Key Points
The posttreatment end point progression of FL within 24 months (POD24) is strongly associated with OS.
A pretreatment clinicogenetic risk model (m7-FLIPI) predicts POD24 and OS and identifies the smallest subgroup with highest unmet need.
Abstract
Follicular lymphoma (FL) is a clinically and molecularly heterogeneous disease. Posttreatment surrogate end points, such as progression of disease within 24 months (POD24) are promising predictors for overall survival (OS) but are of limited clinical value, primarily because they cannot guide up-front treatment decisions. We used the clinical and molecular data from 2 independent cohorts of symptomatic patients in need of first-line immunochemotherapy (151 patients from a German Low-Grade Lymphoma Study Group [GLSG] trial and 107 patients from a population-based registry of the British Columbia Cancer Agency [BCCA]) to validate the predictive utility of POD24, and to evaluate the ability of pretreatment risk models to predict early treatment failure. POD24 occurred in 17% and 23% of evaluable GLSG and BCCA patients, with 5-year OS rates of 41% (vs 91% for those without POD24, P < .0001) and 26% (vs 86%, P < .0001), respectively. The m7–FL International Prognostic Index (m7-FLIPI), a prospective clinicogenetic risk model for failure-free survival, had the highest accuracy to predict POD24 (76% and 77%, respectively) with an odds ratio of 5.82 in GLSG (P = .00031) and 4.76 in BCCA patients (P = .0052). A clinicogenetic risk model specifically designed to predict POD24, the POD24-PI, had the highest sensitivity to predict POD24, but at the expense of a lower specificity. In conclusion, the m7-FLIPI prospectively identifies the smallest subgroup of patients (28% and 22%, respectively) at highest risk of early failure of first-line immunochemotherapy and death, including patients not fulfilling the POD24 criteria, and should be evaluated in prospective trials of precision medicine approaches in FL.
Introduction
Follicular lymphoma (FL) is among the most common malignant lymphomas worldwide and remains incurable for most patients.1 FL is a highly heterogeneous disease,2 with a subgroup of patients experiencing remarkably poor outcome. Several recent studies have suggested that posttreatment surrogate end points are powerful predictors for overall survival (OS).3,4 For example, 19% to 26% of patients receiving first-line immunochemotherapy with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) experienced progression of disease within 24 months (“early progression of disease,”4 herein referred to as POD24) and had a 5-year OS of only 34% to 50% compared with a 5-year OS of 90% to 94% for patients without POD24.4 Independent validation of these results is needed, also in the context of different treatment regimens. Furthermore, the length of first remission was calculated differently across studies, either from date of diagnosis4 (for database reasons) or after treatment.3
Although conceptually similar results are emerging for event-free survival at 12 and 24 months (EFS12 and EFS24)5,6 and complete response rate at 30 months (CR30),7 retrospective evaluation of treatment outcome is of limited clinical utility, because it cannot be used to guide up-front treatment decision. Furthermore, the molecular determinants of poor patient outcome remain to be defined. To develop precision medicine treatment strategies, it is essential to establish pretreatment strategies for risk assessment that include clinically relevant biomarkers.
We have previously shown that a clinicogenetic risk model called the m7–FL International Prognostic Index (m7-FLIPI), which includes the mutation status of 7 genes (EZH2, ARID1A, MEF2B, EP300, FOXO1, CREBBP, and CARD11), the FLIPI, and the Eastern Cooperative Oncology Group (ECOG) performance status at the time of treatment initiation improves risk stratification for failure-free survival (FFS) in patients with FL receiving first-line immunochemotherapy.8 An online tool for calculating the m7-FLIPI is available at: http://www.glsg.de/m7-flipi/.
In this study, we aimed to independently validate the predictive utility of posttreatment evaluation by POD24 in 2 independent cohorts of patients who received different immunochemotherapy regimens as first-line treatment. Furthermore, we evaluated and compared the ability of pretreatment risk models, including the m7-FLIPI, to predict POD24, and explored additional pretreatment risk models specifically designed to predict POD24.
Methods
We fully reanalyzed the clinical and molecular data from 2 independent cohorts of patients with symptomatic, advanced stage, or bulky FL considered ineligible for curative radiotherapy. All patients had an available biopsy specimen obtained within 12 months before the initiation of first-line therapy that was previously sequenced to determine the mutational status of 74 genes.8
Briefly, the GLSG cohort consisted of 151 patients who needed treatment as defined by the presence of B-symptoms, bulky disease (mediastinal lymphomas >7.5 cm or other lymphomas >5 cm), impairment of normal hematopoiesis (hemoglobin level <100 g/L, granulocyte count <1.5 × 109/L, or thrombocyte count <100 × 109/L), compression of internal organs, or disease progression (>50% increase of lymphoma manifestations within <6 months). All patients received R-CHOP and interferon-α (IFN-α) maintenance as part of the randomized GLSG2000 trial of the German Low-Grade Lymphoma Study Group (GLSG).9 Median age of GLSG patients was 57 years (range 27-77); 77 (51%) had high-risk FLIPI. With a median follow-up of 7.7 years, 5-year FFS and OS rates were 66% and 83%, respectively.8
The BCCA cohort consisted of 107 patients from a population-based registry of the British Columbia Cancer Agency (BCCA) who received rituximab, cyclophosphamide, vincristine, and prednisone (R-CVP), followed by R-maintenance by intention to treat in 93 patients (87%). Median age of BCCA patients was 62 years (range 37-83); 53 (50%) had high-risk FLIPI. With a median follow-up of 6.7 years, 5-year FFS and OS rates were 58% and 74%, respectively.8
Progression of disease within 24 months was defined as progression or relapse of the disease within the first 24 months after diagnosis (original definition)4 or after first-line treatment initiation (modified definition). Patients were not evaluable for POD24 if they were censored (eg, lost to follow-up) or died within 24 months without POD.
