To the editor:
The central nervous system (CNS) is a key site of extramedullary disease in pediatric acute lymphoblastic leukemia (ALL),1 and prior to the development of contemporary risk-adapted treatment strategies, CNS involvement was inevitable in most cases.1 However, the biology of those leukemic cells that reside in this site is poorly understood, and indeed, the extent to which genetic, transcriptional, and phenotypic variegation exists between leukemic cells located in different organs such as the CNS is unknown. To address these questions, we have undertaken a detailed analysis of primary B-precursor acute lymphoblastic leukemia (B-ALL) cells isolated from the CNS and bone marrow (BM) from both affected children and mouse xenograft recipients.
Leukemic cells isolated from the BM of patients presenting with B-ALL were transplanted into recipient mice by tail-vein injection (supplemental Table 1, available on the Blood Web site).2 We confirmed BM engraftment in recipients by flow cytometry, and using both histological and radiological techniques, we could demonstrate that they had patterns of CNS involvement mimicking those seen in patients with CNS disease (supplemental Figures 1 and 2). B-ALL cells have an inherent capacity to infiltrate the CNS,3 and we observed that by 6 months, recipient mice had CNS disease, regardless of whether the patients whose leukemic cells they received had CNS disease at the time the cells were donated for research. Thus, this xenograft model faithfully recapitulates the natural clinical history of untreated B-ALL, and we used such mice, which had been allowed to develop CNS disease for our studies.
We compared gene expression profiles of leukemic cells isolated from the CNS (CNS-derived leukemic cells) and BM (BM-derived leukemic cells) of recipient mice (supplemental Table 2). Initially, we screened BM-derived and CNS-derived leukemic cells from recipients of 4 different ALLs (ALL1-ALL4) using microarrays. Gene set enrichment analysis showed that in CNS-derived leukemic cells, gene sets related to cell cycle and oxidative phosphorylation were downregulated (supplemental Figure 3A; supplemental Table 3).
We next asked whether these differences were evident in the leukemic cells in BM and cerebrospinal fluid from 3 patients with B-ALL who had both CNS and BM involvement (ALL 6, 7, and 8). Despite containing varying numbers of leukemic cells, we found that genes related to cell cycle and oxidative phosphorylation in CNS-derived leukemic cells were similarly downregulated (supplemental Figure 3B; supplemental Table 4).
During the course of this work, a patient (ALL9) presented with B-ALL and radiological evidence of leukemic meningitis. We transplanted this patient’s BM sample into xenograft recipients in order to study the physiology and transcriptional profiles of the BM- and CNS- derived leukemic cells. Leukemic cells from the 2 sites had similar surface immunophenotypes (supplemental Figure 4). However, fewer CNS cells were in the S/G2/M phase of the cell cycle (Figure 1A-B), and consistent with this, a lower proportion of leukemic cells in the CNS were actively proliferating, as assessed by Ki67 staining (Figure 1C; supplemental Figure 5). In addition, CNS-derived leukemic cells consumed less oxygen in in vitro assays, indicating diminished mitochondrial activity (Figure 1D). These observations supported the transcriptional observations made in our initial microarray screen and the 3 primary patient samples.
