Abstract
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive malignancy caused by the accumulation of genomic lesions that affect the development of T cells. For many years, it has been established that deregulated expression of transcription factors, impairment of the CDKN2A/2B cell-cycle regulators, and hyperactive NOTCH1 signaling play prominent roles in the pathogenesis of this leukemia. In the past decade, systematic screening of T-ALL genomes by high-resolution copy-number arrays and next-generation sequencing technologies has revealed that T-cell progenitors accumulate additional mutations affecting JAK/STAT signaling, protein translation, and epigenetic control, providing novel attractive targets for therapy. In this review, we provide an update on our knowledge of T-ALL pathogenesis, the opportunities for the introduction of targeted therapy, and the challenges that are still ahead.
Introduction
The characterization of chromosomal abnormalities, such as 9p deletions resulting in inactivation of CDKN2A (p16) and CDKN2B (p15) and translocations affecting the T-cell receptor (TCR) genes, has been fundamental in providing initial insights into the genetic defects present in T-cell acute lymphoblastic leukemia (T-ALL). The incorporation of gene expression profiling has provided an additional view on the subgroups present in T-ALL, each characterized by the presence of specific chromosomal aberrations leading to ectopic expression of 1 particular transcription factor such as TAL1, TLX1, TLX3, or others. Sequencing approaches focusing on candidate oncogenes or, more recently, genome-wide sequencing (exome sequencing, whole-genome sequencing, or transcriptome sequencing), have identified >100 genes that can be mutated in T-ALL. Only 2 of these genes, NOTCH1 and CDKN2A/2B, are mutated in >50% of T-ALL cases, and a large variety of genes are mutated at lower frequency (Table 1). Based on all available genomic data, we can conclude that each T-ALL case contains probably >10 biologically relevant genomic lesions, each contributing to the transformation of normal T cells into an aggressive leukemia cell with impaired differentiation, improved survival and proliferation characteristics, and altered metabolism, cell cycle, and homing properties. These changes help the T-ALL cells proliferate and survive by changing their own behavior as well as interact effectively with normal cells, which further supports their stem cell characteristics and the survival of these high numbers of abnormal T cells.
Mutation frequencies in adult vs pediatric T-ALL
| Gene . | Type of genetic aberration . | Frequency . | |
|---|---|---|---|
| Pediatric . | Adult . | ||
| NOTCH1 signaling pathway | |||
| FBXW7 | Inactivating mutations | 14 | 14 |
| NOTCH1 | Chromosomal rearrangements/activating mutations | 50 | 57 |
| Cell cycle | |||
| CDKN2A | 9p21 deletion | 61 | 55 |
| CDKN2B | 9p21 deletion | 58 | 46 |
| RB1 | Deletions | 12* | |
| Transcription factors | |||
| BCL11B | Inactivating mutations/deletions | 10 | 9 |
| ETV6 | Inactivating mutations/deletions | 8 | 14 |
| GATA3 | Inactivating mutations/deletions | 5 | 3 |
| HOXA (CALM-AF10, MLL-ENL and SET-NUP214) | Chromosomal rearrangements/inversions/expression | 5 | 8 |
| LEF1 | Inactivating mutations/deletions | 10 | 2 |
| LMO2 | Chromosomal rearrangements/deletions/expression | 13 | 21 |
| MYB | Chromosomal rearrangements/duplications | 7 | 17 |
| NKX2.1/NKX2.2 | Chromosomal rearrangements/expression | 8* | |
| RUNX1 | Inactivating mutations/deletions | 8 | 10 |
| TAL1 | Chromosomal rearrangements/5′ super-enhancer mutations/deletions/expression | 30 | 34 |
| TLX1 | Chromosomal rearrangements/deletions/expression | 8 | 20 |
| TLX3 | Chromosomal rearrangements/expression | 19 | 9 |
| WT1 | Inactivating mutation/deletion | 19 | 11 |
| Signaling | |||
| AKT | Activating mutations | 2 | 2 |
| DNM2 | Inactivating mutations | 13 | 13 |
| FLT3 | Activating mutations | 6 | 4 |
| JAK1 | Activating mutations | 5 | 7 |
| JAK3 | Activating mutations | 8 | 12 |
| IL7R | Activating mutations | 10 | 12 |
| NF1 | Deletions | 4 | 4 |
| KRAS | Activating mutations | 6 | 0 |
| NRAS | Activating mutations | 14 | 9 |
| NUP214-ABL1/ ABL1 gain | Chromosomal rearrangement/duplication | 8* | |
| PI3KCA | Activating mutations | 1 | 5 |
| PTEN | Inactivating mutations/deletion | 19 | 11 |
| PTPN2 | Inactivating mutations/deletion | 3 | 7 |
| STAT5B | Activating mutations | 6 | 6 |
| Epigenetic factors | |||
| DNMT3A | Inactivating mutations | 1 | 14 |
| EED | Inactivating mutations/deletions | 5 | 5 |
| EZH2 | Inactivating mutations/deletions | 12 | 12 |
| KDM6A/UTX | Inactivating mutations/deletions | 6 | 7 |
| PHF6 | Inactivating mutations/deletions | 19 | 30 |
| SUZ12 | Inactivating mutations/deletions | 11 | 5 |
| Translation and RNA stability | |||
| CNOT3 | Missense mutations | 3 | 8 |
| mTOR | Activating mutations | 5* | |
| RPL5 | Inactivating mutations | 2 | 2 |
| RPL10 | Missense mutations | 8 | 1 |
| RPL22 | Inactivating mutations/deletion | 4 | 0 |
| Gene . | Type of genetic aberration . | Frequency . | |
|---|---|---|---|
| Pediatric . | Adult . | ||
| NOTCH1 signaling pathway | |||
| FBXW7 | Inactivating mutations | 14 | 14 |
| NOTCH1 | Chromosomal rearrangements/activating mutations | 50 | 57 |
| Cell cycle | |||
| CDKN2A | 9p21 deletion | 61 | 55 |
| CDKN2B | 9p21 deletion | 58 | 46 |
| RB1 | Deletions | 12* | |
| Transcription factors | |||
| BCL11B | Inactivating mutations/deletions | 10 | 9 |
| ETV6 | Inactivating mutations/deletions | 8 | 14 |
| GATA3 | Inactivating mutations/deletions | 5 | 3 |
| HOXA (CALM-AF10, MLL-ENL and SET-NUP214) | Chromosomal rearrangements/inversions/expression | 5 | 8 |
| LEF1 | Inactivating mutations/deletions | 10 | 2 |
| LMO2 | Chromosomal rearrangements/deletions/expression | 13 | 21 |
| MYB | Chromosomal rearrangements/duplications | 7 | 17 |
| NKX2.1/NKX2.2 | Chromosomal rearrangements/expression | 8* | |
| RUNX1 | Inactivating mutations/deletions | 8 | 10 |
| TAL1 | Chromosomal rearrangements/5′ super-enhancer mutations/deletions/expression | 30 | 34 |
| TLX1 | Chromosomal rearrangements/deletions/expression | 8 | 20 |
| TLX3 | Chromosomal rearrangements/expression | 19 | 9 |
| WT1 | Inactivating mutation/deletion | 19 | 11 |
| Signaling | |||
| AKT | Activating mutations | 2 | 2 |
| DNM2 | Inactivating mutations | 13 | 13 |
| FLT3 | Activating mutations | 6 | 4 |
| JAK1 | Activating mutations | 5 | 7 |
| JAK3 | Activating mutations | 8 | 12 |
| IL7R | Activating mutations | 10 | 12 |
| NF1 | Deletions | 4 | 4 |
| KRAS | Activating mutations | 6 | 0 |
| NRAS | Activating mutations | 14 | 9 |
| NUP214-ABL1/ ABL1 gain | Chromosomal rearrangement/duplication | 8* | |
| PI3KCA | Activating mutations | 1 | 5 |
| PTEN | Inactivating mutations/deletion | 19 | 11 |
| PTPN2 | Inactivating mutations/deletion | 3 | 7 |
| STAT5B | Activating mutations | 6 | 6 |
| Epigenetic factors | |||
| DNMT3A | Inactivating mutations | 1 | 14 |
| EED | Inactivating mutations/deletions | 5 | 5 |
| EZH2 | Inactivating mutations/deletions | 12 | 12 |
| KDM6A/UTX | Inactivating mutations/deletions | 6 | 7 |
| PHF6 | Inactivating mutations/deletions | 19 | 30 |
| SUZ12 | Inactivating mutations/deletions | 11 | 5 |
| Translation and RNA stability | |||
| CNOT3 | Missense mutations | 3 | 8 |
| mTOR | Activating mutations | 5* | |
| RPL5 | Inactivating mutations | 2 | 2 |
| RPL10 | Missense mutations | 8 | 1 |
| RPL22 | Inactivating mutations/deletion | 4 | 0 |
Genes targeted by genetic alterations in >2% of T-ALL cases are shown. Calculation of the different frequencies is based on several previously published independent studies that analyzed pediatric and/or adult T-ALL patient cohorts. For frequency calculations, chromosomal rearrangements, copy-number variations and mutations were considered. For HOXA, TLX1, TLX3, TAL1, NKX2.1, NKX2.2, and LMO2, gene expression was also considered. The HOXA group includes cases that are positive for CALM-AF10, SET-NUP214, or MLL-ELN chromosomal rearrangements.