Failure-free survival was defined as time from treatment initiation until less than a partial remission (PR) at the end of induction, relapse, progression, or death from any cause. Overall survival was calculated from risk-defining event for POD24 (ie, survival from time of POD for the POD24 cohort, or from 2 years after initial treatment of patients without POD24),4 and from treatment initiation for all other survival analyses.
Clinical and molecular data from the GLSG cohort were used to calculate a risk model that specifically predicts POD24 (POD24 Prognostic Index [POD24-PI]) by applying a previously described statistical approach.8 Briefly, the mutation status of genes that were mutated in >5 patients and the clinical risk factors FLIPI >2 (ie, high-risk FLIPI) and poor performance status (ECOG-PS >1) were used for multivariable L1-penalized logistic regression. Two different risk models were calculated. In the first model, the coefficients for high-risk FLIPI and ECOG-PS >1 were not penalized, forcing these variables into the model. In the second model, all coefficients were penalized. Internal validation by the bootstrap procedure was used to select the best model. The final risk score was calculated as the sum of clinical and molecular predictors weighted by their individual Lasso coefficients. We determined the optimal cutoff value to maximize the Wald statistic, and dichotomized patients into high-risk and low-risk subgroups. The BCCA cohort was used as an independent validation cohort.
Logistic regression analyses were performed to assess whether risk models were predictive of POD24, and Cox regression analysis was used for FFS and OS. All calculations were carried out with the statistical software R (version 3.1.2). The accuracy of pretreatment risk models to predict POD24 was calculated as the number of correctly classified patients ([number of true positives + number of true negatives] ÷ [number of all evaluable patients]). The R-package penalized (version 0.9-45) was used for penalized logistic regression, and the survival package (version 2.37-7) for survival analyses.
This study was covered by approvals of the Ludwig-Maximilians-University Munich Institutional Review Board (#056-00) and the University of British Columbia–BCCA Research Ethics Board (#H13- 01765).
Results
Validation of POD24 to identify high-risk patients
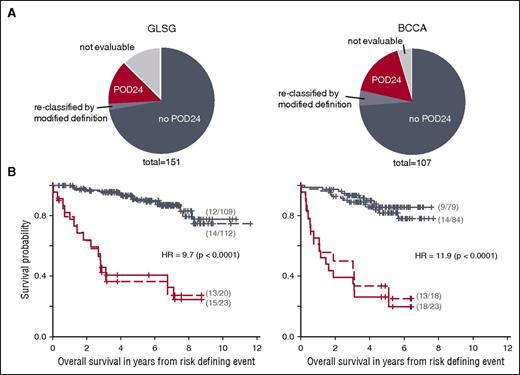
We first aimed to assess the prognostic impact of POD24 on OS in 2 independent cohorts of patients with FL receiving first-line immunochemotherapy. Nineteen (13%) and 5 patients (5%) from the entire GLSG and BCCA cohorts were not evaluable for analysis of POD24 because they were censored or died within 24 months without prior POD (Figure 1A). POD24, originally defined as relapse or progression of FL within 24 months of diagnosis,4 occurred in 15% (20/132) and 18% (18/102) of evaluable patients from the GLSG and the BCCA cohorts (Figure 1A). When calculated from time of first-line treatment initiation to overcome the lead-time bias (ie, the time between diagnosis and symptomatic disease requiring treatment), the size of POD24 subgroups increased to 17% (23/132) and 23% (23/102), respectively (Figure 1A; Table 1). Only 1 of the 8 reclassified patients was still alive at 7.6 years, and the median OS of this subgroup was only 3.1 years (range 1.4-9.5; P < .0001 compared with all other patients). The number of reclassified patients is small (the time between diagnosis and treatment was <1 year by inclusion criteria), but the poor outcome of patients with POD within 24 months of treatment, but not from diagnosis, suggests that these patients should also be considered early progressors. Thus, the modified definition of POD24 was used for the remainder of the study.
Progression of disease within 24 months (POD24) is an accurate predictor of poor overall survival (OS). (A) Distribution of patients from the GLSG and BCCA cohorts according to the POD24 classifier. (B) Kaplan Meier curves for OS from risk-defining events for patients with or without POD24 of diagnosis (dashed lines) or 24 months of treatment initiation (solid lines) from the GLSG (left) and BCCA cohorts (right). Displayed statistics refer to POD24 status calculated from time of treatment initiation (ie, modified definition).
Progression of disease within 24 months (POD24) is an accurate predictor of poor overall survival (OS). (A) Distribution of patients from the GLSG and BCCA cohorts according to the POD24 classifier. (B) Kaplan Meier curves for OS from risk-defining events for patients with or without POD24 of diagnosis (dashed lines) or 24 months of treatment initiation (solid lines) from the GLSG (left) and BCCA cohorts (right). Displayed statistics refer to POD24 status calculated from time of treatment initiation (ie, modified definition).