Leukemic cells in the CNS show adaptation to hypoxia, such as reduced proliferation and oxidative phosphorylation, increased glycolysis, and upregulation of vascular endothelial growth factor A (VEGFA). (A) Representative cell cycle analysis of 4′,6-diamidino-2-phenylindole–stained CNS- and BM-derived leukemic cells from xenograft recipients of ALL9 leukemic cells. The fraction of cells in S/G2/M (8.6% vs 26.0%) is indicated. (B) Mean percentages of CNS- and BM-derived leukemic cells that are in S/G2/M from 5 separate recipients of ALL9 leukemic cells. (C) Immunohistochemical analysis of leukemic cell proliferation in BM and brains from xenograft recipients of ALL9 leukemic cells. The percentage of hCD19+Ki67+ cells out of hCD19+ cells are indicated. n = 3 biological replicates. (D) Mean oxygen consumption rates (RFU/h) in CNS- and BM-derived leukemic cells from recipients of ALL9 leukemic cells. n = 3 technical replicates, and data are representative of 2 independent experiments. Data show mean ± standard error of the mean, analyzed with 2-sided, paired Student t test: *P < .05. (E) Gene set enrichment analysis of RNA-sequencing data illustrating enrichment of hypoxia (normalized enrichment score [NES] = 2.34, false discovery rate [FDR] = 0) and glycolysis (NES = 2.25, FDR = 0) associated genes in CNS-derived leukemic cells from recipients of ALL9 cells. (F) Heatmap indicating levels of glycolytic metabolites in paired CNS- and BM-derived leukemic cells from 4 xenograft recipients (1-4) of ALL9 leukemic cells measured by mass spectrometry. 2PG, 2 phosphoglycerate; 3PG, 3 phosphoglycerate; F6P, fructose 6 phosphate; G1P, glucose 1 phosphate; G6P, glucose 6 phosphate. Red indicates high concentration and green low concentration. (G) Quantification of VEGFA (probe ID A_23_P70398) transcripts in CNS- and BM-derived leukemic cells from xenograft recipients of ALL1-ALL4 leukemic cells with microarray. ALL3 contains data from 2 mice (3-1, 3-2). (H) Quantification of VEGFA in RNA sequencing from primary tissues of ALL6-ALL8. (I) VEGFA expression by RQPCR in CNS- and BM-derived leukemic cells from recipients of ALL9 leukemic cells; n = 4 biological replicates. Data show mean ± standard error of the mean, analyzed with 2-sided, paired Student t test: *P < .05.
Leukemic cells in the CNS show adaptation to hypoxia, such as reduced proliferation and oxidative phosphorylation, increased glycolysis, and upregulation of vascular endothelial growth factor A (VEGFA). (A) Representative cell cycle analysis of 4′,6-diamidino-2-phenylindole–stained CNS- and BM-derived leukemic cells from xenograft recipients of ALL9 leukemic cells. The fraction of cells in S/G2/M (8.6% vs 26.0%) is indicated. (B) Mean percentages of CNS- and BM-derived leukemic cells that are in S/G2/M from 5 separate recipients of ALL9 leukemic cells. (C) Immunohistochemical analysis of leukemic cell proliferation in BM and brains from xenograft recipients of ALL9 leukemic cells. The percentage of hCD19+Ki67+ cells out of hCD19+ cells are indicated. n = 3 biological replicates. (D) Mean oxygen consumption rates (RFU/h) in CNS- and BM-derived leukemic cells from recipients of ALL9 leukemic cells. n = 3 technical replicates, and data are representative of 2 independent experiments. Data show mean ± standard error of the mean, analyzed with 2-sided, paired Student t test: *P < .05. (E) Gene set enrichment analysis of RNA-sequencing data illustrating enrichment of hypoxia (normalized enrichment score [NES] = 2.34, false discovery rate [FDR] = 0) and glycolysis (NES = 2.25, FDR = 0) associated genes in CNS-derived leukemic cells from recipients of ALL9 cells. (F) Heatmap indicating levels of glycolytic metabolites in paired CNS- and BM-derived leukemic cells from 4 xenograft recipients (1-4) of ALL9 leukemic cells measured by mass spectrometry. 2PG, 2 phosphoglycerate; 3PG, 3 phosphoglycerate; F6P, fructose 6 phosphate; G1P, glucose 1 phosphate; G6P, glucose 6 phosphate. Red indicates high concentration and green low concentration. (G) Quantification of VEGFA (probe ID A_23_P70398) transcripts in CNS- and BM-derived leukemic cells from xenograft recipients of ALL1-ALL4 leukemic cells with microarray. ALL3 contains data from 2 mice (3-1, 3-2). (H) Quantification of VEGFA in RNA sequencing from primary tissues of ALL6-ALL8. (I) VEGFA expression by RQPCR in CNS- and BM-derived leukemic cells from recipients of ALL9 leukemic cells; n = 4 biological replicates. Data show mean ± standard error of the mean, analyzed with 2-sided, paired Student t test: *P < .05.
