| 7p15-7p14 | Gain | 24659178 | 37825117 | 41 | TAX1BP1, HOXA10, CREB5, HERPUD2, JAZF1, LOC441212, HNRPA2B1, C7orf41, LOC401320, KBTBD2 |
| 9p21 | Loss | 20351121 | 22479496 | 41 | CDKN2A, MTAP, LOC554202 |
| 17q21 | Gain | 39091531 | 44639847 | 41 | FMNL1, NMT1, NPEPPS, SKAP1, DBF4B, LOC641522, KPNB1, NFE2L1, ARL17P1, GPATCH8, LRRC37A2, TMUB2, ARL17, CCDC43, MAPT, EFTUD2, OSBPL7, ACBD4 |
| 8q24.3 | Gain | 142790550 | Telomere | 36 | HSF1, RECQL4, PLEC1, PPP1R16A, NFKBIL2, LRRC14, SCRIB, CPSF1, SIAHBP1, CPSF1, RPL8, GPAA1, MGC70857, GPR172A, ZNF7, GPR172A, C8orf33, FBXL6, BOP1, GPAA1, PYCRL, EXOSC4, C8orf30A, CYHR1, SHARPIN, ZNF707, JRK, CYC1, EEF1D, KIFC2, MAF1, COMMD5 |
| 13q14-13q31 | Loss | 42301191 | 83766576 | 36 | RB1, KLF12, TPT1, LMO7, HUWE1, RBM26, UTP14C, FNDC3A, DNAJC15, RNASEH2B, NDFIP2, INTS6, RPL13A, PTMA, COG3, DLEU1, |
| 17q25 | Gain | 69639765 | Telomere | 36 | CBX4, RECQL5, HGS, SPHK1, MIF4GD, B3GNTL1, UBE2O, NT5C, LOC124512, FLJ21865, SAP30BP, NUP85, C17orf56, NPLOC4, ACTG1, RAB40B, TRIM65, C17orf70, H3F3B, MIF4GD, FLJ30594, KIAA0195, PRPSAP1, MXRA7, FLJ35220, EXOC7, MFSD11, WDR45L, RHBDF2, TSEN54, TIMP2, TNRC6C |
| 8q24.2 | Gain | 132799581 | 137773461 | 32 | ST3GAL1, PHF20L1, KIAA0143 |
| 9p13-9p11.1 | Loss | 39990599 | 42614658 | (no genes with CNA-associated expression) | |
| 9p21 | Loss | 22579721 | (no genes with CNA-associated expression) | ||
| 9q21 | Loss | 78213759 | 80495074 | 32 | CEP78, VPS13A |
| 9q21 | Loss | 79930787 | 84622895 | (no genes with CNA-associated expression) | |
| 9q21 | Loss | 84783002 | 86180561 | 32 | UBQLN1, C9orf103, GKAP1, LOC389765 |
| 9q22-9q31 | Loss | 101410218 | 105214273 | 32 | TEX10, MRPL50, TXNDC4, RNF20, ZNF189 |
| 17q22-17q23 | Gain | 48664511 | 59626448 | 32 | SUPT4H1, DHX40, PTRH2, AKAP1, FLJ44342, TLK2, RPS6KB1, TUBD1, HEATR6, C17orf71, INTS2, MRPS23, COIL, GDPD1, METTL2A, DDX42, FTSJ3, LOC51136, ICAM2, MKS1, MSI2 |
| Cytogenetic band . | Copy number alteration . | Clone position . | Affected patients, % . | Candidate genes . | |
|---|---|---|---|---|---|
| Start . | Stop . | ||||
| 7q36 | Gain | 148089302 | 151558264 | 59 | FASTK, NUB1, LOC791120 |
| 7q21-7q22 | Gain | 97314794 | 102514284 | 55 | AP1S1, SMURF1, ZKSCAN1, C7orf38, CLDN15, ZNF789, RASA4, ZNF498, ZNF789, ARMC10, POLR2J2, ZNHIT1, ZCWPW1, MGC40499 |
| 7q32-7q35 | Gain | 130270796 | 143852574 | 55 | TRIM24, CNOT4, PTN, C7orf49, KIAA0738, LUC7L2 |
| 7q11.2 | Gain | 71274704 | 76190020 | 50 | GTF2IRD1, ABHD11, NSUN5, NSUN5B, NSUN5C, ELN, WBSCR22, TRIM73 |
| 1p36.2 | Gain | 11098993 | 12351219 | 45 | MFN2 |
| 1q31-1q32 | Gain | 198714422 | 205087972 | 45 | MDM4, NAV1, RBBP5, IPO9, CSRP1, KIF21B, PPP1R15B, NUCKS1, TIMM17A, SNRPE, KIF14 |
| 5q13 | Loss | 67677068 | 70179512 | 45 | TAF9, SERF1A, SERF1B, SMN1, TAF9, GUSBP1 |
