Abstract
Adult T-cell leukemia (ATL) is an aggressive T-cell malignancy caused by human T-cell leukemia virus type 1 (HTLV-1) that develops through a multistep carcinogenesis process involving 5 or more genetic events. We provide a comprehensive overview of recently uncovered information on the molecular basis of leukemogenesis in ATL. Broadly, the landscape of genetic abnormalities in ATL that include alterations highly enriched in genes for T-cell receptor–NF-κB signaling such as PLCG1, PRKCB, and CARD11 and gain-of function mutations in CCR4 and CCR7. Conversely, the epigenetic landscape of ATL can be summarized as polycomb repressive complex 2 hyperactivation with genome-wide H3K27 me3 accumulation as the basis of the unique transcriptome of ATL cells. Expression of H3K27 methyltransferase enhancer of zeste 2 was shown to be induced by HTLV-1 Tax and NF-κB. Furthermore, provirus integration site analysis with high-throughput sequencing enabled the analysis of clonal composition and cell number of each clone in vivo, whereas multicolor flow cytometric analysis with CD7 and cell adhesion molecule 1 enabled the identification of HTLV-1–infected CD4+ T cells in vivo. Sorted immortalized but untransformed cells displayed epigenetic changes closely overlapping those observed in terminally transformed ATL cells, suggesting that epigenetic abnormalities are likely earlier events in leukemogenesis. These new findings broaden the scope of conceptualization of the molecular mechanisms of leukemogenesis, dissecting them into immortalization and clonal progression. These recent findings also open a new direction of drug development for ATL prevention and treatment because epigenetic marks can be reprogrammed. Mechanisms underlying initial immortalization and progressive accumulation of these abnormalities remain to be elucidated.
Introduction
Adult T-cell leukemia (ATL) is an aggressive T-cell malignancy caused by human T-cell leukemia virus type 1 (HTLV-1) infection and often occurs in HTLV-1–endemic areas such as southwestern Japan, the Caribbean Islands, Central and South America, intertropical Africa, and the Middle East.1,2 Approximately 1.1 million individuals are carriers of HTLV-1 in Japan.3,4 The total number of HTLV-1 carriers globally is estimated to be between 5 000 000 and 10 000 000,5 an apparent underestimation because of the lack of epidemiological information in developing countries. Annually, ∼1000 people die of ATL in Japan, based on the national vital statistics data, whereas information on mortality rates in other countries are limited. Age of onset differs across geographic areas: the average age at diagnosis in Central and South America, which is ∼40 years, is lower than that in Japan (∼60 years).6-8 Estimated lifetime risk of ATL in HTLV-1 carriers is 6% to 7% for males and 2% to 3% for females in Japan. Additionally, ATL preferentially develops in individuals infected with HTLV-1 during childhood and seldom occurs in those infected in adulthood.1 A higher proviral load (>4 copies/100 peripheral blood mononuclear cells) is an independent risk factor for progression to ATL.9 Routes of viral transmission are breast milk, sexual intercourse, and blood transfusion. However, whether ATL develops after horizontal transmission by sexual contact or blood transfusion remains unclear, albeit cases of ATL following organ transplantation were previously reported.10,11 Our recent study revealed >4000 cases of new HTLV-1 infection in adolescent and old individuals,12 underlying the importance of studies on HTLV-1–associated diseases among carriers with horizontally transmitted HTLV-1.
Difference in the age of ATL onset depending on the area in the world, and evidence for family clustering of ATL, strongly suggests involvement of environmental and genetic factors in disease outcome.7,13 As for genetic background, many genes have been implicated as determinant factors of disease susceptibility such as HLA class 1, tumor necrosis factor α and killer immunoglobulin-like receptor.14,15 However, because of the lack of large-scale studies, clear conclusions have not been reached as to these issues.
Like other retroviruses, the integrated HTLV-1 proviral genome constitutes of 2 long terminal repeats, gag, pol, and env. The genome also has an extra sequence designated pX, which has 4 partially overlapping open reading frames encoding the proteins p12, p13, and p30, Rex, and Tax. The regulatory proteins, p12, p13, and p30, have important roles in the establishment and maintenance of HTLV-1 infection in vivo.16 The other regulatory protein, Tax and Rex, act in combination to regulate HTLV-1 gene expression and replication in both positive and negative pathways.17 Tax is the viral transcription activator protein and is also a modulator of cellular gene expression involved in the proliferation of T lymphocytes mainly via the activation of the NF-κB and AP-1 pathways. Tax-expressing cells bypass cell-cycle checkpoints, affects mechanisms involved in the DNA damage response and apoptosis pathways, and result in the accumulation of genetic and epigenetic alterations and RNA stability modifications. Tax has transforming activity in rodent fibroblasts and in primary human lymphocytes, and Tax transgenic mice using a variety of promoters develop neoplasia including T-cell lymphoma. Therefore, Tax is considered to be a main viral factor involved in immortalization and transformation of the infected cells.18-22
HTLV-1 also expresses a minus-strand RNA that can encode a basic leucine zipper factor, HTLV-1 bZIP factor (HBZ).23,24 HBZ messenger RNA (mRNA) was shown to promote the proliferation of ATL cells.25 Through the interaction of cellular proteins, HBZ shows a variety of functions, some of which antagonize those of Tax. HBZ is considered to play an important role in the oncogenic process because of its ability to drive infected cell proliferation, to increase hTERT transcription, to inhibit apoptosis, and to disrupt host genomic integrity depending on several HBZ-induced microRNAs (miRNAs).17,26-28 CD4+ T-lymphocyte–specific expression of the HBZ transgene in mice induces T-cell lymphoma in one-third as well as systemic inflammation.21,29 HBZ mRNA is expressed in all HTLV-1–infected cells, including ATL cells ex vivo, suggesting its essential roles in transformation, proliferation, and maintenance of HTLV-1–infected T cells.