Patient and disease characteristics according to POD24 status
| . | GLSG . | BCCA . | ||||
|---|---|---|---|---|---|---|
| . | POD24 . | No POD24 . | P . | POD24 . | no POD24 . | P . |
| No. of evaluable patients | 23 | 109 | 23 | 79 | ||
| First-line treatment | R-CHOP (151/151, 100%) | R-CVP (107/107, 100%) | ||||
| Maintenance treatment by ITT | IFN (151/151, 100%) | Rituximab (93/107, 87%) | ||||
| Median follow-up in years | 8.4 | 8.2 | 7.1 | 6.7 | ||
| Age (y), median (range) | 61 (27-74) | 56 (29-77) | .195 | 62 (43-83) | 61 (37-83) | .398 |
| Male gender | 11/23 (48%) | 55/109 (50%) | >.99 | 15/23 (65%) | 42/79 (53%) | .618 |
| High-risk FLIPI | 18/23 (78%) | 48/109 (44%) | .0059 | 16/23 (70%) | 33/79 (42%) | .035 |
| Age >60 y | 12/23 (52%) | 39/109 (36%) | .218 | 12/23 (52%) | 42/79 (53%) | >.99 |
| No. of nodal sites >4 | 19/23 (83%) | 71/109 (65%) | .165 | 20/23 (87%) | 55/79 (70%) | .164 |
| LDH elevated | 11/23 (48%) | 31/109 (28%) | .117 | 6/21 (29%) | 15/77 (19%) | .548 |
| Hb <120 g/L | 10/23 (43%) | 17/109 (16%) | .0064 | 4/21 (19%) | 7/79 (9%) | .350 |
| ECOG-PS ≥2 | 0/23 (0%) | 5/109 (5%) | .655 | 6/23 (26%) | 9/79 (11%) | .157 |
| . | GLSG . | BCCA . | ||||
|---|---|---|---|---|---|---|
| . | POD24 . | No POD24 . | P . | POD24 . | no POD24 . | P . |
| No. of evaluable patients | 23 | 109 | 23 | 79 | ||
| First-line treatment | R-CHOP (151/151, 100%) | R-CVP (107/107, 100%) | ||||
| Maintenance treatment by ITT | IFN (151/151, 100%) | Rituximab (93/107, 87%) | ||||
| Median follow-up in years | 8.4 | 8.2 | 7.1 | 6.7 | ||
| Age (y), median (range) | 61 (27-74) | 56 (29-77) | .195 | 62 (43-83) | 61 (37-83) | .398 |
| Male gender | 11/23 (48%) | 55/109 (50%) | >.99 | 15/23 (65%) | 42/79 (53%) | .618 |
| High-risk FLIPI | 18/23 (78%) | 48/109 (44%) | .0059 | 16/23 (70%) | 33/79 (42%) | .035 |
| Age >60 y | 12/23 (52%) | 39/109 (36%) | .218 | 12/23 (52%) | 42/79 (53%) | >.99 |
| No. of nodal sites >4 | 19/23 (83%) | 71/109 (65%) | .165 | 20/23 (87%) | 55/79 (70%) | .164 |
| LDH elevated | 11/23 (48%) | 31/109 (28%) | .117 | 6/21 (29%) | 15/77 (19%) | .548 |
| Hb <120 g/L | 10/23 (43%) | 17/109 (16%) | .0064 | 4/21 (19%) | 7/79 (9%) | .350 |
| ECOG-PS ≥2 | 0/23 (0%) | 5/109 (5%) | .655 | 6/23 (26%) | 9/79 (11%) | .157 |
ECOG-PS, Eastern Cooperative Oncology Group Performance Status; Hb, hemoglobin; IFN, interferon-α; ITT, intention-to-treat; LDH, lactate dehydrogenase; POD24, progression of disease within 24 months; R-CHOP, rituximab, cyclophosphamide, doxorubicin, vincristine, prednisone; R-CVP, rituximab, cyclophosphamide, vincristine, prednisone.
Differences in OS, calculated from risk-defining event (ie, survival from time of POD for early progressors, or from 24 months after initial treatment for non–early progressors) were highly significant between patients with and without POD24 (irrespective of whether original or modified definitions were used; Figure 1B). Six and 4 patients with POD24 from the GSLG and BCCA cohorts were still alive at 5 years, for a 5-year OS of 41% vs 91% (hazard ratio [HR] 9.72, 95% confidence interval [CI] [4.51; 20.96], P < .0001) and 26% vs 86% (HR 11.93, 95% CI [5.31; 26.76], P < .0001), respectively (Figure 1B). This confirms that retrospective evaluation of treatment response at 24 months is strongly associated with OS in patients receiving first-line immunochemotherapy.
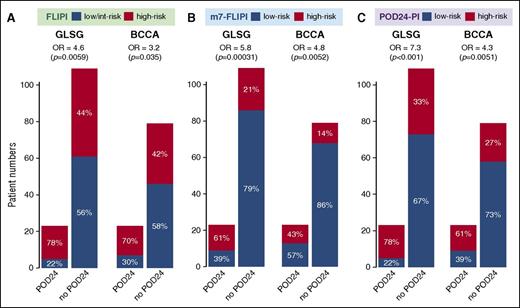
The clinical characteristics of the POD24 and non-POD24 subgroups are summarized in Table 1. The POD24 subgroups were enriched for high-risk FLIPI (78% vs 44%, P = .0059, and 70% vs 42%, P = .035, for GLSG and BCCA patients, respectively; Table 1; Figure 2A). Another 44% and 42% of patients without POD24 had high-risk FLIPI, respectively (Figure 2A). However, patients with high-risk FLIPI and no POD24 did not have inferior FFS compared with those with low-risk FLIPI and no POD24 (Figure 3A). This suggests that the FLIPI, which uses only clinical factors and classified 51% of GLSG and 50% of BCCA patients as high-risk, overestimates the number of patients with poor outcome.
Accuracy of 3 pretreatment risk models to predict POD24 status. (A) Follicular Lymphoma International Prognostic Index (FLIPI), (B) m7-FLIPI, and (C) POD24 Prognostic Index (POD24-PI).
Accuracy of 3 pretreatment risk models to predict POD24 status. (A) Follicular Lymphoma International Prognostic Index (FLIPI), (B) m7-FLIPI, and (C) POD24 Prognostic Index (POD24-PI).