| 7p14-7p13 | Gain | 43272694 | 45048103 | 45 | CAMK2B, POLR2J4 |
| 7p22-7p21 | Gain | 1081263 | 7947777 | 45 | WIPI2, LOC222967, FTSJ2, MICALL2 |
| 1q21-1q22 | Gain | 146342686 | 156056126 | 41 | MCL1, CLK2, PRCC, ARHGEF11, HDGF, GPATCH4, JTB, MSTO1, FLAD1, CRTC2, SMG5, ADAR, MRPL24, KRTCAP2, SETDB1, C1orf2, SF3B4, PRPF3, SEMA4A, MTX1, ISG20L2, SNAPAP, ENSA, PLEKHO1, ISG20L2, DAP3, GON4L, C1orf85, APOA1BP, C1orf43, RUSC1, UBAP2L, CDC42SE1, MAPBPIP, SCAMP3, C1orf77, PYGO2, PSMD4, GATAD2B, PEAR1, FDPS, VPS72, MRPL9, IQGAP3, DENND4B, TNFAIP8L2, UBQLN4, SLC39A1, TPM3, PRUNE |
| 7p15-7p14 | Gain | 24659178 | 37825117 | 41 | TAX1BP1, HOXA10, CREB5, HERPUD2, JAZF1, LOC441212, HNRPA2B1, C7orf41, LOC401320, KBTBD2 |
| 9p21 | Loss | 20351121 | 22479496 | 41 | CDKN2A, MTAP, LOC554202 |
| 17q21 | Gain | 39091531 | 44639847 | 41 | FMNL1, NMT1, NPEPPS, SKAP1, DBF4B, LOC641522, KPNB1, NFE2L1, ARL17P1, GPATCH8, LRRC37A2, TMUB2, ARL17, CCDC43, MAPT, EFTUD2, OSBPL7, ACBD4 |
| 8q24.3 | Gain | 142790550 | Telomere | 36 | HSF1, RECQL4, PLEC1, PPP1R16A, NFKBIL2, LRRC14, SCRIB, CPSF1, SIAHBP1, CPSF1, RPL8, GPAA1, MGC70857, GPR172A, ZNF7, GPR172A, C8orf33, FBXL6, BOP1, GPAA1, PYCRL, EXOSC4, C8orf30A, CYHR1, SHARPIN, ZNF707, JRK, CYC1, EEF1D, KIFC2, MAF1, COMMD5 |
| 13q14-13q31 | Loss | 42301191 | 83766576 | 36 | RB1, KLF12, TPT1, LMO7, HUWE1, RBM26, UTP14C, FNDC3A, DNAJC15, RNASEH2B, NDFIP2, INTS6, RPL13A, PTMA, COG3, DLEU1, |
| 17q25 | Gain | 69639765 | Telomere | 36 | CBX4, RECQL5, HGS, SPHK1, MIF4GD, B3GNTL1, UBE2O, NT5C, LOC124512, FLJ21865, SAP30BP, NUP85, C17orf56, NPLOC4, ACTG1, RAB40B, TRIM65, C17orf70, H3F3B, MIF4GD, FLJ30594, KIAA0195, PRPSAP1, MXRA7, FLJ35220, EXOC7, MFSD11, WDR45L, RHBDF2, TSEN54, TIMP2, TNRC6C |
| 8q24.2 | Gain | 132799581 | 137773461 | 32 | ST3GAL1, PHF20L1, KIAA0143 |
| 9p13-9p11.1 | Loss | 39990599 | 42614658 | (no genes with CNA-associated expression) | |
| 9p21 | Loss | 22579721 | (no genes with CNA-associated expression) | ||
| 9q21 | Loss | 78213759 | 80495074 | 32 | CEP78, VPS13A |
| 9q21 | Loss | 79930787 | 84622895 | (no genes with CNA-associated expression) | |
| 9q21 | Loss | 84783002 | 86180561 | 32 | UBQLN1, C9orf103, GKAP1, LOC389765 |
| 9q22-9q31 | Loss | 101410218 | 105214273 | 32 | TEX10, MRPL50, TXNDC4, RNF20, ZNF189 |
| 17q22-17q23 | Gain | 48664511 | 59626448 | 32 | SUPT4H1, DHX40, PTRH2, AKAP1, FLJ44342, TLK2, RPS6KB1, TUBD1, HEATR6, C17orf71, INTS2, MRPS23, COIL, GDPD1, METTL2A, DDX42, FTSJ3, LOC51136, ICAM2, MKS1, MSI2 |
Genes reported as cancer-related according to the EBI cancer gene database are shown in bold; the remaining genes are ordered according to statistical significance of differential expression. mRNA expression levels of genes underlined are determined by Q-RT-PCR.