Clinical features and treatment of ATL
ATL shows a marked diversity in its clinical manifestations: leukemic manifestations, generalized lymphadenopathy, hepatomegaly, splenomegaly, skin involvement, hypercalcemia, and infiltration of other organs such as central nervous system and gastrointestinal tract. Symptoms and signs of ATL include abdominal pain, diarrhea, ascites, jaundice, pleural effusion, cough, sputum, fever, and unconsciousness because of hypercalcemia and/or opportunistic infections. ATL is classified into 4 clinical subtypes (ie, smoldering, chronic, acute, and lymphoma) based on the diagnostic criteria proposed by Shimoyama et al30 : site of infiltration, presence and degree of leukemic manifestation, lactate dehydrogenase level, and hypercalcemia.
ATL cells in the majority of cases express CD3, CD4, and CD25, but lack CD7.31,32 In ∼10% to 15% cases, ATL cells express both CD4 and CD8. The origin of the ATL cells was suggested to be regulatory T cells based on the expression of FoxP3 in some tumor cells,33 which, however, remains a subject under debate.34
International consensus for ATL treatment strategy is based on the clinical subtype, prognostic factors, and response to initial therapy.35,36 For patients with aggressive ATL (acute and lymphoma types and chronic type with poor prognostic factors), intensive chemotherapy regimens such as VCAP-AMP-VECP (vincristine, cyclophosphamide, doxorubicin, and prednisone; doxorubicin, ranimustine, and prednisone; vindesine, etoposide, carboplatin, and prednisone) or zidovudine/interferon α (AZT/IFN-α) combination therapy, except for lymphoma-type patients, is recommended. For indolent ATL (smoldering type and chronic type without poor prognostic factors), watchful waiting or AZT/IFN-α combination therapy is recommended until disease progression. In the United States and Europe, antiviral therapy using AZT∕ IFN-α is the standard treatment of leukemic-type ATL. In Europe, chemotherapy is the first-line therapy for lymphoma-type ATL because overall survival (OS) with AZT∕ IFN-α alone is very short.37 In Japan, AZT/IFN-α therapy has not been introduced because the national health insurance system has not yet approved the use of these 2 drugs in the treatment of ATL patients. A randomized phase 3 clinical trial is now underway for treating indolent-type ATL with AZT/IFN-α vs watchful waiting. This clinical trial will provide conclusive information regarding the optimal standard treatment of indolent-type ATL. Furthermore, efficacy of the combination of AZT/IFN-α and arsenic trioxide in chronic ATL patients has been reported in a small-scale study,38 and addition of arsenic trioxide was shown to induce Tax degradation in vitro.39 A large-scale clinical trial will provide a conclusive evaluation to this new strategy. Other strategies are up-front allogeneic hematopoietic stem cell transplantation for relatively young patients with aggressive ATL and defucosylated humanized anti-CC chemokine receptor 4 (CCR4) monoclonal antibody (mogamulizumab) for relapsed/refractory CCR4-positive aggressive ATL.36,40
The prognosis of ATL is still very poor. Recently, a Japanese group investigating treatment and survival outcomes among 1594 ATL patients reported that the median survival of acute, lymphoma, chronic, and smoldering ATL subtypes was 8.3, 10.6, 31.5, and 55.0 months, respectively, and that the 4-year OS rates were 11%, 16%, 36%, and 52%, respectively.41 They also found that ∼20% of patients with acute or lymphoma-type ATL underwent allogeneic hematopoietic stem cell transplantation, with median survival and 4-year OS rates of 5.9 months and 26%, respectively. Thus, prognosis of patients with acute and lymphoma-type ATL was still unsatisfactory despite recent advances in treatment modalities; however, they also found that the 4-year OS was better than that reported in a previous survey. Conversely, the prognosis of smoldering ATL was worse than expected, as only 25% of transplanted patients experienced long survival times.
Abnormalities in gene expression in ATL
mRNA expression
ATL cells express cytokines, chemokines and chemokine receptors, and adhesion molecules, most of which are targeted by the viral transactivator protein Tax.33,42-44 Cell adhesion molecule 1 (CADM1), a cell surface molecule originally isolated as a tumor suppressor in non–small cell lung cancer,45 was highly expressed in ATL cells and in HTLV-1–infected nontransformed T cells in carriers.46 CADM1 is currently used for the identification of HTLV-1–infected T cells by multicolor fluorescence-activated cell sorting analysis.47,48 Parathyroid hormone-related protein is another protein that is highly expressed in ATL cells and is responsible for humoral hypercalcemia of malignancy observed during the clinical course of more than half of ATL patients.49 Other factors that are highly expressed in ATL cells, such as interleukin-1, transforming growth factor β, tumor necrosis factor β, and receptor activator of nuclear factor κ-B ligand, have been implicated to be involved in hypercalcemia in ATL patients,50-54 suggesting a complex mechanism is underlying this characteristic paraneoplastic syndrome. Overexpression of cell adhesion molecules and chemokine receptors appear to facilitate organ infiltration of ATL cells.42-44,55-57 One intriguing aspect of these findings was the expression of genes that were not normally expressed in T cells,58,59 which was subsequently confirmed by expression array studies. The lineage-independent ectopic expression of many genes characterized ATL cells and strongly suggested abnormalities in epigenetic regulation that determined tissue-specific gene expression.