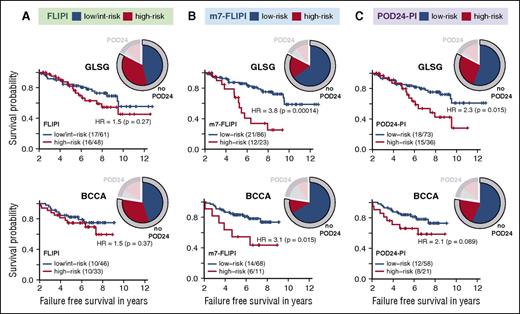
Risk stratification for failure-free survival (FFS) in patients without POD24 according to 3 pretreatment risk models. Kaplan-Meier curves for FFS beyond 2 years after treatment initiation for patients without POD24 according to (A) the FLIPI, (B) the m7-FLIPI, and (C) the POD24-PI. Numbers in parentheses indicate patients with event/number of patients per subgroup. Pie charts illustrate distribution of risk status of the respective risk classifier and POD24 status.
Risk stratification for failure-free survival (FFS) in patients without POD24 according to 3 pretreatment risk models. Kaplan-Meier curves for FFS beyond 2 years after treatment initiation for patients without POD24 according to (A) the FLIPI, (B) the m7-FLIPI, and (C) the POD24-PI. Numbers in parentheses indicate patients with event/number of patients per subgroup. Pie charts illustrate distribution of risk status of the respective risk classifier and POD24 status.
The m7-FLIPI is predictive of POD24
We have previously shown that by integrating the mutation status of 7 genes with clinical risk factors, the clinicogenetic risk model m7-FLIPI results in reclassification of approximately one half of high-risk FLIPI patients into the low-risk group.8 Now, we assessed the performance of the m7-FLIPI to prospectively distinguish patients with and without POD24.
Unlike the POD24 classifier, all patients were evaluable for the m7-FLIPI (Table 2). Forty-three GLSG (28%) and 24 BCCA patients (22%) were classified as high risk by the m7-FLIPI, with a 5-year OS from treatment initiation of 65% vs 90% (HR 3.4, P < .0001) and 42% vs 84% (HR 4.9, P < .0001), respectively (Table 2). High-risk m7-FLIPI patients were significantly more likely to develop POD24 with an odds ratio (OR) of 5.82 (95% CI [2.27; 15.63]; P = .00031) and 4.76 (95% CI [1.68; 13.72]; P = .0052) in GLSG and BCCA patients (Figure 2B; supplemental Figure 1A). Compared with the FLIPI, the specificity of the m7-FLIPI to identify POD24 (ie, the true negative rate) increased from 56% to 79%, and 58% to 86%, respectively (Figure 2B). However, 21% of GLSG and 14% of BCCA patients who did not experience POD24 were still assigned into the high-risk m7-FLIPI subgroup (Figure 2B). To determine whether these cases have an inferior prognosis even though they do not progress within 24 months, we analyzed the impact of high-risk m7-FLIPI in patients without POD24. In both cohorts, high-risk m7-FLIPI was still associated with a shorter FFS (Figure 3B) and OS (supplemental Figure 2B) among patients who did not have POD24. Thus, the accuracy of the FLIPI to predict POD24 is substantially improved by adding the ECOG-PS and the mutation status of 7 genes (Table 2). Furthermore, the m7-FLIPI is also predictive for treatment outcome in patients not fulfilling the criteria of POD24.
Features and test metrics of tested risk classifiers for previously untreated patients with symptomatic, advanced stage FL from 2 independent cohorts
| . | POD24* . | FLIPI . | m7-FLIPI . | POD24-PI . |
|---|---|---|---|---|
| Type | Posttreatment | Pretreatment | Pretreatment | Pretreatment |
| Primary endpoint | OS | OS | FFS | POD24 |
| Validated on independent cohort | Yes | Yes | Yes | Yes |
| Clinical predictors | POD24 | Age >60 y, no. of nodal sites >3, elevated serum LDH, hemoglobin <12 g/L, Ann Arbor stage III/IV | High-risk FLIPI, ECOG performance status >2 | High-risk FLIPI |
| Molecular predictors | None | None | Nonsilent mutations in 7 genes (ARID1A, CARD11, CREBBP, EP300, EZH2, FOXO1, MEF2B) at VAFs ≥10% | Nonsilent mutations in 3 genes (EP300, EZH2, FOXO1) at VAFs ≥10% |
| Calculation of risk score | Single variable | Cumulative sum of predictor values, all predictors have weight = 1 | Cumulative sum of predictor values, predictors have individual weights† | Cumulative sum of predictor values, predictors have individual weights (Figure 4A) |
| Number of risk groups | 2 (no POD24 = low-risk, POD24 = high-risk) | 3 (0-1 = low-risk, 2 = interm-risk, 3-5 = high-risk) | 2 (<0.8 = low-risk, >0.8 = high-risk) | 2 (<0.71 = low-risk, >0.71 = high-risk) |
| High-risk cases among patients with symptomatic, advanced stage FL (%) | GLSG: 23/132 (17) | GLSG: 77/151 (51) | GLSG: 43/151 (28) | GLSG: 63/151 (42) |
| BCCA: 23/102 (22) | BCCA: 53/107 (50) | BCCA: 24/107 (22) | BCCA: 39/107 (36) | |