Confirmation of gene-expression data by quantitative real-time PCR and immunohistochemistry
To validate the results of microarray analysis, we selected several candidate oncogenes and tumor suppressor genes, located in MCRs affecting at least 35% of patients and predicted to show CNA-associated dysregulation (Table 3). Expression levels of these genes were analyzed using qPCR and compared between MF samples with and without CNA of the chromosomal region harboring these genes (Figure 3). Expression levels of the FASTK, SKAP1, and MCL1 genes, located in MCRs with gain on 7q36, 17q21, and 1q21-22, respectively, were analyzed. In addition, expression of the tumor suppressor genes RB1 and DLEU1, located in a MCR on 13q14-13q31 lost in 36% of patients, was investigated. The FASTK gene and SKAP1 gene, also known as SKAP55, were selected because of their essential role in T-cell apoptosis and T-cell activation, respectively.30,32 MCL1 and RB1 are reported to be cancer-related according to the European Bioinformatics Institute cancer gene database. The putative tumor suppressor gene Deleted in Lymphocytic Leukemia 1 (DLEU1) was selected for confirmatory PCR analysis because it has also been found to be affected by promoter hypermethylation in MF.33 The mean expression intensity of FASTK was significantly higher for patients with a corresponding gain of DNA content than for those without gain (fold difference = 1.7, t test, P = .03). Similarly, the SKAP1 gene was thus confirmed to demonstrate a CNA-associated expression pattern (fold difference = 2.6, P = .01). Expression of the MCL1 gene was higher in samples with gain, but the difference did not reach statistical significance. Expression of RB1 and DLEU1 was significantly diminished in patients demonstrating loss of the chromosomal region on 13q14 (fold difference = −1.8 and −2.4; P = .02 and .01, respectively). These results indicate that gene dosage influences transcript abundance of these tumor-related genes. The relatively large SE apparent in the data in Figure 3 on the one hand reflects heterogeneity in expression levels within the group of samples but may also suggest that gene-expression levels are influenced by other factors such as multiple copy gain and promoter hypermethylation.
Relative mRNA expression in MF tumor samples as measured by qPCR. Data (mean ± SEM) of 3 independent qPCR experiments are depicted relative to the reference genes RPS11 and U1A.  indicate qPCR results using cDNA synthesized from RNA isolated from samples with no CNAs;
indicate qPCR results using cDNA synthesized from RNA isolated from samples with no CNAs;  , qPCR results from samples with copy number gains; and □, qPCR results from samples with copy number loss. *Statistically significant differential expression (P < .05).
, qPCR results from samples with copy number gains; and □, qPCR results from samples with copy number loss. *Statistically significant differential expression (P < .05).
Relative mRNA expression in MF tumor samples as measured by qPCR. Data (mean ± SEM) of 3 independent qPCR experiments are depicted relative to the reference genes RPS11 and U1A.  indicate qPCR results using cDNA synthesized from RNA isolated from samples with no CNAs;
indicate qPCR results using cDNA synthesized from RNA isolated from samples with no CNAs;  , qPCR results from samples with copy number gains; and □, qPCR results from samples with copy number loss. *Statistically significant differential expression (P < .05).
, qPCR results from samples with copy number gains; and □, qPCR results from samples with copy number loss. *Statistically significant differential expression (P < .05).