It was not possible to have separate numbers for pediatric and adult cases.
Transcription factors
Careful gene expression profiling of T-ALL cases has led to the identification of subgroups of T-ALL, each characterized by a specific transcriptional profile and the ectopic expression of 1 particular transcription factor, often as a consequence of a chromosomal defect. The largest subgroup is defined by ectopic TAL1 expression (in some cases together with LMO1/LMO2), whereas other major subgroups show mutual exclusive expression of TLX1, TLX3, HOXA9/10, LMO2, or NKX2-1 (Table 2).1-3 Furthermore, the early T-cell precursor subgroup of ALL (ETP-ALL) corresponds to immature T-ALLs expressing ETP/stem cell genes and is characterized by aberrant expression of LYL12 ; hematopoietic transcription factors such as RUNX1 and ETV6 are frequently mutated in this genetically heterogeneous subgroup.1,4
Chromosomal rearrangements resulting in ectopic expression of transcription factors or the generation of fusion genes with transcriptional/epigenetic activity
| Chromosomal rearrangement or mutation . | Partner gene 1 (oncogene) . | Chromosome location . | Partner gene 2 . | Chromosome location . | Consequence of the rearrangement/mutation . |
|---|---|---|---|---|---|
| Major transcription factors | |||||
| del(1)(p32p32) | TAL1 | 1p32 | STIL | 1p32 | Ectopic TAL1 expression driven by STIL promoter |
| t(1;14)(p32;q11) | TCRα | 14q11 | Ectopic TAL1 expression driven by TCR enhancer | ||
| t(1;14)(p32;q11) | TCRδ | 14q11 | Ectopic TAL1 expression driven by TCR enhancer | ||
| Small insertion | — | — | Ectopic TAL1 expression driven by de novo enhancer | ||
| t(10;14)(q24;q11) | TLX1 | 10q24 | TCRα | 14q11 | Ectopic TLX1 expression driven by TCR enhancer |
| t(7;10)(q34;q24) | TCRβ | 7q34 | Ectopic TLX1 expression driven by TCR enhancer | ||
| t(10;14)(q24;q11) | TCRδ | 14q11 | Ectopic TLX1 expression driven by TCR enhancer | ||
| t(5;14)(q35;q11) | TLX3 | 5q35 | TCRδ | 14q11 | Ectopic TLX3 expression driven by TCR enhancer |
| t(5;14)(q35;q32) | BCL11B | 14q32 | Ectopic TLX3 expression driven by BCL11B | ||
| inv(7)(p15;q34) | HOXA9/HOXA10 | 7p15 | TCRβ | 7q34 | Ectopic expression of HOXA genes, predominantly HOXA9 and HOXA10, driven by TCR enhancer |
| t(10;11)(p13;q14) | PICALM (CALM) | 11q14 | MLLT10 (AF10) | 10p13 | PICALM-MLLT10 fusion transcript, resulting in upregulation of HOXA genes |
| del(9)(q34;q34) | SET | 9q34 | NUP214 | 9q34 | SET-NUP214 fusion transcript, resulting in upregulation of HOXA genes |
| inv(14)(q11;q13) | NKX2-1 | 14q13 | TCRα | 14q11 | Ectopic NKX2-1 expression driven by TCR enhancer |
| T(14;20)(q11;p11) | NKX2-2 | 20p11 | TCRδ | 14q11 | Ectopic NKX2-2 expression driven by TCR enhancer |
| t(11;14)(p15;q11) | LMO1 | 11p15 | TCRα | 14q11 | Ectopic LMO1 expression, mostly together with LYL1 or TAL1 expression |
| TCRδ | 14q11 | ||||
| t(11;14)(p13;q11) | LMO2 | 11p13 | TCRα | 14q11 | Ectopic LMO2 expression, mostly together with LYL1 or TAL1 expression |
| TCRδ | 14q11 | ||||
| del(5)(q14;q14) | MEF2C | 5q14 | — | — | Small deletions close to MEF2C leading to upregulation of MEF2C expression |
| t(7;19)(q34;p13) | LYL1 | 19p13 | TCRβ | 7q34 | Ectopic expression of LYL1 driven by TCR enhancer |
| t(11;14)(p11;q32) | SPI1 | 11p11 | BCL11B | 14q32 | Ectopic expression of SPI1 driven by BCL11B |
| Additional transcription factor aberrations | |||||
| dup(6)(q23;q23) | MYB | 6q23 | — | — | Increased MYB expression due to extra copy of MYB |
| t(6;7)(q23;q34) | TCRβ | 7q34 | Ectopic expression of MYB driven by TCR enhancer | ||
| Various translocations | KMT2A (MLL) | 11q23 | Many different partners | KMT2A fusion genes | |
| Mutation/deletion | BCL11B | 14q32 | — | — | Inactivation of BCL11B |
| Mutation/deletion | ETV6 | 12p13 | — | — | Inactivation of ETV6 |
| Mutation/deletion | RUNX1 | 21q22 | — | — | Inactivation of RUNX1 |
| Mutation/deletion | LEF1 | 4q25 | — | — | Inactivation of LEF1 |
| Mutation/deletion | WT1 | 11p13 | — | — | Inactivation of WT1 |
| Chromosomal rearrangement or mutation . | Partner gene 1 (oncogene) . | Chromosome location . | Partner gene 2 . | Chromosome location . | Consequence of the rearrangement/mutation . |
|---|---|---|---|---|---|
| Major transcription factors | |||||
| del(1)(p32p32) | TAL1 | 1p32 | STIL | 1p32 | Ectopic TAL1 expression driven by STIL promoter |
| t(1;14)(p32;q11) | TCRα | 14q11 | Ectopic TAL1 expression driven by TCR enhancer | ||
| t(1;14)(p32;q11) | TCRδ | 14q11 | Ectopic TAL1 expression driven by TCR enhancer | ||
| Small insertion | — | — | Ectopic TAL1 expression driven by de novo enhancer | ||
| t(10;14)(q24;q11) | TLX1 | 10q24 | TCRα | 14q11 | Ectopic TLX1 expression driven by TCR enhancer |
| t(7;10)(q34;q24) | TCRβ | 7q34 | Ectopic TLX1 expression driven by TCR enhancer | ||
| t(10;14)(q24;q11) | TCRδ | 14q11 | Ectopic TLX1 expression driven by TCR enhancer | ||
| t(5;14)(q35;q11) | TLX3 | 5q35 | TCRδ | 14q11 | Ectopic TLX3 expression driven by TCR enhancer |
| t(5;14)(q35;q32) | BCL11B | 14q32 | Ectopic TLX3 expression driven by BCL11B | ||
| inv(7)(p15;q34) | HOXA9/HOXA10 | 7p15 | TCRβ | 7q34 | Ectopic expression of HOXA genes, predominantly HOXA9 and HOXA10, driven by TCR enhancer |
| t(10;11)(p13;q14) | PICALM (CALM) | 11q14 | MLLT10 (AF10) | 10p13 | PICALM-MLLT10 fusion transcript, resulting in upregulation of HOXA genes |
| del(9)(q34;q34) | SET | 9q34 | NUP214 | 9q34 | SET-NUP214 fusion transcript, resulting in upregulation of HOXA genes |