| Predictive for POD24 | n/a | Yes | Yes | Yes |
| GLSG: evaluable patients 132/151 | OR = 4.6 (P = .0059) | OR = 5.8 (P = .00031) | OR = 7.3 (P = .00016) | |
| Sens 78%, Spec 56%, AUC 0.67 | Sens 61%, Spec 79%, AUC 0.70 | Sens 78%, Spec 67%, AUC 0.73 | ||
| Accuracy 60%, PPV 27%, NPV 92% | Accuracy 76%, PPV 38%, NPV 91% | Accuracy 71%, PPV 33%, NPV 94% | ||
| BCCA: evaluable patients 107/107 | OR = 3.2 (P = .035) | OR = 4.8 (P = .0052) | OR = 4.3 (P = .0051) | |
| Sens 70%, Spec 58%, AUC 0.64 | Sens 43%, Spec 86%, AUC 0.65 | Sens 61%, Spec 73%, AUC 0.67 | ||
| Accuracy 61%, PPV 33%, NPV 87% | Accuracy 77%, PPV 48%, NPV 84% | Accuracy 69%, PPV 40%, NPV 87% | ||
| Predictive for FFS | n/a | Yes, but only as a binary classifier (low-/interm-risk vs high-risk) | Yes | Yes |
| GLSG: evaluable patients 151/151 | 5-y FFS 57% vs 76% | 5-y FFS 38% vs 77% | 5-y FFS 50% vs 77% | |
| HR = 2.11 (P = .0034) | HR = 4.14 (P = 6.313·10−9) | HR = 3.06 (P = 5.989·10−6) | ||
| BCCA: evaluable patients 107/107 | 5-y FFS 47% vs 70% | 5-y FFS 25% vs 68% | 5-y FFS 36% vs 72% | |
| HR = 2.18 (P = .0075) | HR = 3.58 (P = 4.924·10−6) | HR = 3.01 (P = 7.178·10−5) | ||
| Predictive for OS | Yes‡ | Yes | Yes | Yes |
| GLSG: evaluable patients 151/151 | 5-y OS 40% vs 96% | 5-y OS 75% vs 91% | 5-y OS 65% vs 90% | 5-y OS 71% vs 91% |
| HR = 10.91 (P = 5.262·10−14) | HR = 2.59 (P = .0083), C-Index 0.75 | HR = 3.38 (P = .00031), C-Index 0.78 | HR = 3.55 (P = .00026), C-Index 0.79 | |
| BCCA: evaluable patients 107/107 | 5-y OS 26% vs 93% | 5-y OS 60% vs 89% | 5-y OS 42% vs 84% | 5-y OS 48% vs 89% |
| HR = 13.602 (P = 6.661·10−16) | HR = 3.90 (P = .00034), C-Index 0.81 | HR = 4.89 (P = 7.661·10−7), C-Index 0.84 | HR = 5.35 (P = 9.996·10−7), C-Index 0.86 | |
| Comments | Risk classification not possible for patients who died/were censored within 24 mo of first-line treatment. Only predictive for OS. First described in 2015. | Most widely used and best validated pretreatment classifier. Contains only clinical variables not necessarily directly reflecting disease biology. First described in 2004, not widely used to guide treatment decisions. | Highest accuracy and specificity to predict POD24. Most discriminative classifier for patients without POD24 into high- and low-risk groups. Sequencing of 7 genes required. First described in 2015, requires further validation. | Highest sensitivity to predict POD24. Sequencing of 3 genes required. Requires further validation. |
| . | POD24* . | FLIPI . | m7-FLIPI . | POD24-PI . |
|---|---|---|---|---|
| Type | Posttreatment | Pretreatment | Pretreatment | Pretreatment |
| Primary endpoint | OS | OS | FFS | POD24 |
| Validated on independent cohort | Yes | Yes | Yes | Yes |
| Clinical predictors | POD24 | Age >60 y, no. of nodal sites >3, elevated serum LDH, hemoglobin <12 g/L, Ann Arbor stage III/IV | High-risk FLIPI, ECOG performance status >2 | High-risk FLIPI |
| Molecular predictors | None | None | Nonsilent mutations in 7 genes (ARID1A, CARD11, CREBBP, EP300, EZH2, FOXO1, MEF2B) at VAFs ≥10% | Nonsilent mutations in 3 genes (EP300, EZH2, FOXO1) at VAFs ≥10% |
| Calculation of risk score | Single variable | Cumulative sum of predictor values, all predictors have weight = 1 | Cumulative sum of predictor values, predictors have individual weights† | Cumulative sum of predictor values, predictors have individual weights (Figure 4A) |
| Number of risk groups | 2 (no POD24 = low-risk, POD24 = high-risk) | 3 (0-1 = low-risk, 2 = interm-risk, 3-5 = high-risk) | 2 (<0.8 = low-risk, >0.8 = high-risk) | 2 (<0.71 = low-risk, >0.71 = high-risk) |
| High-risk cases among patients with symptomatic, advanced stage FL (%) | GLSG: 23/132 (17) | GLSG: 77/151 (51) | GLSG: 43/151 (28) | GLSG: 63/151 (42) |
| BCCA: 23/102 (22) | BCCA: 53/107 (50) | BCCA: 24/107 (22) | BCCA: 39/107 (36) | |
| Predictive for POD24 | n/a | Yes | Yes | Yes |
| GLSG: evaluable patients 132/151 | OR = 4.6 (P = .0059) | OR = 5.8 (P = .00031) | OR = 7.3 (P = .00016) | |
| Sens 78%, Spec 56%, AUC 0.67 | Sens 61%, Spec 79%, AUC 0.70 | Sens 78%, Spec 67%, AUC 0.73 | ||
| Accuracy 60%, PPV 27%, NPV 92% | Accuracy 76%, PPV 38%, NPV 91% | Accuracy 71%, PPV 33%, NPV 94% | ||
| BCCA: evaluable patients 107/107 | OR = 3.2 (P = .035) | OR = 4.8 (P = .0052) | OR = 4.3 (P = .0051) | |
| Sens 70%, Spec 58%, AUC 0.64 | Sens 43%, Spec 86%, AUC 0.65 | Sens 61%, Spec 73%, AUC 0.67 | ||