In addition, protein expression of RB1 and SKAP1 was evaluated by immunohistochemical staining of tissue sections of 10 MF tumor samples. We found that RB1 was expressed by tumor cells of samples without loss of the locus harboring this gene. However, expression of RB1 was absent in the majority of tumor cells in 2 of the 5 tumor samples demonstrating loss of the locus harboring this gene, indicative of loss or epigenetic silencing of the other allele. Loss of RB1 protein expression in CTCL has been reported previously.34,35 SKAP1 showed strong cytoplasmic staining in lymphoid cells in all MF samples. Although tumor samples in which gain of the SKAP1 locus had been detected appeared to display slightly more intense staining, no significant difference in staining intensity between samples with and without gain of the locus harboring the gene could be discerned. Results of exemplary stainings are shown in Figure S4.
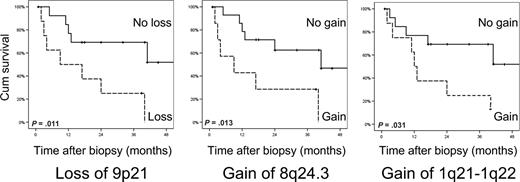
Chromosomal alterations with prognostic significance
Next, we determined possible relationships between the occurrence of specific chromosomal alterations and the clinical behavior of these MF patients. For each of the 24 MCRs with CNA affecting at least 35% of patients, we compared the disease-specific survival rate in the group of patients harboring this CNA to survival in the group of patients not affected by the particular CNA. Patients whose tumor cells showed loss of 9p21 (Mb position 20351121-22479496), gain of 8q24.3, or gain of 1q21-1q22 had a statistically significantly lower survival rate (log-rank test, P = .011, .013, and .031, respectively). Figure 4 shows survival curves of patients with and without these 3 CNAs with prognostic significance. These loci may contain genes that modify the biologic behavior or treatment response of MF. Loss of the 9p21 locus, harboring the CDKN2A tumor suppressor gene, has been reported to predict more aggressive disease behavior in cutaneous B-cell lymphoma and nodal lymphomas previously.36,37
Chromosomal alterations associated with lower disease-specific survival rates in MF. Patients were divided based on the loss of an MCR on 9p21, or gain of MCRs on 8q24.3 or gain of 1q21-22. Actuarial survival rates were calculated from the date of biopsy using the Kaplan-Meier technique. The log-rank test was used to analyze differences between survival rates.
Chromosomal alterations associated with lower disease-specific survival rates in MF. Patients were divided based on the loss of an MCR on 9p21, or gain of MCRs on 8q24.3 or gain of 1q21-22. Actuarial survival rates were calculated from the date of biopsy using the Kaplan-Meier technique. The log-rank test was used to analyze differences between survival rates.
Discussion
Our study provides a genome-wide analysis of recurrent chromosomal alterations in a panel of 22 well-defined tumor-stage MF cases. A primary goal of this investigation was to compare the patterns of chromosomal alterations observed in MF with those recently identified in Sz. Both conditions are malignancies originating from activated, skin-homing, memory T cells with cerebriform nuclei. In 1975, based on the morphologic and immunophenotypic similarities between MF and Sz and related lymphoid malignancies, Lutzner et al proposed the encompassing term CTCL for this group of diseases.38 In many subsequent studies on CTCL, no distinction has been made between MF and Sz. In reviews and textbooks, Sz is often designated as a leukemic phase or variant of MF, suggesting that differences between both conditions are mainly a matter of stage of disease.13,16-20 However, Sz presents with erythroderma and lymph node and blood involvement and has a poor prognosis, whereas MF generally behaves as a low-grade lymphoma with limited, skin-confined disease for years or decades.1,2 There are also histopathologic differences between both conditions. Whereas infiltration of the epidermal basal layers is the hallmark of early MF, in Sz the atypical cells are predominantly found around the dermal blood vessels, although a variable degree of epidermotropism may be present as well.39 Consistent with its leukemic nature, involved lymph nodes in Sz are typically overrun by a monotonous infiltrate of Sézary cells, whereas dermatopathic lymphadenopathy as seen in early involvement by MF tumor cells is often absent.40 MF and Sz have been reported to share several chromosomal alterations, analyzed using conventional CGH, such as loss of chromosomal regions on 1p, 10q, and 17p.4,5 However, in line with our findings differences in CNA patterns of MF and Sz, including a higher frequency of gain of 17q in Sz, have been recognized.18 More recently, expression of CDO1 and DNM3, genes specifically expressed in Sz, could not be demonstrated in MF.41