| inv(14)(q11;q13) | NKX2-1 | 14q13 | TCRα | 14q11 | Ectopic NKX2-1 expression driven by TCR enhancer |
| T(14;20)(q11;p11) | NKX2-2 | 20p11 | TCRδ | 14q11 | Ectopic NKX2-2 expression driven by TCR enhancer |
| t(11;14)(p15;q11) | LMO1 | 11p15 | TCRα | 14q11 | Ectopic LMO1 expression, mostly together with LYL1 or TAL1 expression |
| TCRδ | 14q11 | ||||
| t(11;14)(p13;q11) | LMO2 | 11p13 | TCRα | 14q11 | Ectopic LMO2 expression, mostly together with LYL1 or TAL1 expression |
| TCRδ | 14q11 | ||||
| del(5)(q14;q14) | MEF2C | 5q14 | — | — | Small deletions close to MEF2C leading to upregulation of MEF2C expression |
| t(7;19)(q34;p13) | LYL1 | 19p13 | TCRβ | 7q34 | Ectopic expression of LYL1 driven by TCR enhancer |
| t(11;14)(p11;q32) | SPI1 | 11p11 | BCL11B | 14q32 | Ectopic expression of SPI1 driven by BCL11B |
| Additional transcription factor aberrations | |||||
| dup(6)(q23;q23) | MYB | 6q23 | — | — | Increased MYB expression due to extra copy of MYB |
| t(6;7)(q23;q34) | TCRβ | 7q34 | Ectopic expression of MYB driven by TCR enhancer | ||
| Various translocations | KMT2A (MLL) | 11q23 | Many different partners | KMT2A fusion genes | |
| Mutation/deletion | BCL11B | 14q32 | — | — | Inactivation of BCL11B |
| Mutation/deletion | ETV6 | 12p13 | — | — | Inactivation of ETV6 |
| Mutation/deletion | RUNX1 | 21q22 | — | — | Inactivation of RUNX1 |
| Mutation/deletion | LEF1 | 4q25 | — | — | Inactivation of LEF1 |
| Mutation/deletion | WT1 | 11p13 | — | — | Inactivation of WT1 |
—, not applicable.
Almost all subgroups of T-ALL are characterized by the clear ectopic expression of 1 of these transcription factors, except for some immature T-ALL cases where MEF2C expression, and other sporadic translocations where SPI1, could be important.1 The oncogenic roles of TAL1 and TLX1 have been nicely illustrated by studies in the mouse, in cell lines, and in patient samples. Overexpression of TAL1 or TLX1 in developing thymocytes in the mouse results in the development of T-ALL with long latency.5-8 For TAL1, coexpression of LMO1 and ICN1 dramatically decreased disease latency, in part by increasing the number of leukemia-initiating cells.9 Mouse leukemias that eventually developed in the context of TLX1 expression often harbored mutations in Bcl11b or Notch1, as also observed in human T-ALL.7,8 These data confirm that ectopic expression of transcription factors such as TAL1 or TLX1 alone can be an initiating step in T-ALL development, but that additional mutations are required to fully transform normal T cells to leukemia cells. The ectopic expression of these transcription factors could have a strong effect on the differentiation of the cells, as suggested by the correlation with immunophenotype. Moreover, for TLX1, it was recently shown that TLX1 can bind the TCR enhancer and in this way can suppress TCRα expression and T-cell differentiation.10
There are now 4 major mechanisms known to cause aberrant expression of transcription factors in T-ALL: (1) chromosomal translocations involving 1 of the TCR genes, (2) chromosomal rearrangements with other regulatory sequences, (3) duplication/amplification of the transcription factor, and (4) mutations or small insertions generating novel regulatory sequences acting as enhancers (Table 2). The latter mechanism was only recently identified by studies that revealed changes in chromatin structure close to the TAL1 gene, indicative for the presence of a new enhancer region. Detailed analysis of this region led to the identification of mutations that created a de novo binding site for MYB, thereby resulting in recruitment of additional transcriptional regulators and activation of TAL1 expression in cis.11,12 These data illustrate how noncoding mutations can have a strong effect on leukemia development and we can expect that more of these mutations will be identified in the future as more information comes available on the noncoding part of the genome.
Oncogenic NOTCH1 signaling
The NOTCH1 signaling pathway is essential for the commitment of multipotent hematopoietic progenitors to the T-cell lineage and for further development of thymocytes.13,14 Activation of NOTCH1 signaling constitutes the most predominant oncogenic event involved in the pathogenesis of T-ALL with activating mutations in more than half of T-ALL cases (Table 1).15 Loss of function of negative regulators of NOTCH1 is an alternative mechanism leading to aberrant activation of the pathway. Mutations of FBXW7 are present in 10% to 15% of T-ALL cases and lead to increased NOTCH1 protein stability.16,17 The role of the NOTCH1 cascade in the context of T-ALL is discussed in more detail in a companion review article.18
Increased kinase signaling
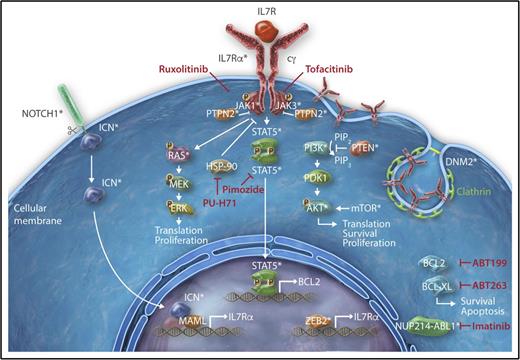
Interleukin 7 (IL7) signaling is essential for normal T-cell development and is triggered by the interaction of IL7 with the heterodimeric IL7 receptor (IL7R). This interaction induces reciprocal JAK1 and JAK3 phosphorylation and subsequent recruitment and activation of STAT5. Upon phosphorylation, STAT5 dimerizes and translocates to the nucleus where it regulates the transcription of many target genes, including the antiapoptotic B-cell lymphoma 2 (BCL-2) family member proteins.19,20 In addition to the JAK/STAT pathway, the RAS-MAPK and phosphatidylinositol 3-kinase (PI3K) pathways are also activated by IL2, IL7, and stem cell factor (SCF) that act on the developing T cells (Figure 1).