miRNA expression
Studies on miRNA expression profiles in HTLV-1/ATL cell lines and primary ATL cells demonstrated both up- and downregulation occurring in a number of miRNAs. In HTLV-1–transformed cell lines, miR-21, miR-24, miR-146a, and miR-155 were upregulated, whereas miR-223 was downregulated.60 In primary ATL cells, miR-181a, miR-132, and miR-125a were downregulated, whereas miR-155 and miR-142-3p were upregulated by microarray analysis.61 Yet, in another study, among a total of 6 miRNAs that were upregulated, miR-93 and miR-130b were shown to target TP53INP1 tumor suppressor.62 Our study on miRNA expression signature of primary ATL cells demonstrated that the vast majority of differentially expressed miRNAs were downregulated, with the most severe suppression observed in miR-31. Intriguingly, we showed that miR-31 targeted MAP3K14 (NIK), which is overexpressed in ATL cells, leading to constitutive activation of the NF-κB pathway.63
Signal transduction
A large body of literature has accumulated showing abnormalities in signal transduction pathways in HTLV-1–infected cell lines and primary ATL cells. It is widely accepted that HTLV-1 Tax activates various signaling pathways through direct interactions with signaling molecules.64 However, only certain aspects of these signaling pathways were confirmed to be activated in primary ATL cells in the absence of Tax.64 Thus, Tax-mediated activation of signaling pathways appears to be involved in the earlier stages of transformation, and transformed cells retain some of the constitutively activated pathways that are essential for proliferation and apoptosis resistance of ATL cells.64,65 Among these, the best-known example is the NF-κB pathway, which is constitutively active in ATL cells.66 One of the mechanisms for constitutive NF-κB activation in ATL cells in the absence of Tax is NIK overexpression caused by miR-31 silencing.63,67 Inhibition of NF-κB was shown to induce ATL cell apoptosis and specifically reduce the number of HTLV-1–infected T cells in asymptomatic carriers.68 Phosphatidylinositol 3-kinase/AKT cascade, which was also constitutively activated in ATL cells, induced the formation of multilobulated nuclei characteristic of ATL cells.69 Furthermore, Notch signaling pathway was constitutively activated, as activating mutations in Notch were found in >30% of ATL patients. Mutations in F-box and WD repeat domain-containing 7 (FBXW7) found in 25% of patients, hindered interaction between FBXW7 and intracellular Notch (NICD), resulting in increased protein stability and constitutive Notch1 signaling.70 Finally, Jak/STAT pathway, another signaling pathway activated in ATL cells, was associated with cell proliferation.71 Although there are a number of other signaling pathways exhibiting abnormalities in the presence of Tax, they were not well characterized in primary ATL cells.
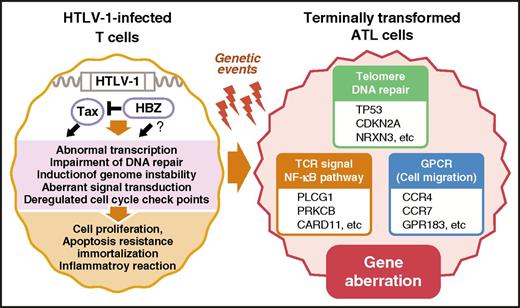
Abnormal gene expression and signal transduction observed in ATL cells have much in common with HTLV-1–infected and virus-expressing T cells, although viral genes, except for antisense transcripts, are not expressed in ATL cells (Figure 1).
Comparison of molecular abnormalities between HTLV-1–infected T cells and transformed ATL cells. Many aspects of the ATL cell phenotype share common characteristics with that of untransformed HTLV-1–infected T cells expressing viral proteins, including Tax. Tax was shown to induce the majority of molecular changes observed in HTLV-1–infected cells, most of which are preserved in ATL cells that do not express Tax. Thus, this phenomenon is sometimes referred to as the signature of Tax. CARD11, caspase recruitment domain-containing protein 11; GPR183, G-protein coupled receptor 183; NRXN3, neurexin-3; PLCG1, phospholipase C, γ 1; PRKCB, protein kinase C β.
Comparison of molecular abnormalities between HTLV-1–infected T cells and transformed ATL cells. Many aspects of the ATL cell phenotype share common characteristics with that of untransformed HTLV-1–infected T cells expressing viral proteins, including Tax. Tax was shown to induce the majority of molecular changes observed in HTLV-1–infected cells, most of which are preserved in ATL cells that do not express Tax. Thus, this phenomenon is sometimes referred to as the signature of Tax. CARD11, caspase recruitment domain-containing protein 11; GPR183, G-protein coupled receptor 183; NRXN3, neurexin-3; PLCG1, phospholipase C, γ 1; PRKCB, protein kinase C β.
Genomic abnormalities in ATL
Cytogenetic characteristics
ATL cells exhibit a variety of cytogenetic abnormalities in almost all patients; however, specific recombination or amplification/deletion events have not been identified.72 Certain chromosomal aberrations were shown to correlate with 1 or more of the clinical features as well as the clinical severity.73 Comparative genomic hybridization analysis also revealed a correlation of the levels of aberrations with the clinical course. The most frequent aberrations included gains at chromosomes 14q, 7q, and 3p and losses at chromosomes 6q and 13q. Chromosomal imbalances, losses, and gains were more frequently observed in aggressive ATL cases than in indolent ATL cases. Analysis of sequential samples during progression suggested clonal changes at crisis; clonal diversity was common during progression to ATL, and chromosomal alterations were associated with the clinical course.74
Abnormalities in specific genes
Inactivation of tumor suppressors by somatic mutations or epigenetic mechanisms is frequently reported in a wide range of cancers and is suggested to be involved in tumor initiation and development. In ATL, main genetic events have been reported to cluster around cyclin dependent kinase inhibitors such as p15 (INK4A), p16 (INK4B), p18 (INK4C), p19 (INK4D), p21 (WAF1), p27 (KIP1), and p57 (KIP2), as well as p53 and retinoblastoma (Rb); all these genes play major regulatory roles during the G1/S transition of cell cycle.75 Frequent alterations were found in p15 (20%) and p16 (28% to 67%) in aggressive-type (acute and lymphoma) ATL, whereas abnormalities were fewer in p15 (0% to 13%) and p16 (5% to 26%) in indolent-type (chronic and smoldering) ATL. Most of these changes were gene deletions; mutations occurred less frequently. Patients with deleted p15 and/or p16 had significantly shorter survival than those individuals with both genes preserved. Genetic alterations in p18, p19, p21, and p27 have rarely been reported.
Conversely, p53 gene was mutated in 10% to 50% of aggressive-type ATL cases, whereas its frequency was lower in indolent-type ATL.76,77 These data clearly implicate that mutations in these cell cycle-related genes are more likely to be associated with progression to more severe stages of ATL than with earlier clinical stages of this malignancy.
The leukemic cells of most ATL patients and HTLV-1–transformed cell lines contain elevated levels of functionally inactive wild-type p53 protein. HTLV-1 Tax oncoprotein alone was shown to be sufficient for abrogating the transactivating function of p53 and for its stabilization in the absence of direct binding between Tax and p53.78 In addition, HBZ was shown to inhibit p53 function through repression of the histone acetyltransferase activities of p300 and HBO1.79 Given the constant expression of HBZ in all HTLV-1–infected cells, these data may provide a clue to explain the underlying mechanisms of p53 inactivation in ATL cells in the absence of Tax expression in majority of cases.22
The Rb tumor suppressor gene is infrequently altered in structure77,80 ; however, ∼50% of ATL cases exhibit loss of Rb protein.81 Additionally, low levels of Rb expression correlated with poor prognosis and shorter survival.82
Notably, alterations in any one of the cyclin dependent kinase inhibitors, p53, or Rb appear to obviate the need for inactivation of other genes in the same pathway. In summary, tumor suppressor genes, which were shown to be frequently altered in aggressive ATL, are the likely driving force fueling the clonal progression of tumor cells.
Comprehensive analysis of genomic abnormalities in ATL
Recently, results of an integrated genomic and transcriptomic analysis of a cohort of 426 ATL cases were reported.83 Massive genomic, methylomic, and transcriptomic data, coupled with cell-based experiments in this study, provided comprehensive and detailed information to provide insight into ATL pathogenesis and confirmed the presence of deletions and mutations in the integrated proviral genome and the lack of expression of the sense strand, including mRNA encoding Tax, in contrast to the constitutive expression of antisense transcript HBZ.
Whole-exome sequencing of 81 ATL cases, combined with targeted resequencing of 370 of the samples, identified 50 genes that were recurrently and significantly mutated; 13 of these genes were affected in >10% of the cases. The most frequently mutated genes were PLCG1 (36%), PRKCB (33%), CARD11 (24%), VAV1 (18%), and IRF4 (14%), all of which are implicated in T-cell receptor (TCR)–NF-κB signaling. In addition, CCR4 or CCR7 were mutated in 29% and 11% of the cases, respectively. Furthermore, CCR4 Tyr331 and CCR7 Trp355 were shown to be sites of gain-of function mutations.83
Single nucleotide polymorphism array–based copy number analysis of 426 ATL cases in the same study revealed 50 copy number decrease and 26 amplification events. Some of the genes with copy number abnormalities overlapped with gene mutation sites. To characterize structural abnormalities, whole-genome sequencing was performed on 48 paired samples. On average, 60 structural variations (SVs) per sample were identified, which included accumulated deletions in common fragile sites such as 14q31.1 (NRXN3), 7q31.1 (IMMP2L), 1p21.3 (DPYD), and 3p14.2 (FHIT). NRXN3 deletion was demonstrated in >60% of ATL cases. These results further reflected the genomic instability of ATL cells.83
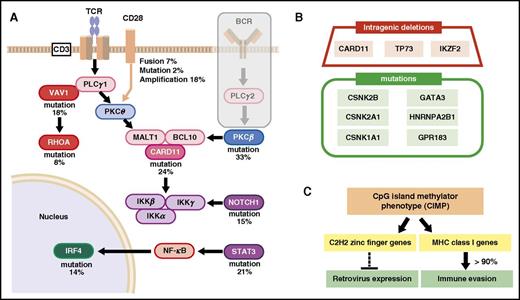
Accumulation of additional mutations affecting the TCR and NF-κB pathways, together with the inactivation of p53, p16, and other mutations, eventually transforms T cells into fully malignant cells. Data also indicate that ATL cells delete, mutate, or hypermethylate genes that encode components of the class I major histocompatibility complex (MHC-I), death receptors, and proteins involved in cellular adhesion or immune checkpoints as a strategy to escape detection by the immune system83 (Figure 2A-B).
Schematic summary of genetic abnormalities in ATL cells. (A) Accumulation of mutations in TCR signaling and NF-κB pathway. (B) examples of minor mutations. Intragenic deletions and mutations in genes other than those involved in TCR signaling. (C) possible effects of cytosine guanine dinucleotide (CpG) island methylator phenotype (CIMP). CSNK2B, casein kinase II subunit β; CSNK2A1, casein kinase 2 α 1; CSNK1A1, casein kinase 1 α 1; HNRNPA2B1, heterogeneous nuclear ribonucleoproteins A2/B1; IKZF2, zinc finger protein Helios.
Schematic summary of genetic abnormalities in ATL cells. (A) Accumulation of mutations in TCR signaling and NF-κB pathway. (B) examples of minor mutations. Intragenic deletions and mutations in genes other than those involved in TCR signaling. (C) possible effects of cytosine guanine dinucleotide (CpG) island methylator phenotype (CIMP). CSNK2B, casein kinase II subunit β; CSNK2A1, casein kinase 2 α 1; CSNK1A1, casein kinase 1 α 1; HNRNPA2B1, heterogeneous nuclear ribonucleoproteins A2/B1; IKZF2, zinc finger protein Helios.
In a study investigating SVs, the 3′ region of the programmed cell death 1 ligand (PD-L1) gene was found to be disrupted by SVs in 27% of ATL cases.84 These SVs invariably led to marked elevations in aberrant PD-L1 transcripts that were stabilized by truncation of the 3′-untranslated region. This is a unique genetic mechanism of immune escape triggered by SVs.
Deep sequencing of 203 ATL cases by Nagata et al found RHOA mutations in 30 cases that were widely distributed across the entire coding sequence but were almost always located at the guanosine triphosphate–binding pocket.85 Depending on the mutation type and position, these mutants showed variegated functional consequences, suggesting the involvement of both loss- and gain-of-RHOA functions in ATL leukemogenesis.
Another report showed mutations in SUZ12, DNMT1, DNMT3A, DNMT3B, TET1, TET2, IDH1, IDH2, MLL, MLL2, MLL3, and MLL4.86 TET2 was the most frequently mutated gene, occurring in 32% of the samples (10/31). Next-generation sequencing revealed nonsense mutations accompanied by loss of heterozygosity in TET2 and MLL3, suggesting important consequences of MLL3 and TET2 inactivation in the leukemogenesis of HTLV-1–induced ATL.
Epigenetic abnormalities in ATL
DNA methylation abnormalities
The CDKN2 locus encodes proteins such as p16, p14, and p15 and is involved in cell cycle regulation. It is also considered a hot spot of genomic and epigenomic abnormalities. In our copy number variation analysis, genomic deletions at this locus were found in 46 out of 168 cases (27%).63 Additionally, CpG methylation analysis showed hypermethylation of this locus in 47% and 73% of acute-type and lymphoma-type ATL cases, respectively, whereas the methylation levels in both chronic- and smoldering-type ATL cases were 17%.88 In another report, CpG methylation levels of CDK2B were higher than those of CDK2A.89 In contrast, our expression profiling of ATL samples did not show any downregulation in the expression of CDKN2 family members. This underscores the importance of detailed analyses of expression levels and functional consequences of cell cycle regulators in ATL cells. Progressive accumulation of CpG methylations of HCAD, SHP1, DAPK, and other genes with disease progression was also reported.90 CDKN1A (p21waf/Cip1) was reported to be downregulated by DNA methylation,91 which was in line with our data.63 These results suggest a relationship between abnormal DNA methylation and cell cycle regulation and the possible breakdown of their functions in ATL.
In addition to cell cycle regulators, DNA hypermethylation of bone morphogenetic protein-6 (BMP6), adenomatous polyposis coli (APC), and CD26 was also reported.92-94 A screening study using methylated CpG island amplification of representational difference analysis revealed abnormal DNA methylation in 53 genes, including those involved in apoptosis resistance such as KLF4 and EGR3.95
A comprehensive description of CpG methylation in ATL was also reported by Kataoka et al.83 ATL genome was characterized by prominent CpG island DNA hypermethylation; in ATL, a much higher number of genes were significantly hypermethylated with transcriptional silencing. Approximately 40% of the samples showed more extensive hypermethylation, known as the CpG island methylator phenotype (CIMP). CIMP status was associated with aggressive phenotypes but not with the mutation status of epigenetic regulators known to be associated with CIMP in other cancers (TET2, IDH2, and DNMT3A). Additionally, C2H2-type zinc finger genes implicated in the suppression of both endogenous and exogenous retroviruses were hypermethylated and silenced. Furthermore, MHC-I genes were hypermethylated and silenced in many ATL cases, and loss of MHC-I expression was observed in ∼90% of ATL cases, contributing to immune evasion by ATL cells (Figure 2C).
Although these data provide a basic understanding of the DNA methylation status in ATL cells, mechanisms underlying the abnormal DNA methylation and its biological significance remain to be determined.
Abnormal histone modification patterns
Various chemical histone modifications are involved in the modulation of chromatin structure and gene expression. Gene expression profile of ATL cells is characterized by marked deviations from that of normal CD4+ T cells, with upregulated expression of genes encoding various receptors and cytokines as well as lineage-independent aberrant expression of various differentiation-associated genes63 (Figure 1). These data suggest the presence of complex aberrations in histone modifications in ATL cell and provide a rationale for aberrant gene expression profiling. However, in contrast to DNA methylation studies, significant progress in characterization of histone modifications in ATL cells has not been made until recently, mainly because of technical difficulties.
Although histone acetylation abnormalities in ATL cells have not been well characterized, apoptosis induction and inhibition of NF-κB activity by histone deacetylase inhibitors (HDACis) have been reported in ATL cells and HTLV-1–infected cell lines.96,97 Although several clinical trials assessing several HDACis in ATL are ongoing, future studies are paramount to determine the mechanisms of histone deacetylase abnormalities and the efficacy of HDACis.
Trimethylation of H3K27 (H3K27m3) is well known for its role in survival, proliferation, de-differentiation, invasion, and metastasis of many human cancers such as breast and prostate cancer, and B-cell lymphoma.98 H3K27m3 is a marker for the suppression of gene expression in euchromatin region. Polycomb group (PcG) proteins form multiprotein complexes that function as transcriptional repressors of several thousand genes controlling differentiation pathways during development. PcG proteins work as transcriptional repressors through chemical modifications of histones by 2 major PcG protein complexes: polycomb repressive complex 1 (PRC1) and PRC2. PRC2 contains enhancer of zeste 1 (EZH1) and EZH2 that function as methylases for H3K27. PRC1 recognizes H3K27 m3 and induces ubiquitination of H2AK119 (H2K119Ub), stabilizing the chromatin structure.
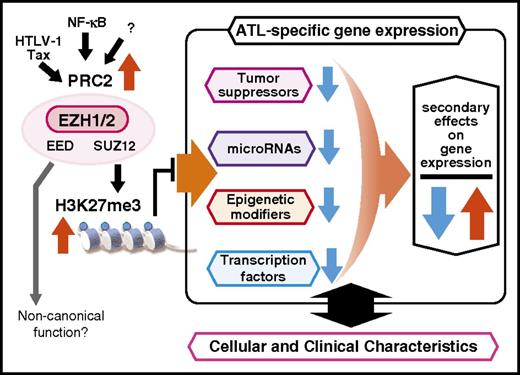
Our recent comprehensive study on the polycomb-dependent epigenetic landscape of ATL cells provided data to decipher the ATL-specific epigenetic code critical for ATL pathogenesis.99 Integrative analyses of the epigenome (n = 3) and transcriptome (n = 58) of primary ATL cells revealed that PRC2 components, including EZH2, were highly expressed and that H3K27m3 was significantly and frequently reprogrammed in over half of the genes (53.8%). Furthermore, H3K27me3 pattern in ATL was distinct from that in other cancer types and PcG-dependent cell lineages. No EZH2 mutation was found among 50 ATL patients included in our study. Progressive downregulation of gene expression was demonstrated with disease progression from indolent to aggressive ATL. Genes that were downregulated included key genes such as miR-31, BCL2L11, EVC1/2, CDKN1A, and NDRG2. Another finding was the presence of diverse outcomes resulting from the remote regulation of a broad spectrum of gene regulators, including various transcription factors, miRNAs, epigenetic modifiers, and developmental genes. Increased EZH1 expression, which compensated for the functions of EZH2, was also confirmed in peripheral T cells. Collectively, PRC2 hyperactivation with genome-wide H3K27m3 accumulation characterized the ATL epigenome. Consequently, simultaneous depletion of EZH1 and EZH2 by shRNAs was demonstrated to significantly reduce cellular H3K27m3 levels and to dramatically inhibit ATL cell growth compared with single depletion of either EZH, suggesting that the compensatory actions of EZH1/2 were critical for ATL (Figures 3 and 4).
Epigenetic landscape of ATL cells. High levels of EZH2 expression is observed in HTLV-1–infected cells as well as in ATL cells. Tax and NF-κB can induce EZH2 expression. ATL cells are characterized by PRC2 overexpression and H3K27 m3 accumulation, the level of which appears to progress with clonal progression.
Epigenetic landscape of ATL cells. High levels of EZH2 expression is observed in HTLV-1–infected cells as well as in ATL cells. Tax and NF-κB can induce EZH2 expression. ATL cells are characterized by PRC2 overexpression and H3K27 m3 accumulation, the level of which appears to progress with clonal progression.
Accumulation of H3K27m3 as the basis of ATL cell phenotype. PRC2-mediated accumulation of H3K27 me3 suppresses important genes such as tumor suppressors, miRNAs, epigenetic modifiers, and transcription factors, culminating in abnormalities in regulation of downstream genes that determine the phenotype of ATL cells.
Accumulation of H3K27m3 as the basis of ATL cell phenotype. PRC2-mediated accumulation of H3K27 me3 suppresses important genes such as tumor suppressors, miRNAs, epigenetic modifiers, and transcription factors, culminating in abnormalities in regulation of downstream genes that determine the phenotype of ATL cells.
Clonal growth and identification of HTLV-1–infected T cells in vivo
Clonality of ATL cells and HTLV-1–infected T cells in carriers
Clonal growth of ATL cells was first demonstrated by Southern blot analysis using HTLV-1 provirus DNA as a probe.8 ATL cells were found to be monoclonally expanded HTLV-1–infected cells except for rare cases with 2 or more provirus copies in ATL cells. Subsequent studies demonstrated that some ATL patients had multiple simultaneously occurring ATL clones at certain time points during the clinical course of disease.100-103 Clonal exchange during the clinical course was also reported.104,105 Array comparative genomic hybridization analysis of paired samples from the peripheral blood and lymph nodes of patients with acute-type ATL revealed multiple subclones of tumor cells with distinct genomic aberrations.106 The data suggested that clonal progression takes place in lymph nodes and a selected subclone among the lymph node subclones appears in the peripheral blood. Collectively, these reports strongly suggested the presence of clones concurrently undergoing transformation in vivo.
Inverse polymerase chain reaction (PCR) detected multiple amplified bands consisting of HTLV-1 proviral and flanking cellular DNA in HTLV-1–infected T cells in asymptomatic carriers, demonstrating the polyclonal nature of HTLV-1–infected cells in asymptomatic carriers.107 This technique has been widely used to characterize the clonality in various clinical settings.108-110 Difference in the stability of clonality among patients was shown to depend on the mode of virus transmission. The clonality of adults who acquired HTLV-1 horizontally from their spouses was more heterogeneous and less stable than that of long-term carriers who appeared to be infected through breastfeeding in infancy.111,112 Linker-mediated PCR was more sensitive, allowing detection of a greater number of integrated proviruses.113 HTLV-1 carriers doubly infected with Strongyloidiasis stercoralis showed oligoclonal expansion of HTLV-1–infected cells and high proviral loads.114 These techniques provided information on the clonality of HTLV-1–infected cells in various clinical conditions, although the findings were descriptive but not quantitative.
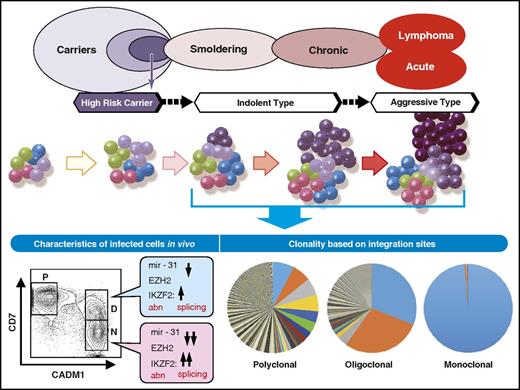
Subsequently, Bangham and colleagues developed a novel, high-throughput protocol using next-generation sequencing to map and quantify provirus integration sites.115 They found that there were between 10 000 and 100 000 clones in the peripheral blood of HTLV-1–infected individuals.115 They also claimed that HTLV-1 integration into the host genome was not random but showed associations with specific transcription factor binding sites116 and that specific sites upstream of certain proto-oncogenes were associated with ATL.117 Although these new findings significantly advanced the understanding of the clonality of HTLV-1–infected T cells in vivo, this technique unfortunately had limited utility in determining the number of infected cells belonging to a specific clone. To successfully measure the number of cells in individual clones, Bangham et al depended on the size variation of sheared genomic DNA fragments that showed the maximum variety of 400, because the size of recovered DNA fragments were distributed between 300 and 700 base pairs after shearing. To overcome this limitation, Firouzi et al introduced oligomer tags of 8 nucleotides that marked sheared DNA before amplification, which enabled the discrimination of >60 000 cells, dramatically expanding the ability to quantify the number of cells in individual clones.118 The results of these analyses provided firm evidence for the hypothesis that HTLV-1 carriers harbored a large number of HTLV-1–infected and immortalized clones and that each clone originated from a single infected T cell (Figure 5). The size of cell populations belonging to specific clones (ie, clone sizes) also varied greatly, and some clones had populations large enough to be detected as clonal expansion by conventional techniques, such as Southern blot hybridization and inverse PCR.
Schematic description of clonal progression and phenotypic changes. HTLV-1–infected T cells and ATL cells in vivo are now available for molecular analyses. Accumulating data indicate that epigenetic abnormalities occur early during leukemogenesis, as untransformed HTLV-1–infected cells show evidence of epigenetic abnormalities that are observed in ATL cells as well. The extent of clonality during HTLV-infection and progression to ATL have been characterized in detail by recent studies that provide information on clonal progression based on integration sites.
Schematic description of clonal progression and phenotypic changes. HTLV-1–infected T cells and ATL cells in vivo are now available for molecular analyses. Accumulating data indicate that epigenetic abnormalities occur early during leukemogenesis, as untransformed HTLV-1–infected cells show evidence of epigenetic abnormalities that are observed in ATL cells as well. The extent of clonality during HTLV-infection and progression to ATL have been characterized in detail by recent studies that provide information on clonal progression based on integration sites.
Furthermore, newly developed techniques to characterize the integration sites of HTLV-1 provirus provided a new set of evidence that encourages reappraisal of the classic promoter insertion model of retrovirus-mediated tumor genesis.119
Identification of HTLV-1–infected cells in vivo
For the identification of HTLV-1–infected T cells in vivo, a technique using multicolor flow cytometry and fluorescence-activated cell sorting was developed. The CD4 cells expressing CADM1 with reduction of CD7 expression represent the transformation status of infected cells and allow for the discrimination of immortalized but not untransformed cells from transformed cells.47,48 This method enabled molecular characterization of HTLV-1–infected T cells in vivo during both immortalization and transformation. Expression array analysis of fractionated CD4+ cells revealed that H3K27m3 target genes were suppressed in immortalized HTLV-1–infected cells as well as in terminally transformed ATL cells; suppression was more extensive in the latter cells47 (Figure 5). These data provided evidence that epigenetic abnormalities were one of the earlier leukemogenic events of HTLV-1–infected T cells. Currently, studies utilizing mutation analyses of cells derived from each fraction are underway to determine the timing of genetic mutations during the development of ATL. Ultimately, these data will provide in-depth information on the chronological landscape of genetic and epigenetic changes accumulated during immortalization and transformation of HTLV-1–infected CD4 T cells.
Recently, a hierarchical structure of ATL cell clones has been reported. CD45RA+ T memory stem (TSCM) cells were identified as the hierarchical apex that is capable of reconstituting identical ATL clones. TSCM cells may be a venue for clonal evolution and reservoir for ATL cells that can be exploited for the development of future molecular targeted therapies.120
Summary and perspectives
Significant progress has been made in the field of ATL research, with a growing body of information on the cellular and molecular characteristics of ATL cells available for better understanding of disease progression. In addition, “microenvironment” of the ATL cells is supposed to be involved in ATL cell survival and resistance to chemotherapy. Several reports suggested importance of interaction with normal epithelial cells and mesenchymal stem cells.121-123 Furthermore, involvement of angiogenesis and fibroblast-derived osteopontin has been suggested in proliferation and survival, as well as possible roles of exosomes produced by ATL cells.124-126 In this regard, a possible role of mutations of Notch1 signaling70,87 appears to deserve further examination. Further studies are expected to understand the mechanisms underlying resistance and survival of ATL cells in vivo. Xenograft models transplanted with ATL-derived cell lines or primary ATL cells using SCID mice have been reported, which is usually used for testing the efficacy of drugs.127 However, several questions remain unanswered. First, information on the mechanisms underlying immortalization after HTLV-1 infection of T cells is limited because of technical difficulties for in vitro infection experiments and the lack of appropriate animal models of HTLV-1 infection. Second, molecular events during clonal progression have not yet been fully elucidated. The reported landscape of genetic and epigenetic abnormalities is mostly that of terminally transformed ATL cells; thus, studies on HTLV-1–infected clones at various stages during transformation will provide crucial data needed to understand the mechanisms of immortalization and clonal expansion (Figure 6). Third, analysis of genetic and epigenetic abnormalities accumulating in individual HTLV-1–infected clones is necessary to understand the mechanism of clonal progression and malignant transformation. Finally, elucidation of factors that determine the provirus load is anticipated, given that proviral load is one of the major risk factors that appear to be determined by host factors. Taken together, clarification of molecular processes involved in the natural history of HTLV-1 infection is paramount for the prevention and treatment of HTLV-1-associated diseases, including ATL.
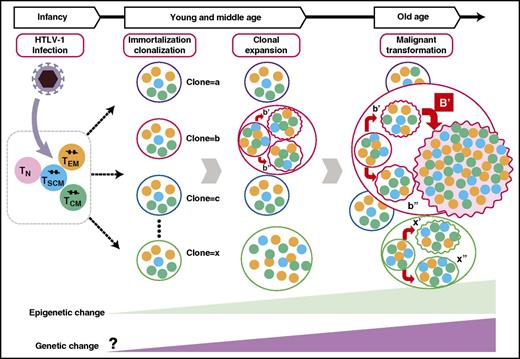
Schematic presentation of the clonal progression and hierarchical structure of HTLV-1–infected T cells. A clone that is defined by the integration site contains subclones with different genetic abnormalities. Among these subclones, there is a hierarchical structure where TSCM cells behave as ATL-initiating stem cells. TCM, CD45RO+ central memory T cells; TEM, CD45RO+ effector memory T cells; TN, CD45RA+ naive T cells.
Schematic presentation of the clonal progression and hierarchical structure of HTLV-1–infected T cells. A clone that is defined by the integration site contains subclones with different genetic abnormalities. Among these subclones, there is a hierarchical structure where TSCM cells behave as ATL-initiating stem cells. TCM, CD45RO+ central memory T cells; TEM, CD45RO+ effector memory T cells; TN, CD45RA+ naive T cells.
Acknowledgments
This work was supported by grants from the Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (KAKENHI) (JP26293226) and the Japan Agency for Medical Research and Development (AMED), Practical Research for Innovative Cancer Control (16ck0106133h0003 and 16ck0106136h0003) (T.W.).
Authorship
Contribution: T.W. collected all the information and prepared the manuscript and figures.
Conflict-of-interest disclosure: Part of the research by T.W. was supported by a research fund from Daiichi Sankyo Company.
Correspondence: Toshiki Watanabe, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae, Kawasaki, Kanagawa 216 8512, Japan; e-mail: tnabe@ims.u-tokyo.ac.jp.