We then asked whether these physiological differences could be explained by any genetic differences between CNS- and BM-derived leukemic cells in the xenograft recipients of ALL9. We used targeted-capture deep sequencing of the diagnostic sample and of CNS- and BM-derived leukemic cells from 2 recipient mice to compare genomic mutations and copy-number abnormalities in cells isolated from the 2 microenvironments, and we found no obvious differences between them (supplemental Figure 6; supplemental Table 5). Therefore, we analyzed their transcriptomes to identify a gene expression signature specific to the CNS-derived cells. We compared leukemic cells from BM, CNS, and spleen from 5 recipients of BM from ALL9 (supplemental Figure 7). Gene sets uniquely upregulated in CNS-derived leukemic cells included those associated with hypoxia and glycolysis (Figure 1E), and as expected, cell cycle gene sets were downregulated compared with BM-derived cells (supplemental Table 6). We observed no obvious expression changes in gene sets encoding cell adhesion molecules. Interestingly, hypoxic genes were also upregulated in CNS leukemic cells isolated from 3 patients (supplemental Figure 3C). Increased expression of the glycolytic enzymes hexokinase 2 (HK2) and pyruvate dehydrogenase kinase 1 (PDK1), which are known to be upregulated in response to hypoxia,4 was confirmed by real-time quantitative polymerase chain reaction (RQPCR) (supplemental Figure 8). Consistent with these differences in gene expression, we found that CNS-derived leukemic cells had much higher levels of glycolytic intermediates than those derived from the BM (Figure 1F). Our results indicate that CNS-derived leukemic cells are not genetically distinct but are both transcriptionally and physiologically adapted to hypoxic conditions.
VEGFA (a hypoxia-responsive gene5 ) was upregulated in both CNS-derived leukemic cells (supplemental Table 7; Figure 1G) and primary leukemic cells isolated from the cerebrospinal fluid of children with CNS involvement (Figure 1H). We confirmed this by RQPCR in CNS leukemic cells of xenograft recipients (Figure 1I) and also observed that they express high levels of VEGFA receptor (FLT-1/VEGFR1) (supplemental Figure 8). FLT-1 is commonly expressed in childhood ALL,6,7 and VEGFA/FLT-1 signaling promotes migration and survival of leukemic cells in vivo and vitro.6,8
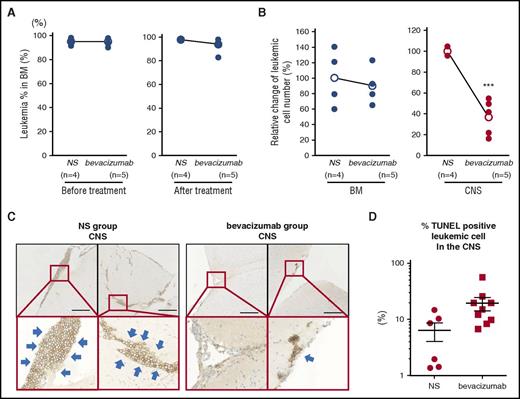
Bevacizumab is a VEGFA-neutralizing antibody used in the treatment of several non-hematological cancers, and we tested its impact on mice with CNS involvement in our xenograft model. Recipients with equivalent levels of BM engraftment (Figure 2A), and therefore presumably similar levels of CNS involvement, were given intraperitoneal injections of either bevacizumab or saline. The CNS leukemic burden was significantly reduced in mice treated with bevacizumab (Figure 2B-C), and terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) staining demonstrated increased apoptosis in CNS-derived leukemic cells (Figure 2D; supplemental Figure 9A). Although some TUNEL-positive apoptotic cells were seen in BM in bevacizumab recipients (supplemental Figure 9B), its impact on BM disease burden was less marked (Figure 2A-B).
Effect of bevacizumab on the leukemic cell burden in the CNS of xenograft recipients. Leukemic mice showing >90% engraftment in the BM received intraperitoneal bevacizumab or normal saline (NS). (A) Mean burden of leukemic cells in the BM of each group at the start and the end of treatment. (B) Mean burden of leukemic cells in the BM and CNS of each group at the end of treatment showing significant lower leukemic cells in the CNS of bevacizumab-treated mice. Data show mean and analyzed with 2-sided, unpaired Student t test; ***P < .001. (C) Representative immunohistochemical staining for human CD19 in tissue sections from the CNS of NS- and bevacizumab-treated mice. Thick blue arrows mark leukemic cells. Scale bar, 250 μm. (D) The percentage of TUNEL-positive leukemic cells in the CNS was counted from 3 distinct areas of the each section of 2 control mice and 3 bevacizumab-treated mice. Graph shows the percentage of TUNEL-positive cells in the CNS.