| Accuracy 61%, PPV 33%, NPV 87% | Accuracy 77%, PPV 48%, NPV 84% | Accuracy 69%, PPV 40%, NPV 87% | ||
| Predictive for FFS | n/a | Yes, but only as a binary classifier (low-/interm-risk vs high-risk) | Yes | Yes |
| GLSG: evaluable patients 151/151 | 5-y FFS 57% vs 76% | 5-y FFS 38% vs 77% | 5-y FFS 50% vs 77% | |
| HR = 2.11 (P = .0034) | HR = 4.14 (P = 6.313·10−9) | HR = 3.06 (P = 5.989·10−6) | ||
| BCCA: evaluable patients 107/107 | 5-y FFS 47% vs 70% | 5-y FFS 25% vs 68% | 5-y FFS 36% vs 72% | |
| HR = 2.18 (P = .0075) | HR = 3.58 (P = 4.924·10−6) | HR = 3.01 (P = 7.178·10−5) | ||
| Predictive for OS | Yes‡ | Yes | Yes | Yes |
| GLSG: evaluable patients 151/151 | 5-y OS 40% vs 96% | 5-y OS 75% vs 91% | 5-y OS 65% vs 90% | 5-y OS 71% vs 91% |
| HR = 10.91 (P = 5.262·10−14) | HR = 2.59 (P = .0083), C-Index 0.75 | HR = 3.38 (P = .00031), C-Index 0.78 | HR = 3.55 (P = .00026), C-Index 0.79 | |
| BCCA: evaluable patients 107/107 | 5-y OS 26% vs 93% | 5-y OS 60% vs 89% | 5-y OS 42% vs 84% | 5-y OS 48% vs 89% |
| HR = 13.602 (P = 6.661·10−16) | HR = 3.90 (P = .00034), C-Index 0.81 | HR = 4.89 (P = 7.661·10−7), C-Index 0.84 | HR = 5.35 (P = 9.996·10−7), C-Index 0.86 | |
| Comments | Risk classification not possible for patients who died/were censored within 24 mo of first-line treatment. Only predictive for OS. First described in 2015. | Most widely used and best validated pretreatment classifier. Contains only clinical variables not necessarily directly reflecting disease biology. First described in 2004, not widely used to guide treatment decisions. | Highest accuracy and specificity to predict POD24. Most discriminative classifier for patients without POD24 into high- and low-risk groups. Sequencing of 7 genes required. First described in 2015, requires further validation. | Highest sensitivity to predict POD24. Sequencing of 3 genes required. Requires further validation. |
AUC, area under the curve; CR30, complete response rate at 30 months; FLIPI, Follicular Lymphoma International Prognostic Index; HR, hazard ratio; NPV, negative predictive value; OR, odds ratio; POD, progression of disease; POD24-PI, early progression prognostic index; PPV, positive predictive value; Sens, sensitivity; Spec, specificity.
The modified definition of POD24 was used for this analysis (see text).
For this analysis only, OS for POD24 was calculated from time of treatment initiation for better comparison (nota bene, numbers differ from the text, wherein OS was calculated from time of risk-defining event).
A clinicogenetic risk classifier specifically designed to predict POD24
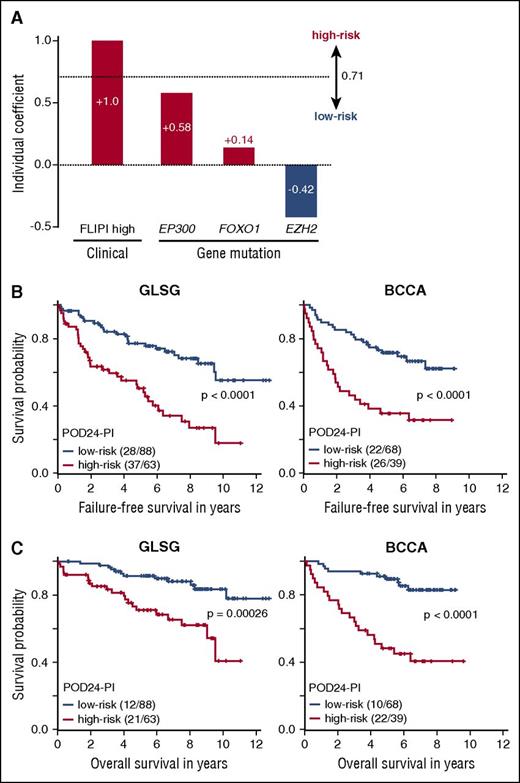
Despite the superior performance of the m7-FLIPI, 6% of GLSG patients (9/151) and 12% of BCCA patients (13/107) were classified as low-risk m7-FLIPI but developed progression of FL within 24 months of treatment (4 and 6 of whom were high-risk FLIPI), for an overall sensitivity of 61% and 43%, respectively, at predicting POD24 (Figure 2B). We aimed to improve that by using the clinical and molecular data from the GLSG cohort to calculate another risk model that specifically predicts POD24. Internal validation showed superiority of the model in which all clinical and molecular coefficients were penalized (bootstrap-corrected coefficient of 0.95 vs 0.23 for the model in which the coefficients for high-risk FLIPI and ECOG-PS >1 were not penalized). We termed this risk model the POD24 Prognostic Index (POD24-PI). The risk score, calculated as the sum of predictor values weighted by Lasso coefficients, contained 4 factors that were all within the m7-FLIPI: high-risk FLIPI (βLasso=1.0), and nonsilent mutations in EP300 (βLasso=0.58), FOXO1 (βLasso=0.14), and EZH2 (βLasso = −0.42) (Figure 4A). The optimal cutoff value to stratify patients into high- and low-risk subgroups was determined to be 0.71 (Figure 4A). The BCCA cohort was used to independently validate the results.
The POD24 Prognostic Index (POD24-PI). (A) The POD24-PI is calculated as the sum of individual clinical and gene mutation predictor values weighted by their individual coefficients. (B) Kaplan-Meier curves for failure-free survival (FFS), and (C) overall survival (OS) for patients from the GLSG and BCCA cohorts according to POD24-PI status. Numbers in parentheses indicate patients with event/number of patients per subgroup.