Deregulation of the JAK-STAT signaling cascade in T-ALL. Representation of the different oncogenic mechanisms that lead to aberrant activation of the IL7 signaling in T-ALL. Interaction of IL7 with the heterodimeric IL7R induces reciprocal JAK1 and JAK3 phosphorylation and subsequent recruitment of STAT5. STAT5 dimerizes and translocates to the nucleus where it induces transcription of the prosurvival factor BCL2. IL7 also activates the RAS-MAPK and PI3K kinase pathways. IL7 signaling can indirectly be enhanced by abnormal NOTCH1 signaling, constitutive expression of ZEB2, or by increased presentation of IL7R on the cell surface of thymocytes due to impaired clathrin-dependent endocytosis caused by DNM2 mutations. Promising therapeutic agents targeting the oncogenic IL7-JAK-STAT cascade are indicated in red. *Proteins that are mutated in T-ALL.
Deregulation of the JAK-STAT signaling cascade in T-ALL. Representation of the different oncogenic mechanisms that lead to aberrant activation of the IL7 signaling in T-ALL. Interaction of IL7 with the heterodimeric IL7R induces reciprocal JAK1 and JAK3 phosphorylation and subsequent recruitment of STAT5. STAT5 dimerizes and translocates to the nucleus where it induces transcription of the prosurvival factor BCL2. IL7 also activates the RAS-MAPK and PI3K kinase pathways. IL7 signaling can indirectly be enhanced by abnormal NOTCH1 signaling, constitutive expression of ZEB2, or by increased presentation of IL7R on the cell surface of thymocytes due to impaired clathrin-dependent endocytosis caused by DNM2 mutations. Promising therapeutic agents targeting the oncogenic IL7-JAK-STAT cascade are indicated in red. *Proteins that are mutated in T-ALL.
Activating mutations in IL7R, JAK1, JAK3, and/or STAT5 are present in 20% to 30% of T-ALL cases (Table 1),4,21,22 with a higher representation within TLX+, HOXA+, and ETP-ALL patient subgroups (Figure 2).4,23 A small percentage of cases also show aberrations in phosphatases like PTPN2 and PTPRC, which, among other substrates, dephosphorylate and inactivate JAK kinases.24,25 Interestingly, the IL7R signaling cascade can also be hyperactivated in patients who do not carry any genetic aberrations in the IL7R, JAK, or STAT5 genes, indicating that still other mechanisms exist to activate this pathway.26,27 Indeed, only recently was it discovered that loss-of-function mutations in DNM2 lead to increased presentation of IL7R on the cell surface of thymocytes due to impaired clathrin-dependent endocytosis.26 In addition, a rare translocation targeting the zinc finger E-box–binding homeobox 2 locus (ZEB2) has been recently described in T-ALL, and Zeb2 overexpression was shown to increase Il7r expression and Stat5 activation, promoting T-ALL cell survival in a mouse model.27 Although most mutations seem to result in activation of JAK1 and JAK3, rare fusion transcripts involving JAK2 have also been described, and wild-type TYK2 may have an important role in activating survival pathways of T-ALL cells.28,29
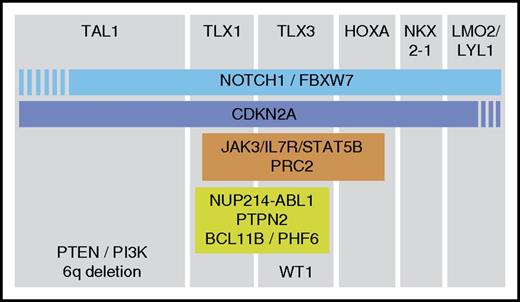
Representation of the cooperation of oncogenic events. The major subclasses of T-ALL are shown based on the expression of the transcription factors TAL1, TLX1, TLX3, HOXA genes, NKX2-1, or LMO2/LYL1. For each subclass, additional genes are shown that are most frequently mutated in that subclass. The PRC2 complex contains EZH2, SUZ12, and EED.
Representation of the cooperation of oncogenic events. The major subclasses of T-ALL are shown based on the expression of the transcription factors TAL1, TLX1, TLX3, HOXA genes, NKX2-1, or LMO2/LYL1. For each subclass, additional genes are shown that are most frequently mutated in that subclass. The PRC2 complex contains EZH2, SUZ12, and EED.
Aberrant activation of the PI3K-AKT pathway results in enhanced cell metabolism, proliferation, and impaired apoptosis.30,31 Hyperactivation of the oncogenic PI3K-AKT pathway in T-ALL is mainly caused by inactivating mutations or deletions of the phosphatase and tensin homolog (PTEN), the main negative regulator of the pathway.22,32-36 In addition, some T-ALL cases show gain-of-function mutations in the regulatory and catalytic subunits of PI3K, respectively, p85 and p110, or in the downstream effectors of the cascade such as AKT and mechanistic target of rapamycin (mTOR) (Table 1).22,35,37 PI3K-AKT signaling activation is also achieved through IL7 stimulation or RAS activation.22,31,38 A recent study on 146 pediatric T-ALL cases described that almost 50% of the patients harbored at least 1 mutation in the JAK-STAT, PI3K-AKT, or RAS-MAPK pathways, underscoring the importance of activation of those cascades for the leukemic cells.22
In addition to the activation of the JAK kinases or the PI3K pathway, activation of the ABL1 kinase is observed in up to 8% of T-ALL cases, with 6% of patients expressing the NUP214-ABL1 fusion protein.39 NUP214-ABL1 is a constitutively active tyrosine kinase that activates STAT5 and the RAS-MAPK pathway, but is much weaker as compared with BCR-ABL1.39,40 Interestingly, the kinase activity of NUP214-ABL1 is dependent on its location to the nuclear pore39,41 and its oncogenic properties rely on its interaction with MAD2L1, NUP155, and SMC4 and on the activity of the LCK, a member of the SRC family kinase.42 All data collected to date suggest that the NUP214-ABL1 fusion kinase is a weak oncogene that cooperates with additional oncogenic events to drive leukemia development. In that sense, it is of interest to note that the NUP214-ABL1 fusion is always found together with TLX1 or TLX3 expression and is often associated with loss of PTPN2, a negative regulator of NUP214-ABL1.25,39
RAS-activating mutations were recently described in 44% of diagnosis-relapse sample pairs, indicating an overrepresentation of these defects in high-risk ALL. Interestingly, KRAS mutations rendered lymphoblasts resistant toward methotrexate, while sensitizing them to vincristine.43
Epigenetic factors
T-ALL is one of the pediatric tumor types with the highest incidence of epigenetic lesions, and 56% of samples from children with T-ALL contain mutations in this gene class.44 Also, in adult T-ALL, mutations in epigenetic factors are highly common (Table 1).45,46 For a complete overview of all epigenetic lesions identified in T-ALL, we refer to recent reviews on this topic.47,48
The plant homeodomain protein 6 (PHF6) protein was postulated as an epigenetic factor because it contains 2 atypical plant homeodomain (PHD)-like zinc fingers. Canonical PHD domains typically bind posttranslationally modified histones. However, the C-terminal PHD-like domain in PHF6 binds double-stranded DNA. Besides DNA, PHF6 binds to a growing list of transcriptional regulators such as the nucleosome remodeling deacetylase (NuRD) complex, the RNA polymerase II–associated factor 1 (PAF1) transcriptional elongation complex, and upstream binding transcription factor, RNA polymerase I (UBTF1), a key transcriptional activator of ribosomal RNA (rRNA). The latter brings us to the nucleolar role of PHF6 and its proposed role in ribosome biogenesis. Besides interacting with UBTF1, PHF6 binds the ribosomal DNA (rDNA) promotor and rDNA-coding sequences and modulation of PHF6 levels alters rRNA synthesis rates. Todd and colleagues propose that the N-terminal PHD-like domain may mediate binding to rRNA whereas the C-terminal domain may bind rDNA. As such, PHF6 may act as a scaffold, bridging rDNA transcriptional elongation with early processing of produced rRNA. Finally, PHF6 seems involved in the DNA damage response and cell-cycle regulation: PHF6 is a substrate of the DNA damage checkpoint kinase ataxia telangiectasia mutated (ATM) and loss of PHF6 results in accumulation of phosphorylated γH2AX, a mark of DNA double-strand breaks, and in G2/M cell-cycle arrest.49 Further studies are, however, required to investigate how inactivation of PHF6 in T-ALL promotes leukemia.