The mapping resolution of the array-based CGH method applied in this study allows a more detailed definition of chromosomal alterations than obtained by fluorescence in situ hybridization and conventional CGH, used in previous studies of CTCL. The detailed genomic profiles of chromosomal imbalances of MF tumor cells displayed marked differences with those previously identified in Sz cells using identical methods. Numerical chromosomal alterations most frequently observed in MF include gain of 7q21-36 and 1p36.2 and loss of 5q13 and 9p21, whereas Sz is characterized by gain of 17q22-25 and 8q22-24 and loss of 17p13 and 10q25. Amplification of the locus containing the MYC gene on 8q24, observed in 75% of patients with Sz and associated with increased expression of this oncogenic transcription factor, was detected in only a minority of patients with MF.15 Notably, several aberrations commonly observed in MF are not or infrequently seen in Sz, arguing against the notion that Sz represents an advanced stage of MF. Gain or loss of chromosomal regions may be associated with altered expression of resident oncogenes or tumor suppressor genes and thereby have a causative role in the development and progression of lymphoma. The pattern of chromosomal alterations, in particular highly recurrent focal gains and losses, is therefore often characteristic of a certain type of malignancy and can be informative of its pathogenesis. Although the chromosomal alterations in MF and Sz show heterogeneity within the group, the overall patterns clearly differ. This strongly suggests that the molecular pathogenesis of these CTCLs follows distinct pathways. By implication, patients with these 2 CTCL subtypes may respond differently to treatment regimens. In current clinical trials, patients with MF and Sz are often included collectively as CTCL or MF/Sz.42 It is conceivable that the efficacy of experimental therapeutics, such as inhibitors of STAT or MYC transcription factors, would differ considerably between MF and Sz. Therefore, patients with MF and Sz should be entered in clinical trials separately or results of such trials should at least be stratified according to CTCL type.
The molecular genetic alterations underlying the development and progression of MF are largely unresolved. The second goal of this study was to identify pathobiologically relevant genes in MF by evaluating the expression of genes residing in smallest overlapping chromosomal regions (MCRs) with highly recurrent CNA. A subset of the 253 genes that demonstrated CNA-associated dysregulated expression is known to be cancer-related, and several other genes have been reported to have essential roles in T-cell activation and proliferation. By integrating array-based genetic maps with gene-expression signatures derived from the same MF tumor biopsy samples, we thus identified several oncogenes and tumor suppressor genes, including RB1, CDKN2A, MCL1, and MDMX as targets of gain and loss in MF. Interestingly, the most frequently observed CNA, gain of an MCR on chromosome 7q36 affecting 59% of MF patients, was associated with increased expression of the FASTK gene. The protein encoded by this gene is a member of the serine/threonine protein kinase family and is normally expressed in human T cells.31 Although some earlier reports suggested that FASTK may be involved in the induction of Fas-induced apoptosis, most evidence indicates that this protein has antiapoptotic properties.30 FASTK attenuates apoptosis induced by ultraviolet radiation and FAS ligation, in part by increasing expression of XIAP and cIAP1. Short interfering RNA (siRNA)–mediated interference with FASTK expression increases apoptosis in human cells.30 Moreover, FASTK regulates splicing of several genes, including FGFR2 and FAS.43 Previously, our group has noted aberrant splicing of the FAS gene in MF.8 It is conceivable that FASTK dysregulation may be related to FAS splicing alterations in MF tumor cells, and this possible relationship may be a subject of further study. Consistent with previous reports, we found recurrent loss of 9p21 and 13q14 and diminished expression of the CDKN2A and RB1 tumor suppressor genes residing in these loci.6,34,35 In a subset of MF patients with loss of the RB1 locus protein, expression of this essential cell cycle regulator was diminished. In a study by Zhang et al, the RB1 protein was found to be functionally inactivated in a subset of patients with advanced MF through hyperphosphorylation.35 In addition, the DLEU1 gene is located on 13q14.3 and shows reduced expression. The promoter of DLEU1 displays frequent hypermethylation in MF, suggesting that genetic and epigenetic mechanisms collectively act to silence this gene.33 In addition to RB1 and DLEU1, the 13q14 region lost in 36% of MF patients also contains the miR-15a and miR-16-1 gene cluster. These microRNA genes have tumor suppressive properties as their expression inhibits translation of the antiapoptotic protein BCL2. Loss of 13q14.3 and concomitant reduced expression of these tumor suppressive microRNA genes, resulting in elevated protein levels of BCL2, are frequent events in chronic lymphocytic leukemia.44 Consistently, malignant T cells in MF skin lesions have been reported to demonstrate high expression of BCL2.45
Finally, we attempted to evaluate the prognostic relevance of registered recurrent CNAs. Patients with MF who demonstrated loss of the MCR on 9p21, gain 8q24.3, or gain of 1q21-1q22 appeared to have significantly lower survival rates than patients whose tumor cells were not affected by these CNAs. The chromosomal region on 9p21 harbors the CDKN2A tumor suppressor gene, which showed reduced expression in patients with loss of this region. Loss of 9p21 and reduced expression of p16 encoded by CDKN2A have been found to predict an unfavorable prognosis in various hematopoietic malignancies.36,37 Consistent with clinical observations, inactivating mutations in CDKN2A promote tumorigenesis and resistance to chemotherapy in experimental lymphoma in murine model systems. As in experimental lymphoma, treatment resistance in MF patients whose tumor cells are affected by loss of CDKN2A may be explained by defects in the induction of apoptosis and senescence in response to therapy. The locus with prognostic significance on 8q24.3 contains 28 genes with gain-associated increased expression, including the HSF1 gene. This heat shock response regulator has been found to be a determinant of chemotherapeutic efficacy in malignancy.46 Gain of chromosome 8q was previously identified as a hallmark of progressive MF associated with shorter survival.47,48 Gain of the chromosomal region on 1q21-22 is associated with significantly higher expression of several genes, including the MCL1 gene. This antiapoptotic gene was recently observed to be part of a gene cluster up-regulated in patients with advanced CTCL.49 Protein levels of MCL1 have been demonstrated to be elevated in advanced skin lesions of patients with CTCL.50 It is tempting to speculate that dysregulated expression of this gene influences the disease course of patients with MF, because MCL1 has been shown to modulate glucocorticoid resistance in lymphoid malignant cells.51 Interestingly, it was reported in that study that the mTOR-inhibitor rapamycin can modulate MCL1 activity and thereby restore glucocorticoid sensitivity, suggesting that addition of rapamycin to chemotherapy of patients with treatment-refractory MF, especially in case of 1q21-22 gain, could potentially enhance therapeutic efficacy. Whereas our study and those of others5,47,48 have focused on genomic alterations associated with an adverse prognosis, Shin et al aimed to identify gene-expression patterns marking patients with aggressive disease.49 Apart from MCL1, no other candidate genes detected in their study as being associated with aggressive CTCL were found to reside in the 3 loci with prognostic significance we identified. The observed associations of specific chromosomal alterations and gene-expression patterns with prognosis require further investigation in independent prospective studies.
In conclusion, we have attempted to provide a comprehensive characterization of recurrent chromosomal alterations of MF, a thus far poorly understood malignancy. The application of array-based CGH has revealed important molecular distinctions between MF and Sz not previously appreciated. These findings may have consequences, not only for our understanding of the pathogenesis of these CTCLs, but also clinically for the design of trials to evaluate the efficacy of novel treatments. The integration of high-resolution copy number and gene-expression data has afforded relevant novel insights into molecular genetic alterations underlying MF. Overexpression of FASTK, MCL1, and SKAP1 associated with chromosomal gain and reduced expression of CDKN2A, RB1, and DLEU1 related to loss are important candidate oncogenic events in MF. Elucidation of the biologic role of the identified candidate oncogenes and tumor suppressor genes in the development and progression of MF should be the focus of further studies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank E. J. Dreef and Dr P. M. Jansen (Department of Pathology, Leiden University Medical Center, Leiden, The Netherlands) for their assistance in performing the immunohistochemical stainings.
This work was supported by a grant from The Netherlands Organisation for Scientific Research (NWO VIDI grant 016.076.347; M.H.V.).
Authorship
Contribution: R.v.D. designed research, contributed vital patient material, analyzed and integrated data, and wrote the paper; M.S.v.K. and C.P.T. designed research, analyzed and integrated data, and wrote the paper; R.D designed and performed research; M.H.V. and R.W. contributed vital patient material, designed research, and wrote the paper; A.A.M. and J.K. performed research; and K.S. and J.M.B. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Remco van Doorn, Department of Dermatology, Leiden University Medical Center, PO Box 9600, 2300 RC, Leiden, The Netherlands; e-mail: rvandoorn@lumc.nl.