Effect of bevacizumab on the leukemic cell burden in the CNS of xenograft recipients. Leukemic mice showing >90% engraftment in the BM received intraperitoneal bevacizumab or normal saline (NS). (A) Mean burden of leukemic cells in the BM of each group at the start and the end of treatment. (B) Mean burden of leukemic cells in the BM and CNS of each group at the end of treatment showing significant lower leukemic cells in the CNS of bevacizumab-treated mice. Data show mean and analyzed with 2-sided, unpaired Student t test; ***P < .001. (C) Representative immunohistochemical staining for human CD19 in tissue sections from the CNS of NS- and bevacizumab-treated mice. Thick blue arrows mark leukemic cells. Scale bar, 250 μm. (D) The percentage of TUNEL-positive leukemic cells in the CNS was counted from 3 distinct areas of the each section of 2 control mice and 3 bevacizumab-treated mice. Graph shows the percentage of TUNEL-positive cells in the CNS.
In conclusion, we have shown that B-ALL cells that infiltrate the CNS are more quiescent, consume less oxygen, and have greater glycolytic activity than those residing in the BM, showing features of enhanced adaptation to hypoxia. This is consistent with the CNS being a harsher, more hypoxic and nutrient-poorer microenvironment than the BM.9 We have previously reported that the small numbers of residual leukemic cells that remain in the BM of B-ALL patients who have undergone treatment are profoundly quiescent and consequently resistant to conventional chemotherapy, and we have proposed that these cells make a contribution to relapse.10 Combined with the observations that hypoxic environment confers resistance to chemotherapy11 and is associated with minimal residual disease of leukemia,12 our data correlate with the clinical observation that residual CNS disease can drive relapse of B-ALL. It is possible that the “hypoxic adaptation” we see in CNS-infiltrating childhood B-ALL is also found in other types of CNS-infiltrating hematological malignancies, including “high-risk” groups with CNS involvement.1 One interesting possibility is that VEGFA may be expressed in the limited leukemic cells adapted to hypoxic microenvironments in the BM, and bevacizumab could target them. Although the number of patients analyzed is limited and larger studies are required, we suggest that these data and the wide clinical experience with bevacizumab, together with its favorable toxicity profile, warrant its inclusion in future clinical trials for B-ALL with CNS involvement.
The online version of this article contains a data supplement.
Authorship
Acknowledgments: The authors thank Kumi Kodama, Yanping Guo, Virginia Turati, Mathew Robson, and Preeta Datta for the support of animal experiments; Naoki Amano for performing the bioinformatics analysis; Shoko Okawada, Hironobu Kitazawa, and Yuki Arakawa for providing patient samples; Katsuyuki Ohmori for critical advice on cell cycle analysis; Kenichi Chiba and Hiroko Tanaka for establishing genetic material analysis; Tony Brooks and his team for performing the transcriptome sequencing; Jason Wray for critical reading of the manuscript; and Ohad Yogev, Rachael Nimmo, Elitza Deltcheva, and all other Stem Cell Laboratory members at the University College London (UCL) Cancer Institute for technical assistance and advice. The authors are grateful to the ALL patients who participated in this study.
This work was supported by Bloodwise (grant 16001) (T.E.), Cancer Research UK (grant A12796) (T.E.), Children with Cancer (grant 14/173) (T.E.), Great Ormond Street Hospital charity (grants W1062 and V1362) (T.E.), the UCL/University College London Hospital Biomedical Research Centre (grant 51517) (T.E.), Japan Agency for Medical Research and Development (grant 15 gm0510014h0004) (T.N.), Grant-in-Aim for Challenging Exploratory (grant 26670498) (T.N.), the Japan Society for the Promotion of Science (grant 25860857) (I.K.), Morinaga Foundation for Health & Nutrition (I.K.), Japan Leukemia Research Fund (I.K.), the Uehara Memorial Foundation (I.K.), and the Waksman Foundation of Japan (I.K.).