Enzymes involved in regulating methylation of histone 3 lysine 27 (H3K27) are also implicated in T-ALL. Loss-of-function defects in the polycomb-repressive complex 2 (PRC2) components EZH2, SUZ12, and EED suggest a tumor suppressor role for this complex in T-ALL,4,46,50 which is supported by accelerated leukemia onset in mice upon EZH2 downregulation.46 PRC2 catalyzes H3K27 trimethylation (H3K27me3) and its tumor suppressor role may be explained by antagonism with NOTCH1: NOTCH1 activation causes eviction of PRC2 and recruitment of the lysine demethylase 6B (KDM6B, also known as JMJD3) at NOTCH1 target gene promoters, resulting in loss of the repressive H3K27me3 chromatin modification and activation of these genes.46,51 Also, the lysine demethylase 6A (KDM6A, also called UTX), which demethylates H3K27me3, shows inactivating lesions in T-ALL,50-52 and its downregulation accelerates NOTCH1-driven leukemia in mice.51,52 Interestingly, KDM6A binds TAL1, and it is recruited to TAL1 target genes to remove repressive H3K27me3 marks and activate genes. In TAL1+ leukemias, KDM6A behaves as a proto-oncogene and is required for leukemia maintenance, which may explain why KDM6A inactivation so far has not been detected in TAL1+ cases.53 The mechanism by which KDM6A plays a tumor suppressor role in TAL1− T-ALL is currently not understood. The KDM6B and KDM6A inhibitor GSKJ4 shows promising preclinical results in T-ALL. Whereas the Brand group reported specificity of this drug for the TAL1+ T-ALL subgroup, the Aifantis group observed sensitivity in all tested T-ALL cell lines and samples.51,53
RNA metabolism and translation
A few new oncogenes and tumor suppressors recently emerged in T-ALL. Somatic mutations in ribosomal protein genes RPL5, RPL10, and RPL22 have been described in ∼20% of T-ALL cases.50,54 Whereas RPL5 and RPL22 show heterozygous inactivating mutations and deletions, RPL10 contains an intriguing mutational hotspot at residue arginine 98 (R98), with 8% of pediatric T-ALL patients harboring an R98S RPL10 missense mutation. Inactivation of Rpl22 accelerates myristoylated Akt-driven T-cell lymphoma in mice.54,55 Mechanistically, heterozygous Rpl22 deletion activates NF-κB and induces stemness factor Lin28B.54 The roles of the RPL5 and RPL10 R98S defects in T-ALL development remain to be determined. Characterization of RPL10 R98S in yeast revealed that it impairs ribosome formation and translation fidelity. Over time, compensatory mutations are acquired, restoring ribosome biogenesis but not translation fidelity, which may drive expression of an oncogenic protein profile.56,57 Translational deregulation in T-ALL may, however, go far beyond mutations in ribosomal proteins: PHF6 has been linked to ribosome biogenesis, major T-ALL oncogenes and tumor suppressors such as PTEN, NOTCH1, and FBXW7 regulate translation, and T-ALL cells are sensitive to translation inhibitors 4EGI-1 and silvestrol.58-60
Another intriguing novel tumor suppressor that is inactivated in 8% of adult T-ALL samples is CCR4-NOT transcription complex subunit 3 (CNOT3).50 The CCR4-NOT complex catalyzes messenger RNA (mRNA) deadenylation and may also link again to control of protein translation. In addition, roles for CNOT3 as transcriptional regulator have been proposed.61 Using studies in a fruitfly eye cancer model that is driven by Notch activation, we confirmed that NOT3 (CNOT3 homolog in the fruitfly) is a tumor suppressor gene, but more studies are required to determine how mutations in CNOT3 contribute to leukemia development. It is interesting to note that somatic mutations in CNOT3 have now also been observed in chronic lymphocytic leukemia (CLL) and in some solid tumors.62
Micro-RNAs and long noncoding RNAs
Inactivation of Dicer1, an essential component of the micro-RNA (miRNA) processing machinery, impairs NOTCH1-driven T-ALL development in mice and can induce regression of established NOTCH1-driven tumors, indicating a role for 1 or more miRNAs in (at least NOTCH1-driven) T-ALL.63 Indeed, many miRNAs are misexpressed in T-ALL, either due to a translocation, as key downstream targets of T-ALL oncogenes such as NOTCH1 or TAL1, or via unknown mechanisms.64-67 Oncogenic miRNAs (onco-miRs) have been identified downregulating the expression of known T-ALL–associated tumor suppressor genes. Examples are miR-19b, miR-20a, miR-26a, miR-92, and miR-223 which cooperatively downregulate IKZF1 (or IKAROS), PTEN, BIM, PHF6, NF1, and FBXW7 transcripts.68 Other miRNAs are underexpressed in T-ALL, leading to overexpression of T-ALL–associated oncogenes. An example here is miR-193b, which regulates the expression of MYB and the antiapoptotic factor MCL1.69 Many additional onco-miRs and tumor-suppressive miRNAs have been described in T-ALL. For a partial overview, we refer to Aster.66
A related field of interest that recently emerged in T-ALL is long noncoding RNAs (lncRNAs), a heterogeneous class of transcripts defined by a minimum length of 200 nucleotides and an apparent lack of protein-coding potential. Diverse molecular mechanisms have been described by which lncNRAs control a wide variety of cellular functions and developmental processes, and promote disease pathogenesis, including cancer.70 Interestingly, different T-ALL subgroups are characterized by distinct lncRNA expression profiles.71 In addition, a series of NOTCH1-regulated lncRNAs were identified.72,73 The best characterized one is leukemia-induced noncoding activator RNA 1 (LUNAR1), which is a NOTCH1-regulated pro-oncogenic lncRNA that is required for efficient growth of T-ALL cells by maintaining high expression of insulin-like growth factor 1 receptor (IGF1R).73
Cooperation of oncogenic events
Recent sequencing studies have suggested that on average 10 to 20 protein-altering mutations are present in T-ALL cells.4,50,74 This accumulation of mutations does not occur randomly, and specific combinations of mutations are often found, suggesting that those activated oncogenes and inactivated tumor suppressor genes are physiologically interconnected and cooperate during the development and progression of the leukemia (Figure 2).75 Initial founder genomic lesions initiate a premalignant process by interacting with the existing machinery of the physiological cell state76 and additional mutations are necessary to drive transformation75,77 and lead to the development of subclonal variegation.
This has been very nicely observed in TAL1/LMO2+ T-ALL cases, where PTEN inactivation occurs most frequently. Interestingly, transplantation of TAL1 rearranged leukemia cells into immune-deficient mice allowed development of leukemic subclones with newly acquired PTEN microdeletions.32 These data suggest that there is an enormous pressure for TAL1-expressing T-ALL cells to inactivate PTEN, and that TAL1 expression and PTEN inactivation cooperate during T-cell transformation. In contrast, components of the IL7R-JAK signaling pathway are frequently mutated in immature T-ALL cases or in TLX/HOXA+ cases, but are underrepresented in TAL1/LMO2+ cases. In addition, we observed that mutations of IL7R-JAK are positively associated with mutations and deletions of members of the PRC2 complex (EZH2, SUZ12 and EED).21 Similarly, WT1 mutations are most prevalent in TLX3+ cases.78 These and other genomic data suggest that initiating lesions leading to ectopic transcription factor expression sensitize the cells to alterations of very specific pathways.
T-ALL treatment
Current treatment of T-ALL consists of high-intensity combination chemotherapy and results in a very high overall survival for pediatric patients.79 Unfortunately, this treatment comes with significant short-term and long-term side effects. Especially for young children, the effects of this high-dose chemotherapy on bone development, the central nervous system, and fertility should not be underestimated.80 A complete overview of the treatment options was recently described.81,82
In addition to the side effects, the occurrence of relapse is another important challenge as it is observed in up to 20% of pediatric and 40% of adult T-ALL.83 Relapsed T-ALL cases are often refractory to chemotherapeutics and are associated with a poor prognosis.84,85 From comparative genetic studies conducted on T-ALL samples taken at diagnosis and relapse, and studies with xenotransplantation in mice, it is clear that relapse is determined by the clonal evolution of either a minor genetic subclone present in the primary leukemia, or from the clonal expansion of an ancestral cell (prediagnosis).86-89 However, some rare T-ALL late-relapsed cases are considered to be secondary malignancies rather than originated by the initial disease.90
The use of whole-exome sequencing has provided a way to take a detailed view of the genomes of relapsed T-ALL cases and led to the identification of activating mutations in cytosolic 5′-nucleotidase II (NT5C2) as one of the causes of relapse.86,87 NT5C2 is an enzyme responsible for the inactivation of nucleoside-analog chemotherapy drugs.91 When NT5C2 mutant proteins were expressed in T-ALL lymphoblast, increased nucleotidase activity was observed and the cells became resistant to 6-mercaptopurine (6-MP) and 6-thioguanine (6-TG), 2 chemotherapeutic drugs used in maintenance treatment of T-ALL, suggesting the important role of the identified NT5C2-activating mutations in therapy resistance.86
To further improve the treatment of T-ALL and to reduce the toxicity of current treatment, we await the introduction of newer targeted agents. Recent drug development in both oncology and immunology has generated a spectrum of new drugs that could be useful for the treatment of specific T-ALL subsets (Table 3).
Promising targeted agents in T-ALL and their stage in clinical testing
| Compound . | Specificity . | Clinical phase . | Disease . |
|---|---|---|---|
| NOTCH1 signaling | |||
| MK-0752 | GSI | Phase 1 | T-ALL and lymphoma |
| PF-03084014 | GSI | Phase 1 | Advanced-stage cancers, T-ALL, lymphoblastic lymphoma |
| BMS-906024 (with dexamethasone) | GSI | Phase 1 | T-ALL or T-cell lymphoblastic lymphoma |
| OMP-52M51 | Notch1 inhibitory antibody | Phase 1 | Dose-escalation study in lymphoid malignancy |
| LY3039478 | Notch1 inhibitor | Phase 1 | Dose-escalation study in advanced-stage cancers |
| BMS-536924 | ATP-competitive IGF-1R/IR inhibitor | Preclinical | |
| JAK-STAT inhibitors | |||
| Ruxolitinib | JAK1/2 inhibitor | Phase 1 | AML |
| Phase 2 | B-ALL | ||
| FDA approved | Myelofibrosis | ||
| Phase 1/2 | Fallopian tube cancer, ovarian cancer, primary peritoneal cancer | ||
| Tofacitinib | JAK3 inhibitor | FDA approved | Rheumatoid arthritis |
| Phase 1 | Systemic lupus erythematosus | ||
| Phase 3 | Juvenile idiopathic arthritis | ||
| Pimozide | STAT5 inhibitor | FDA approved | Schizophrenia, psychotic disorders |
| Phase 2 | Amyotrophic lateral sclerosis | ||
| 17-AAG | HSP90 inhibitor | Phase 3 | Multiple myeloma |
| Phase 2 | Pancreatic cancer | ||
| Phase 2 | Advanced malignancies | ||
| Phase 2 | Ovarian cancer | ||
| PU-H71 | HSP90 inhibitor | Phase 1 | Solid tumor, lymphoma |
| Phase 1 | Metastatic solid tumor, lymphoma, myeloproliferative neoplasms | ||
| Inducers of apoptosis | |||
| ABT-263 (Navitoclax) | Inhibitor of Bcl-xL, Bcl-2, and Bcl-w | Phase 1 | Non-small cell lung cancer |
| Phase 2 | Platinum-resistant or refractory ovarian cancer | ||
| Phase 1 | Hepatocellular carcinoma | ||
| Phase 1/2 | Melanoma | ||
| ABT-199 (Venetoclax) | Bcl-2–selective inhibitor | FDA approved | CLL |
| Phase 3 | Relapsed/refractory multiple myeloma | ||
| PI3K inhibitors | |||
| CAL-130 | Inhibits p110γ and p110δ catalytic domains | Preclinical | |
| Ly294002 | Inhibit PI3Kα/δ/β | Phase 1 | Neuroblastoma |
| Pictilisib (GDC-0941) | Inhibitor of PI3Kα/δ | Phase 2 | Breast cancer |
| Phase 2 | Nonsquamous non-small cell lung cancer | ||
| Apitolisib (GDC-0980, RG7422) | Inhibitor for PI3Kα/β/δ/γ | Phase 2 | Endometrial carcinoma |
| Phase 1/2 | Prostate cancer | ||
| Phase 2 | Renal cell carcinoma | ||
| MEK inhibitors | |||
| CI-1040 | ATP noncompetitive MEK1/2 inhibitor | Phase 2 | Breast cancer, colorectal cancer, lung cancer, pancreatic cancer |
| Selumetinib (AZD6244) | MEK1 inhibitor | Phase 2 | Triple-negative breast cancer |
| Phase 1 | Lung cancer, melanoma, head and neck carcinoma, gastroesophageal cancer, breast cancer, pancreatic adenocarcinoma, colorectal cancer | ||
| Trametinib (GSK1120212) | MEK1/2 inhibitor | Phase 2 | Gastrointestinal stromal tumors |
| Phase 1 | Melanoma | ||
| Phase 1 | Neuroblastoma | ||
| Phase 2 | Non-small cell lung cancer | ||
| ABL inhibitors | |||
| Imatinib | v-Abl, c-Kit, and PDGFR inhibitor | FDA approved | Chronic myeloid leukemia |
| Phase 2 | B-ALL, B lymphoblastic lymphoma, T-ALL, T lymphoblastic lymphoma | ||
| Phase 3 | Philadelphia chromosome–positive adult ALL | ||
| Dasatinib | Abl, Src, and c-Kit inhibitor | FDA approved | Chronic myeloid leukemia |
| Phase 2 | ALL | ||
| Phase 1 | Chronic kidney disease | ||
| Nilotinib | Abl, Src, and c-Kit inhibitor | Phase 2 | Glioma |
| Phase 3 | Philadelphia chromosome–positive adult ALL | ||
| FDA approved | Chronic myeloid leukemia | ||
| Hedgehog inhibitors | |||
| Vismodegib (GDC-0449) | SMO inhibitor | FDA approved | Basal cell carcinoma |
| Phase 2 | Breast cancer | ||
| Phase 2 | Non-Hodgkin lymphoma, multiple myeloma, advanced solid tumors | ||
| GANT61 | GLI1 inhibitor | Preclinical | |
| Translation inhibitors | |||
| Rapamycin | Specific mTOR inhibitor | Phase 2 | Myelodysplastic syndrome, CLL, ALL, T lymphoblastic lymphoma, acute myelogenous leukemia, acute biphenotypic leukemia, acute undifferentiated leukemia |
| Phase 1 | Fallopian tube carcinoma, ovarian carcinoma, primary peritoneal carcinoma | ||
| 4EGI-1 | Competitive eIF4E/eIF4G interaction inhibitor | Preclinical |
| Compound . | Specificity . | Clinical phase . | Disease . |
|---|---|---|---|
| NOTCH1 signaling | |||
| MK-0752 | GSI | Phase 1 | T-ALL and lymphoma |
| PF-03084014 | GSI | Phase 1 | Advanced-stage cancers, T-ALL, lymphoblastic lymphoma |
| BMS-906024 (with dexamethasone) | GSI | Phase 1 | T-ALL or T-cell lymphoblastic lymphoma |
| OMP-52M51 | Notch1 inhibitory antibody | Phase 1 | Dose-escalation study in lymphoid malignancy |
| LY3039478 | Notch1 inhibitor | Phase 1 | Dose-escalation study in advanced-stage cancers |
| BMS-536924 | ATP-competitive IGF-1R/IR inhibitor | Preclinical | |
| JAK-STAT inhibitors | |||
| Ruxolitinib | JAK1/2 inhibitor | Phase 1 | AML |
| Phase 2 | B-ALL | ||
| FDA approved | Myelofibrosis | ||
| Phase 1/2 | Fallopian tube cancer, ovarian cancer, primary peritoneal cancer | ||
| Tofacitinib | JAK3 inhibitor | FDA approved | Rheumatoid arthritis |
| Phase 1 | Systemic lupus erythematosus | ||
| Phase 3 | Juvenile idiopathic arthritis | ||
| Pimozide | STAT5 inhibitor | FDA approved | Schizophrenia, psychotic disorders |
| Phase 2 | Amyotrophic lateral sclerosis | ||
| 17-AAG | HSP90 inhibitor | Phase 3 | Multiple myeloma |
| Phase 2 | Pancreatic cancer | ||
| Phase 2 | Advanced malignancies | ||
| Phase 2 | Ovarian cancer | ||
| PU-H71 | HSP90 inhibitor | Phase 1 | Solid tumor, lymphoma |
| Phase 1 | Metastatic solid tumor, lymphoma, myeloproliferative neoplasms | ||
| Inducers of apoptosis | |||
| ABT-263 (Navitoclax) | Inhibitor of Bcl-xL, Bcl-2, and Bcl-w | Phase 1 | Non-small cell lung cancer |
| Phase 2 | Platinum-resistant or refractory ovarian cancer | ||
| Phase 1 | Hepatocellular carcinoma | ||
| Phase 1/2 | Melanoma | ||
| ABT-199 (Venetoclax) | Bcl-2–selective inhibitor | FDA approved | CLL |
| Phase 3 | Relapsed/refractory multiple myeloma | ||
| PI3K inhibitors | |||
| CAL-130 | Inhibits p110γ and p110δ catalytic domains | Preclinical | |
| Ly294002 | Inhibit PI3Kα/δ/β | Phase 1 | Neuroblastoma |
| Pictilisib (GDC-0941) | Inhibitor of PI3Kα/δ | Phase 2 | Breast cancer |
| Phase 2 | Nonsquamous non-small cell lung cancer | ||
| Apitolisib (GDC-0980, RG7422) | Inhibitor for PI3Kα/β/δ/γ | Phase 2 | Endometrial carcinoma |
| Phase 1/2 | Prostate cancer | ||
| Phase 2 | Renal cell carcinoma | ||
| MEK inhibitors | |||
| CI-1040 | ATP noncompetitive MEK1/2 inhibitor | Phase 2 | Breast cancer, colorectal cancer, lung cancer, pancreatic cancer |
| Selumetinib (AZD6244) | MEK1 inhibitor | Phase 2 | Triple-negative breast cancer |
| Phase 1 | Lung cancer, melanoma, head and neck carcinoma, gastroesophageal cancer, breast cancer, pancreatic adenocarcinoma, colorectal cancer | ||
| Trametinib (GSK1120212) | MEK1/2 inhibitor | Phase 2 | Gastrointestinal stromal tumors |
| Phase 1 | Melanoma | ||
| Phase 1 | Neuroblastoma | ||
| Phase 2 | Non-small cell lung cancer | ||
| ABL inhibitors | |||
| Imatinib | v-Abl, c-Kit, and PDGFR inhibitor | FDA approved | Chronic myeloid leukemia |
| Phase 2 | B-ALL, B lymphoblastic lymphoma, T-ALL, T lymphoblastic lymphoma | ||
| Phase 3 | Philadelphia chromosome–positive adult ALL | ||
| Dasatinib | Abl, Src, and c-Kit inhibitor | FDA approved | Chronic myeloid leukemia |
| Phase 2 | ALL | ||
| Phase 1 | Chronic kidney disease | ||
| Nilotinib | Abl, Src, and c-Kit inhibitor | Phase 2 | Glioma |
| Phase 3 | Philadelphia chromosome–positive adult ALL | ||
| FDA approved | Chronic myeloid leukemia | ||
| Hedgehog inhibitors | |||
| Vismodegib (GDC-0449) | SMO inhibitor | FDA approved | Basal cell carcinoma |
| Phase 2 | Breast cancer | ||
| Phase 2 | Non-Hodgkin lymphoma, multiple myeloma, advanced solid tumors | ||
| GANT61 | GLI1 inhibitor | Preclinical | |
| Translation inhibitors | |||
| Rapamycin | Specific mTOR inhibitor | Phase 2 | Myelodysplastic syndrome, CLL, ALL, T lymphoblastic lymphoma, acute myelogenous leukemia, acute biphenotypic leukemia, acute undifferentiated leukemia |
| Phase 1 | Fallopian tube carcinoma, ovarian carcinoma, primary peritoneal carcinoma | ||
| 4EGI-1 | Competitive eIF4E/eIF4G interaction inhibitor | Preclinical |
ATP, adenosine triphosphate; FDA, US Food and Drug Administration; HSP90, heat shock protein 90; PDGFR, platelet-derived growth factor receptor; SMO, smoothened.
With NOTCH1 being the major oncogene in T-ALL and with drugs available that can interfere with the activation (cleavage) of NOTCH1 by the γ-secretase complex, a number of clinical trials have been initiated with γ-secretase inhibitors (GSIs). However, these trials led to disappointing clinical results due to the dose-limiting toxicity and low response rates. It remains to be determined how second-generation GSIs will perform, but at least some promising anecdotal responses have already been reported and with milder toxicities.92-95 Besides GSIs, other therapeutic approaches for the inhibition of NOTCH1 have been developed. Monoclonal antibodies against the NOTCH1 receptor have antitumor effects in vitro and in vivo with limited gastrointestinal toxicities.96,97 Inhibition of ADAM10 may also facilitate effective inhibition of wild-type and mutant NOTCH receptors.98 An antibody against the γ-secretase complex (A5226A) showed preclinical activity against T-ALL.99 Mastermind-inhibiting peptides that mimic the NOTCH1 interaction with MAML (SAMH1 peptides) are also being tested.100,101
Because both JAK and ABL1 kinases are often activated in T-ALL, available JAK inhibitors (ruxolitinib and tofacitinib) and ABL1 inhibitors (imatinib, dasatinib, nilotinib) could be repurposed for the treatment of T-ALL cases with documented mutations leading to JAK/STAT or ABL1 activation. Several preclinical studies have demonstrated activity of ruxolitinib or tofacitinib for the inhibition of T-ALL cells with IL7R or JAK1/JAK3 mutations, whereas case reports have shown some activity of imatinib or dasatinib for the treatment of NUP214-ABL1+ T-ALL.4,23,102,103 The therapeutic potential of targeting the IL7Rα signaling and the use of JAK inhibitors in ALL has been recently reviewed.104,105
Similarly, recently approved hedgehog pathway inhibitors could show activity in T-ALL. The hedgehog signaling pathway plays an important role in normal T-cell development, which is steered by the secretion of the sonic hedgehog ligand (SHH) by certain thymic epithelial cells.106 In T-ALL, the hedgehog pathway was reported to be aberrantly activated by rare mutations or ectopic expression of hedgehog pathway genes, and those cases showed response to hedgehog inhibitor treatment.50,107,108 Finally, some other attractive novel therapies have recently been described in T-ALL. Selective inhibitors of nuclear export (SINE) were shown to be highly toxic to T-ALL and acute myeloid leukemia (AML) cells in mouse xenograft models, while having little toxic effects on normal mouse hematopoietic cells.109 In addition, it was demonstrated that TCR+ T-ALL cells can be targeted by mimicking thymic-negative selection as obtained by TCR stimulation via antigen/major histocompatibility complex (MHC) presentation or via an antibody against CD3.110 The advantage of these types of therapies is that they seem independent of genetic subtypes, avoiding extensive genetic characterization before therapeutic choices are to be made.
Challenges ahead
In the past decades, enormous progress was made in our understanding of the genetics and biology of T-ALL, but there are still significant gaps in our knowledge. To date, almost all attention has gone to defects affecting protein coding genes. The recent identification of mutations in noncoding regions of the genome that result in aberrant transcription factor expression and the discovery of several miRNAs and lncRNAs with a pathogenic role in T-ALL underscore that the “noncoding genome” should not be neglected and that novel classes of oncogenes and tumor suppressor genes may remain to be discovered.
Genomic screens have identified recurrent novel oncogenes and tumor suppressors, such as PHF6, RPL10, and CNOT3, for which the exact role in the pathogenesis of T-ALL remains poorly understood. Moreover, genome-wide screening has mainly been limited to diagnostic cases and such screens of large patient cohorts are lacking for relapse T-ALL. A better understanding of the biology of relapse is an absolute requirement to improve the dismal prognosis these patients currently are facing.
Analysis of mutational patterns in large patient cohorts revealed that the mutational landscape of T-ALL is not random and that particular defects often co-occur or are mutually exclusive.21 More research is needed to understand the biology behind particular mutational patterns and to characterize the specific clinical behaviors and distinct prognosis associated with these patterns. Related to this issue, the order in which mutations are acquired in T-ALL development is also most likely not random. In addition, the cellular context and exact hematopoietic developmental stage of the cell in which a mutation arises might be very relevant for the oncogenic action of the defect. At this point, not much is known on the order in which mutations are acquired and on the cell of origin in which they need to arise. Extensive modeling in mice and single-cell sequencing will be required to answer these questions.
Finally, the interaction of the leukemia cells with their microenvironment and characterization of the T-ALL niche are also essential to fully capture T-ALL pathogenesis. In this context, recently developed advanced microscopy techniques allow in vivo imaging of interactions of leukemia cells with their environment and an essential role in T-ALL maintenance has been discovered for the CXCL12 chemokine and its receptor CXCR4.111-113
The ultimate goal of the T-ALL research community is to translate the knowledge into highly efficient and low-toxicity targeted therapies. Whereas the current chemotherapy-based regimens in pediatric T-ALL are associated with high survival rates, the toxicities of these therapies should not be underestimated. With many new targeted agents under development for cancer and autoimmune diseases (Table 3), it should be possible to effectively repurpose the best agents for the treatment of T-ALL, and to design effective combination therapies directed against the specific sets of cooperating mutations in T-ALL.
Acknowledgments
The authors thank all researchers and clinicians for their contributions to the field and apologize to those whose work they could not describe or cite.
The laboratory of K.D.K. was supported by a European Research Council (ERC) starting grant (334946), Fonds Wetenschappelijk Onderzoek (FWO)-Vlaanderen funding (G067015N and G084013N), and a Stichting Tegen Kanker grant (2012-176). The laboratory of J.C. was supported by an ERC consolidator grant (617340), and funding from FWO-Vlaanderen (G.0683.12), Stichting Tegen Kanker (2014-120), and Kom Op Tegen Kanker. T.G. was supported by the Emmanuel van der Schueren Kom Op Tegen Kanker fellowship.
Authorship
Contribution: T.G., C.V., J.C., and K.D.K. contributed to data analysis and the writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jan Cools, KU Leuven, Campus Gasthuisberg O&N4, Herestraat 49 (Box 602), 3000 Leuven, Belgium; e-mail: jan.cools@kuleuven.be; and Kim De Keersmaecker, Department of Oncology, KU Leuven, Campus Gasthuisberg O&N1, Herestraat 49 (Box 603), 3000 Leuven, Belgium; e-mail: kim.dekeersmaecker@kuleuven.be.