The POD24 Prognostic Index (POD24-PI). (A) The POD24-PI is calculated as the sum of individual clinical and gene mutation predictor values weighted by their individual coefficients. (B) Kaplan-Meier curves for failure-free survival (FFS), and (C) overall survival (OS) for patients from the GLSG and BCCA cohorts according to POD24-PI status. Numbers in parentheses indicate patients with event/number of patients per subgroup.
Compared with the m7-FLIPI, a higher fraction of patients was classified into the high-risk subgroup by the POD24-PI (Figures 2C and 5), specifically 42% (63/151) and 36% (39/107) of GLSG and BCCA patients, respectively (Table 2). As intended, the POD24-PI had a higher sensitivity to predict POD24 compared with the m7-FLIPI (78% vs 61%, and 61% vs 43% in the GLSG and BCCA cohorts, respectively; Figure 2C), albeit at the cost of a lower specificity and accuracy (Table 2; supplemental Figure 1B). Overall, high-risk POD24-PI was associated with significantly shorter FFS and OS (Figure 4; Table 2): the 5-year FFS rates were 50% vs 77% (HR = 3.06, P < .0001) and 36% vs 72% (HR = 3.01, P < .0001), and the 5-year OS rates were 71% vs 91% (HR = 3.55, P = .00026) and 48% vs 89% (HR = 5.35, P < .0001) in the GLSG and BCCA cohorts, respectively (Figure 4). In patients without POD24, high-risk POD24-PI was still associated with a shorter FFS and OS, but less discriminative compared with the m7-FLIPI (ie, the POD24-PI had lower HRs and inferior P values compared with the m7-FLIPI (Figure 3C; supplemental Figure 2C).
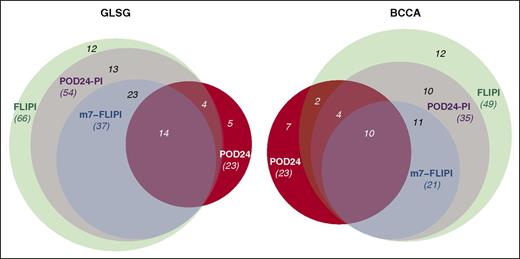
Identification of high-risk patients by 3 pretreatment risk models and POD24 status. Venn diagrams visualize the overlap of high-risk patients as identified by the FLIPI, m7-FLIPI, POD24-PI, and POD24.
Identification of high-risk patients by 3 pretreatment risk models and POD24 status. Venn diagrams visualize the overlap of high-risk patients as identified by the FLIPI, m7-FLIPI, POD24-PI, and POD24.
Table 2 summarizes the specific features of the 2 clinicogenetic risk scores, in context with the FLIPI and the POD24 classifier. Although the m7-FLIPI had the highest accuracy and POD24-PI the highest sensitivity to predict POD24, 22% (5/23) and 30% (7/23) of patients with POD24 from the GLSG and BCCA cohorts were still not correctly identified as high risk by any of the pretreatment risk models (Figure 5). Because mutations in TP53 are not included in any of the clinicogenetic risk models but are known to be associated with inferior OS,8,10 we compared TP53 mutation frequency in patients with or without POD24. In both cohorts, TP53 mutations were in fact enriched in the POD24 subgroup (13% [3/23] vs 3% [3/109] in GLSG patients [P = .11], and 13% [3/23] vs 4% [3/79] in BCCA patients [P = .25]), but failed to reach statistical significance (supplemental Table 1; supplemental Figure 3).
Discussion
Currently applied immunochemotherapy regimens result in long-lasting remissions and excellent OS in a majority (∼80%) of patients with FL requiring systemic treatment. However, our study confirms that a subset of patients (∼20%) experience short remissions and markedly inferior outcome with a median OS of <5 years. Clearly, strategies to guide risk-adapted treatment approaches in FL are needed to avoid overtreatment of low-risk patients, and to prioritize alternative over standard treatment regimens in high-risk patients. Also, clinical trials focusing on high-risk patients are likely to identify higher activity regimens at a much faster rate if study results were not mitigated by patients with highly indolent clinical courses in unselected study cohorts.
Retrospective evaluation of treatment response at 24 months after first-line immunochemotherapy currently represents the strongest predictor of OS, although a subset of patients with POD24 are still alive at >5 years (26% and 41% in our series, up to 50% in a previous series4 ). By its definition, POD24 is not confounded by subsequent therapies (as is OS), or by deaths without prior POD as a result of comorbidity or treatment-related mortality (as is progression-free survival or event-free survival),11 and thus very closely reflects either the aggressiveness of the disease and/or treatment-specific resistance. As such, POD24 will be highly useful to select cases for in-depth molecular characterization to identify the tumor-biological determinants of poor patient outcome.
POD24 will immediately be useful in clinical practice to select high-risk patients for experimental salvage treatments. One such example is the S1608 trial conducted through the National Cancer Institute’s National Clinical Trial Network, which will specifically enroll patients with POD24 after first-line immunochemotherapy. However, as a posttreatment surrogate marker, POD24 cannot guide first-line treatment including consolidation/maintenance regimens in first remission, and by definition is unable to assess patients who die within 24 months without prior documented POD or to identify high-risk patients who do not fail first-line treatment within 24 months.
We propose that comprehensive risk models that integrate established clinical risk factors with disease-specific biomarkers to predict biology-relevant end points are useful in up-front identification of high-risk patients. The previously described m7-FLIPI is the most stringent pretreatment risk model currently available and identifies the smallest subgroup of patients (∼25%) at highest risk of early failure of first-line immunochemotherapy and death. The m7-FLIPI has the highest accuracy and PPV for POD24 among all pretreatment risk models. Also, high-risk m7-FLIPI is associated with inferior outcome in patients who do not fail treatment within 24 months, a subset currently missed by the POD24 classifier. As such, high-risk m7-FLIPI prospectively defines the subgroup of patients with the highest clinical need in FL before initiation of first-line treatment, and supports clinical trials with alternative up-front regimens with highest antitumor activity, potentially accepting higher toxicity profiles as deemed acceptable for the majority of patients with low-risk disease. Furthermore, among all pretreatment risk scores, the m7-FLIPI has the highest specificity for POD24 (ie, it identifies the highest percentage of non–early progressors correctly as low-risk). This indicates that the m7-FLIPI might also be useful in up-front identification of low-risk patients with excellent outcome with currently applied immunochemotherapy regimens, and a subset might actually qualify for treatment de-escalation strategies.
The POD24-PI, specifically designed to improve the sensitivity to predict POD24, classified more patients into the high-risk subgroup (∼40%), which was less enriched for poor outcome compared with high-risk m7-FLIPI. Despite its inferior performance by most test metrics, the POD24-PI may still be considered a valuable predictor in certain clinical situations; eg, when testing very-well-tolerated regimens (eg, post-remission vaccines) investigators may want to minimize the risk of excluding high-risk patients while accepting some that have been falsely identified as such. Furthermore, the fact that the POD24-PI contains the 4 highest weighted components of the m7-FLIPI likely explains the performance of the latter to predict POD24, and provides clues about how the biology of high-risk tumors may be different from others. Of note, a subset of patients with POD24 was not distinguishable by any of the 2 clinicogenetic risk models, suggesting that further improvements and probably integration of additional biomarkers are needed to capture these cases.
Based on the results from the PRIMA trial,12 many patients now receive maintenance treatment with rituximab after first-line immunochemotherapy. Interestingly and similar to previous studies,4 the percentage of patients progressing within 24 months was in the 20% range in both of our cohorts, despite IFN maintenance in GLSG patients and rituximab maintenance for the majority of BCCA patients, implying no major impact of these approaches on POD24. Thus, substantial improvement of treatment results is most likely to be expected from innovative, risk-adapted first-line regimens, eventually combined with minimal residual disease–guided consolidation/maintenance strategies.13-17
In this study, we analyzed stringently selected patients with advanced stage or bulky disease in need of systemic treatment from both a prospective clinical trial (the GLSG cohort), which might not necessarily reflect routine clinical practice,18 and a population-based registry (the BCCA cohort), a retrospective cohort that might be more prone to confounding and bias, but also more closely reflects real-life patients. Remarkably, analyzing these 2 different cohorts yielded highly consistent results. As such, the m7-FLIPI establishes solid grounds for up-front patient stratification by actual risk; however, several challenges still remain to be addressed before it can be applied in clinical trials and practice. Standardization of molecular technologies and analysis pipelines will be needed to ensure widely reproducible results. The m7-FLIPI will have to be validated and compared with other posttreatment surrogate markers (eg, EFS12, EFS24, and CR30)5-7 and pretreatment risk models (eg, the FLIPI-2)19 in additional and larger cohorts with longer follow-up, and evaluated in the context of specific treatments, such as the now widely used bendamustine plus rituximab regimen.20,21 Integrating gene mutations into risk assessment for molecular-targeting approaches will be particularly informative (eg, for BCL2 and EZH2 inhibitors),22,23 and will ultimately pave the way from risk-adapted to biology-directed treatment algorithms in FL. Other potentially targetable candidate genes captured by the m7-FLIPI include the acetyltransferases EP300 and the structurally and functionally related CREBBP, because mutations in these genes are primarily disruptive and may sensitize tumors to histone deacetylase inhibition.24 Likewise, N-terminally clustered mutations in FOXO125 might affect response to inhibitors of the phosphatidylinositol 3′ OH kinase (PI3K) pathway, given that FOXO transcription factors and PI3K often function as antagonists in the biology of B cells.26 Eventually, the relative impact of individual molecular predictors will have to be adjusted to specific molecular targeting approaches; for example, CARD11 mutations have a relatively small m7-FLIPI coefficient in the context of immunochemotherapy, but they might well increase the risk of treatment failure in patients receiving BTK inhibitors by activating NF-κB signaling downstream of BTK, as has been shown for ibrutinib for relapsed/refractory diffuse large B-cell lymphoma.27 Several large and collaborative efforts are underway to address these questions.
In summary, the m7-FLIPI currently represents the most promising predictor for treatment outcome of patients receiving first-line immunochemotherapy, including patients with early treatment failure but not fulfilling the POD24 criteria, and should be evaluated in prospective trials of precision medicine approaches in FL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by the Max-Eder Program of the Deutsche Krebshilfe e.V. (110659) (O.W.), the Deutsche Forschungsgemeinschaft (DFG-SFB/CRC-1243, TP-A11) (O.W.), and in part by a Program Project Grant from the Terry Fox Research Institute (1023) (J.M.C. and R.D.G.).
G.O. receives funding from the Robert-Bosch-Foundation. D.M.W. is a Leukemia and Lymphoma Scholar.
Authorship
Contribution: O.W., D.M.W., A.D.Z., P.M.B., J.W.F., S.A., R.D.G., and W.H. contributed to study design; V.J., M.U., and O.W. performed data analysis and review, and interpretation of data; V.J. and O.W. created figures and wrote the initial manuscript; and R.K., A.M.S., M.S., H.H., M.H.D., A.R., G.O., W.K., L.H.S., J.M.C., and R.D.G. provided patient samples and data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.S. is Department of Internal Medicine II, Hematology Laboratory Kiel, Schleswig-Holstein University Hospital, Campus Kiel, Kiel, Germany.
Correspondence: Oliver Weigert, University Hospital of the Ludwig-Maximilians-University Munich, Medical Department III, Laboratory for Experimental Leukemia and Lymphoma Research (ELLF), Max-Lebsche Platz 30, 81377 Munich, Germany; e-mail: oliver.weigert@med.uni-muenchen.de.