Contribution: I.K., Y.N., A.N., and H.O. designed and performed experiments and analyzed data; M. Morita performed animal experiments; M.N., D.W., and A.W. performed bioinformatics analyses and discussion; A.U.A. and T.M. performed pathological analysis and discussion; K.Y., H.U., M.N., A.W., Y. Shiozawa, Y. Shiraishi, and S.M. analyzed genetic materials; M. Mori, R.G., K.U., K.W., K.K., and S.A. provided clinical samples; T.E., T.N., T.H., M.K.S., S.O., and M.S. supervised the project and experimental design; and I.K., R.G., and T.E. wrote the paper with input from Y.N., A.N., H.O., M.N., A.W., K.Y., D.W., and T.M.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tariq Enver, UCL Cancer Institute, Paul O’Gorman Building, 72 Huntley St, London WC1E 6BT, United Kingdom; e-mail: t.enver@ucl.ac.uk; and Tatsutoshi Nakahata, Center for iPS Cell Research and Application, Kyoto University, 53 Kawahara-cho, Shogoin, Sakyo, Kyoto 606-8507, Japan; e-mail: tnakaha@cira.kyoto-u.ac.jp.

![Figure 1. Leukemic cells in the CNS show adaptation to hypoxia, such as reduced proliferation and oxidative phosphorylation, increased glycolysis, and upregulation of vascular endothelial growth factor A (VEGFA). (A) Representative cell cycle analysis of 4′,6-diamidino-2-phenylindole–stained CNS- and BM-derived leukemic cells from xenograft recipients of ALL9 leukemic cells. The fraction of cells in S/G2/M (8.6% vs 26.0%) is indicated. (B) Mean percentages of CNS- and BM-derived leukemic cells that are in S/G2/M from 5 separate recipients of ALL9 leukemic cells. (C) Immunohistochemical analysis of leukemic cell proliferation in BM and brains from xenograft recipients of ALL9 leukemic cells. The percentage of hCD19+Ki67+ cells out of hCD19+ cells are indicated. n = 3 biological replicates. (D) Mean oxygen consumption rates (RFU/h) in CNS- and BM-derived leukemic cells from recipients of ALL9 leukemic cells. n = 3 technical replicates, and data are representative of 2 independent experiments. Data show mean ± standard error of the mean, analyzed with 2-sided, paired Student t test: *P < .05. (E) Gene set enrichment analysis of RNA-sequencing data illustrating enrichment of hypoxia (normalized enrichment score [NES] = 2.34, false discovery rate [FDR] = 0) and glycolysis (NES = 2.25, FDR = 0) associated genes in CNS-derived leukemic cells from recipients of ALL9 cells. (F) Heatmap indicating levels of glycolytic metabolites in paired CNS- and BM-derived leukemic cells from 4 xenograft recipients (1-4) of ALL9 leukemic cells measured by mass spectrometry. 2PG, 2 phosphoglycerate; 3PG, 3 phosphoglycerate; F6P, fructose 6 phosphate; G1P, glucose 1 phosphate; G6P, glucose 6 phosphate. Red indicates high concentration and green low concentration. (G) Quantification of VEGFA (probe ID A_23_P70398) transcripts in CNS- and BM-derived leukemic cells from xenograft recipients of ALL1-ALL4 leukemic cells with microarray. ALL3 contains data from 2 mice (3-1, 3-2). (H) Quantification of VEGFA in RNA sequencing from primary tissues of ALL6-ALL8. (I) VEGFA expression by RQPCR in CNS- and BM-derived leukemic cells from recipients of ALL9 leukemic cells; n = 4 biological replicates. Data show mean ± standard error of the mean, analyzed with 2-sided, paired Student t test: *P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/23/10.1182_blood-2016-06-721712/4/m_blood721712f1.jpeg?Expires=1767731001&Signature=wyvmIkx5QEzW9rKUfFELwqjYtNJmZZc3O4H79JkujZtBfbFNjpGIbV-YmVDRNgdjW809t~Nan5KvcoLCZSmB39gnDXWb7kV4gF4~W2E4IXzfxIlhN67BIjYyTnh9R7UaZTZyseF3GYsEwVkbPoWVOaIRM1KxBgTY5kYOQtU-uJ-3f2nzPv3xM0ciwAnAlRTOEROaVM3pS6Yex32waOgr9uBJD3zrpU2sPHMsjHcYaJFYZkbaIywtuHxsI2c7MuvdVaRhocPJBsDJmg0gNXVr~WyM8X8DgKBoDKgzy6WKe69PqonahJiWZTQeqLjGVTEM7u9fEl7p9ZbYsvcP3BdW2w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal