Key Points
Mutational trajectories are defined by complex patterns of molecular heterogeneity in MDS, including lower-risk cases.
Therapeutic intervention dynamically reshapes mutational patterns often resulting in branched or independent evolution of MDS clones.
Abstract
Clonal evolution is believed to be a main driver for progression of various types of cancer and implicated in facilitating resistance to drugs. However, the hierarchical organization of malignant clones in the hematopoiesis of myelodysplastic syndromes (MDS) and its impact on response to drug therapy remain poorly understood. Using high-throughput sequencing of patient and xenografted cells, we evaluated the intratumoral heterogeneity (n= 54) and reconstructed mutational trajectories (n = 39) in patients suffering from MDS (n = 52) and chronic myelomonocytic leukemia-1 (n = 2). We identified linear and also branching evolution paths and confirmed on a patient-specific level that somatic mutations in epigenetic regulators and RNA splicing genes frequently constitute isolated disease-initiating events. Using high-throughput exome- and/or deep-sequencing, we analyzed 103 chronologically acquired samples from 22 patients covering a cumulative observation time of 75 years MDS disease progression. Our data revealed highly dynamic shaping of complex oligoclonal architectures, specifically upon treatment with lenalidomide and other drugs. Despite initial clinical response to treatment, patients’ marrow persistently remained clonal with rapid outgrowth of founder-, sub-, or even fully independent clones, indicating an increased dynamic rate of clonal turnover. The emergence and disappearance of specific clones frequently correlated with changes of clinical parameters, highlighting their distinct and far-reaching functional properties. Intriguingly, increasingly complex mutational trajectories are frequently accompanied by clinical progression during the course of disease. These data substantiate a need for regular broad molecular monitoring to guide clinical treatment decisions in MDS.
Introduction
Myelodysplastic syndromes (MDS) are malignant bone marrow (BM) disorders with a highly variable disease course, characterized by ineffective production of mature blood cells and risk of progression to acute myeloid leukemia (AML). This clinical heterogeneity is underlined by a complex genetic make-up involving a plethora of somatically acquired mutations that occur in diverse combinations and recurrently affect genes involved in various cellular processes, such as RNA splicing, DNA repair, epigenetic pathways, and others.1-9 It is believed that these mutations are progressively acquired over many years in hematopoietic stem cells (HSCs) that gain a selective advantage and outcompete normal hematopoiesis. An increase of cellular clonality with age was first demonstrated by analyzing X-chromosome inactivation in hematologically healthy aging females.10,11 More recently, hematopoietic cells from aging individuals have been shown to carry an increased frequency of somatic mutations affecting genes involved in myeloid neoplasia, such as TET2, DNMT3A, and ASXL112-15 but also, to a lesser extent, cytogenetic lesions.16,17 Hence, it has been speculated that these lesions may represent early recurrent events shifting the pool of long-lived HSCs into a “primed-state” that can acquire further somatic events and give rise to myeloid malignancies.
To test this hypothesis, we focused on MDS, which is a slow growing disease that mainly affects the elderly. We used a multisampling approach combined with next-generation sequencing (NGS) to decipher the mutational trajectories leading to the diseased state, and quantify the extent of clonal diversity on a patient-specific level. Furthermore, we performed whole exome sequencing (WES) of multiple time points to interrogate evolutionary dynamics and clonal diversity during disease course and in response to therapy with drugs typically applied in MDS such as lenalidomide (LEN). This workflow allowed the identification of recurrent founding events in MDS and revealed both linear and variegated branching evolution with increasing complexity that is frequently accompanied by progressive disease.
Methods
Detailed procedures can be found in supplemental Methods available on the Blood Web site.
Patient sample acquisition
Diagnostic BM aspirates were obtained from 52 MDS and 2 chronic myelomonocytic leukemia-1 (CMML-I) patients, further collectively denoted as the MDS cohort, treated in the Department of Hematology and Oncology of the Medical Faculty Mannheim, University of Heidelberg, Germany, after obtaining written informed consent. The use of human material within this study was approved by the Institutional Review Board of the Medical Faculty Mannheim, in accordance with the Declaration of Helsinki. Detailed patient characteristics are listed in supplemental Table 1.
WES and targeted ultra-deep sequencing (UDS)
For mutational screening, genomic DNA was isolated from patient BM/CD34+ cells and patient-matched BM-derived in vitro expanded mesenchymal stromal cells (MSCs, n = 45), or CD3+ peripheral blood (PB) cells (n = 5) as a control. Subsequently, paired DNA samples were subjected to WES with an average coverage of 100× on an Illumina HiSeq 2000 sequencing device. In addition, a custom amplicon-based MiSeq panel was applied to BM samples for sensitive interrogation of 24 commonly mutated genes in MDS (ASXL1, EZH2, DNMT3A, IDH1, IDH2, TET2, SF3B1, SRSF2, U2AF1, ZRSR2, STAG2, TP53, BCOR, ETV6, NPM1, RUNX1, CBL, FLT3, JAK2, KRAS, NRAS, GNAS, MPL, and KIT). Briefly, sequence reads were mapped using Burrows-Wheeler Aligner (version 0.7.9a), deduplicated and recalibrated with Picard (version 1.1) and Genome Analysis Tool Kit (version 3.1), followed by mutational analysis via application of Genome Analysis Tool Kit Haplotypecaller, VarScan2 (version 2.3) and Pindel (version 0.2.5) detection tools.
Molecular validation analyses
For validation of mutational events, we performed targeted quantitative pyrosequencing for point and small insertion/deletion (INDEL) mutations and capillary fragment analysis for longer INDEL variants. In addition, cytogenetic lesions were quantified using interphase fluorescence in situ hybridization (FISH) and a newly developed multiplex microsatellite assay for measurement of loss of heterozygosity at aberrant genomic loci.18
Flow cytometry analysis and cell sorting
BM cells were isolated from primary patients or xenografted mice19 and stained in phosphate buffered saline + 2% fetal calf serum (5 × 106 cells/100 μL) in the presence of human Fcγ receptor blockers (Miltenyi) using the antibodies described in detail in supplemental Methods. The indicated fractions were fluorescence-activated cell sorter (FACS) purified using a BD FACSAriaII or BD FACSAriaIII. The lineage cocktail used in this study contained: CD4, CD8, and CD20 (BD Biosciences), and CD19 and CD235a (eBiosciences) antibodies conjugated to APC. Dead cells were gated out using 7-aminoactinomycin D (#A1310; Life Technologies) or propidium iodide (#349483; BD).
Mouse experiments
Six- to 10-week-old NOD/LtSz-scid IL2Rγc−/− (NSG) (stock #00557) and NSGS (stock #013062) mice were purchased from The Jackson Laboratories and bred in house at the “Deutsches Krebsforschungzentrum” Heidelberg under pathogen-free conditions. Only females were used for experiments.20 Xenotransplantation experiments were carried out as previously described.19 Briefly, mice were sublethally irradiated (200 cGy) 24-hours before transplantation. All xenografts were performed with 5 × 105 MSCs alongside with 105 CD34+ cells, unless otherwise indicated. Engrafted cells were FACS sorted 16 to 28 weeks after transplantation.
All animal experiments were carried out in accordance with institutional guidelines approved by the Animal Health Care Committee of the Regierungspräsidium Karlsruhe under authorization numbers G74/12 and G210/12.
Results
Use of an integrative multisampling approach allows to retrospectively define the molecular trajectories leading to MDS
In order to reconstruct mutational hierarchies in MDS, we performed de novo screening of somatic mutations in primary BM via WES or a MiSeq 24-gene panel that interrogated the most frequently mutated genes in MDS (n = 52 MDS and n = 2 CMML-I) patients (supplemental Table 1). Identified mutations were validated and the allelic burden quantified in all additional available samples from each patient using pyrosequencing, UDS, interphase FISH, microsatellite analysis, and capillary electrophoresis (supplemental Figure 1A-B). Their somatic origin was validated by parallel assessment of patient-matched nonhematopoietic MSCs as germ line controls.
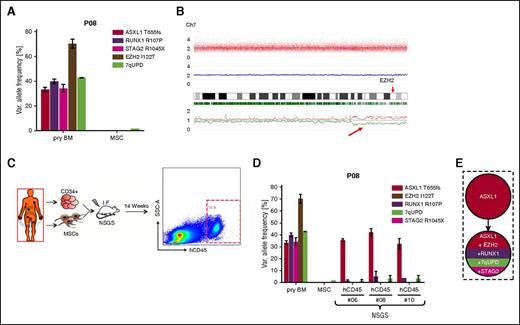
We frequently identified multiple mutations in single samples suggestive of an oligoclonal composition. Expectedly, the molecular requantification using patient-specific assays often revealed only minor differences of variant allele frequencies (VAFs) for the single mutations, likely reflecting the clonal dominance of an advanced subclone as demonstrated for patient 08 (P08) (Figure 1A). Therefore, in the majority of cases, mutational VAFs in single samples were not discriminative enough to unequivocally determine their relative order of historic acquisition. Moreover, ostensibly pre-eminent VAFs (>>50%) determined by quantitative sequencing alone often had to be critically reevaluated in the context of cytogenetic aberrations such as deletions or uniparental disomy (UPD). Exemplarily, case P08 displayed a highly elevated VAF of 70% for an EZH2 mutation (Figure 1A) within a region of UPD on chromosome 7q (Figure 1B). This mutation could have been misinterpreted as a founder event if the chromosomal profile had been neglected.
High-throughput genomic analysis using array- and sequencing-based methods. (A) For P08, EZH2 I122T appears to have the highest but unexpected VAF (>>50%), which can be explained by the presence of a telomeric copy number neutral UPD of chromosome 7q as identified by single nucleotide polymorphism (SNP)-array analysis (B). (C) Experimental setup of xenotransplantation for P08. After 14 weeks of long-term engraftment, human CD45+ (hCD45+) cells were FACS sorted for subsequent mutational analysis (exemplarily shown for NSGS mouse #6). (D) Three independent xenografts from P08 were subjected to quantitative profiling of mutations detected in primary BM and revealed exclusive expansion of an ancestral ASXL1-only mutated clone (see supplemental Figure 1D for mutational raw data). (E) Reconstructed mutational hierarchy for P08. Error bars indicate standard deviation (SD). I.F., intrafemoral; SSC-A, side scatter area.
High-throughput genomic analysis using array- and sequencing-based methods. (A) For P08, EZH2 I122T appears to have the highest but unexpected VAF (>>50%), which can be explained by the presence of a telomeric copy number neutral UPD of chromosome 7q as identified by single nucleotide polymorphism (SNP)-array analysis (B). (C) Experimental setup of xenotransplantation for P08. After 14 weeks of long-term engraftment, human CD45+ (hCD45+) cells were FACS sorted for subsequent mutational analysis (exemplarily shown for NSGS mouse #6). (D) Three independent xenografts from P08 were subjected to quantitative profiling of mutations detected in primary BM and revealed exclusive expansion of an ancestral ASXL1-only mutated clone (see supplemental Figure 1D for mutational raw data). (E) Reconstructed mutational hierarchy for P08. Error bars indicate standard deviation (SD). I.F., intrafemoral; SSC-A, side scatter area.
To circumvent these limitations, we analyzed an extensive set of diverse patient-derived cell fractions by quantifying mutational and cytogenetic lesions (supplemental Figure 1C; supplemental Table 2). This was based on the hypothesis that the successive acquisition of lesions would impair the ability of HSCs to contribute to different mature cell lineages, thereby leading to a differential representation of each subclone in highly purified hematopoietic lineages (myeloid, lymphoid, erythroid, and stem cell enriched fractions). These were either isolated directly from primary BM, PB, or from patient-derived xenografts in NSG or NSGS mice previously established by our group.19 Moreover, we also analyzed serial samples collected from chronological BM biopsies.
Only the analysis of such sample collectives allowed to reconstruct individual mutational hierarchies. In order to consider mutation “A” being acquired earlier than mutation “B” we required that at least 1 of 2 conditions was fulfilled: (1) at least one investigated sample was requested to show a higher burden for mutation “A” and a difference in frequency of ≥10% relative to mutation “B” or (2) at least 3 investigated samples were requested to exhibit a higher mutational frequency for mutation “A” and a difference in frequency of ≥5% relative to mutation “B” with non-overlapping error intervals. Moreover, the sum of the allelic frequency of both heterozygous mutations was required to exceed 55% (theoretically 51% would be sufficient) in at least 1 sample in order to be confident that both mutations arose within the same cell and do not designate independent clones.
To demonstrate usage of MDS xenograft samples for reconstructing mutational pedigrees, we isolated long-term xenoengrafted human CD45+ cells from patient P08 (Figure 1C), which revealed the exclusive propagation of an early MDS clone only carrying an ASXL1 mutation (Figure 1D; supplemental Figure 1D). This identified ASXL1 as the founder lesion and determined RUNX1, STAG2, EZH2, and 7qUPD as secondary hits (Figure 1E). Similarly, usage of FACS-purified BM and xenograft fractions, as well as sequentially acquired chronological samples was essential for the reconstruction of P15’s mutational hierarchy (Figure 2A). In this case, the patient’s BM obtained in the year 2010 was almost completely dominated by an intermediate clone carrying concomitant ASXL1, CSNK1A1, NF1, and EZH2 (N130T) mutations as well as del(5q) (Figure 2B). Molecular profiling of primary and xenografted FACS-purified BM fractions from 2013 revealed highly differential VAFs within these fractions. Erythroid (CD235a+) and lymphoid (CD19+) cells predominantly originated from a very early clone harboring a single ASXL1 mutation, specifically identifying del(5q) as a secondary event. In contrast, stem/progenitor (lin−CD34+38−) and myeloid cells (CD33+) displayed strong representation of a highly advanced clone harboring additional mutations in EZH2 (L617V) and ETV6, as well as UPD of chromosome 5q (Figure 2B). These findings enabled us to resolve the first 5 steps in clonal evolution of this patient’s MDS cells (Figure 2C), and revealed the coexistence of early and late clones that specifically contributed to distinct hematopoietic lineages.
Molecular dissection of mutational hierarchies using an integrative multisample screening approach. (A) Experimental setup and sorting strategies for dissection of mutational hierarchies for P15. (B) UDS-based profiling of somatic mutations for P15 in a BM sample obtained in 2010, various FACS-sorted subfractions from a BM sample from 2013, as well as xenografted erythroid progenitor cells revealed highly variable VAF patterns (top), representing differential oligoclonal contribution to these compartments (bottom). Cytogenetic lesions were quantified using microsatellite marker analysis. (C) Mutational hierarchy based on sequencing results in (B) showing the initial 5 steps of molecular evolution for P15. (D) Patient-derived xenotransplanted cells (PDX) from P11 showed the presence of an advanced MDS clone in hCD45+ cells resembling the patient’s primary BM but a TET2-only mutated early clone in engrafted hCD19+ cells. (E) Proportion of patients with detectable contribution of MDS-specific mutations to any lymphocytic compartment (primary or xenografted hCD19+ or hCD3+ cells), summarized in supplemental Table 2. Error bars indicate SD. FSC-A, forward scatter area; PI-A, propidium iodide area; PI neg, propidium iodide–negative.
Molecular dissection of mutational hierarchies using an integrative multisample screening approach. (A) Experimental setup and sorting strategies for dissection of mutational hierarchies for P15. (B) UDS-based profiling of somatic mutations for P15 in a BM sample obtained in 2010, various FACS-sorted subfractions from a BM sample from 2013, as well as xenografted erythroid progenitor cells revealed highly variable VAF patterns (top), representing differential oligoclonal contribution to these compartments (bottom). Cytogenetic lesions were quantified using microsatellite marker analysis. (C) Mutational hierarchy based on sequencing results in (B) showing the initial 5 steps of molecular evolution for P15. (D) Patient-derived xenotransplanted cells (PDX) from P11 showed the presence of an advanced MDS clone in hCD45+ cells resembling the patient’s primary BM but a TET2-only mutated early clone in engrafted hCD19+ cells. (E) Proportion of patients with detectable contribution of MDS-specific mutations to any lymphocytic compartment (primary or xenografted hCD19+ or hCD3+ cells), summarized in supplemental Table 2. Error bars indicate SD. FSC-A, forward scatter area; PI-A, propidium iodide area; PI neg, propidium iodide–negative.
Importantly, the use of nonhematopoietic MSCs as germ line controls allowed the discrimination of somatically acquired lesions at high fidelity and demonstrated the frequent observation of MDS-typical mutations in the lymphoid lineage (Figure 2D; supplemental Figure 2A-B). We ultimately detected somatically acquired mutations in primary and/or xenografted B and T lymphocytes from 65% of the patients analyzed (20/31) (Figure 2E; supplemental Table 3).
MDS-initiating “founder” mutations frequently affect RNA splicing and epigenetic modifier genes, whereas mutations in signaling and transcription regulator genes as well as cytogenetic lesions occur subsequently
Using the above-established approach, we successfully reconstructed mutational hierarchies for 41 out of 54 cases analyzed (76%) and deciphered the relative timing of at least 2 and up to 11 distinct mutational events (Figure 3; supplemental Figure 3). Additional data supporting the reported hierarchies is provided in supplemental Figure 4. Our results showed that “founder” lesions recurrently affected genes involved in the regulation of DNA methylation such as TET2, DNMT3A (n = 10), chromatin remodeling such as ASXL1 or EZH2 (n = 6), or RNA splicing (eg, SF3B1 or ZRSR2) (n = 6) (Figure 3). In 5 additional cases, 2 “founder” gene mutations were unresolvable from each other, but both clearly preceded the acquisition of other subclonal events (Figure 3, lower panel). In 3 patients, we identified mutated TP53 as an initiating event. Importantly, despite being enriched in “founder” clones, mutations in genes from our “founder” categories could also arise as subclonal events in a subset of individuals.
Reconstruction of mutational hierarchies in MDS reveals recurrent patterns of molecular evolution with distinct disease-initiating events. Individual mutational hierarchies for MDS patients (n = 30) show that candidate “founder” mutations are enriched in gene groups affecting DNA methylation (blue background), RNA splicing (yellow background), and chromatin remodeling (red background). A “founder” clone consisting of 2 mutated genes from these groups was detected in 5 patients (orange background). In addition, 3 cases with TP53 mutations as isolated “founder” lesion have been identified (lilac background). Each tree represents a single patient. Dashed lines indicate more than one possible (sub)clonal dependency. Below each tree, the earliest and latest known clinical disease status, as assessed using the WHO criteria, is indicated. For clinically nonprogressive cases, the single known disease status is depicted. Additional patients with resolved oligoclonal hierarchies are found in supplemental Figure 3. MPN, myeloproliferative neoplasm; RA, refractory anemia; RAEB-1, refractory anemia with excess blasts subtype 1; RARS, refractory anemia with ring sideroblasts; RARS-T, RARS with thrombocytopenia; RCMD, refractory cytopenia with multilineage dysplasia.
Reconstruction of mutational hierarchies in MDS reveals recurrent patterns of molecular evolution with distinct disease-initiating events. Individual mutational hierarchies for MDS patients (n = 30) show that candidate “founder” mutations are enriched in gene groups affecting DNA methylation (blue background), RNA splicing (yellow background), and chromatin remodeling (red background). A “founder” clone consisting of 2 mutated genes from these groups was detected in 5 patients (orange background). In addition, 3 cases with TP53 mutations as isolated “founder” lesion have been identified (lilac background). Each tree represents a single patient. Dashed lines indicate more than one possible (sub)clonal dependency. Below each tree, the earliest and latest known clinical disease status, as assessed using the WHO criteria, is indicated. For clinically nonprogressive cases, the single known disease status is depicted. Additional patients with resolved oligoclonal hierarchies are found in supplemental Figure 3. MPN, myeloproliferative neoplasm; RA, refractory anemia; RAEB-1, refractory anemia with excess blasts subtype 1; RARS, refractory anemia with ring sideroblasts; RARS-T, RARS with thrombocytopenia; RCMD, refractory cytopenia with multilineage dysplasia.
Mutations in genes involved in signaling cascades (eg, CBL, NF1) and transcription regulation (eg, RUNX1, ETV6) were detected in 27 (50%) patients and identified as unequivocal secondary events in 93% of these cases (25/27) (Figure 3; supplemental Figure 3). Apart from point and small INDEL mutations, 34 patients carried MDS typical cytogenetic lesions (eg, monosomy 7, trisomy 8, del(5q)). In 59% of these cases (20/34), including 15 cases with del(5q), such lesions occurred as secondary events or constituted minor independent clones (Figure 3; supplemental Figure 3). In 24% of cases (8/34), the clonal relationship of cytogenetic lesions was unresolvable (supplemental Figure 3B). Of note, only 18% cases (6/34) harbored cytogenetic lesions as potential “founder” events (supplemental Figure 3C).
Complex branched and independent evolution promotes dynamic clonal heterogeneity in MDS leading to escape from drug response
The above data demonstrated that cytogenetic aberrations appear predominantly as late events in MDS development. This implies that conventional cytogenetic diagnostic methodology, currently in wide use for molecular monitoring of MDS patients, bears the risk of registering advanced MDS clones but missing initiating or occult independent clones. To evaluate this in a clinical setting, we performed comprehensive molecular characterization of serially acquired MDS patient samples.
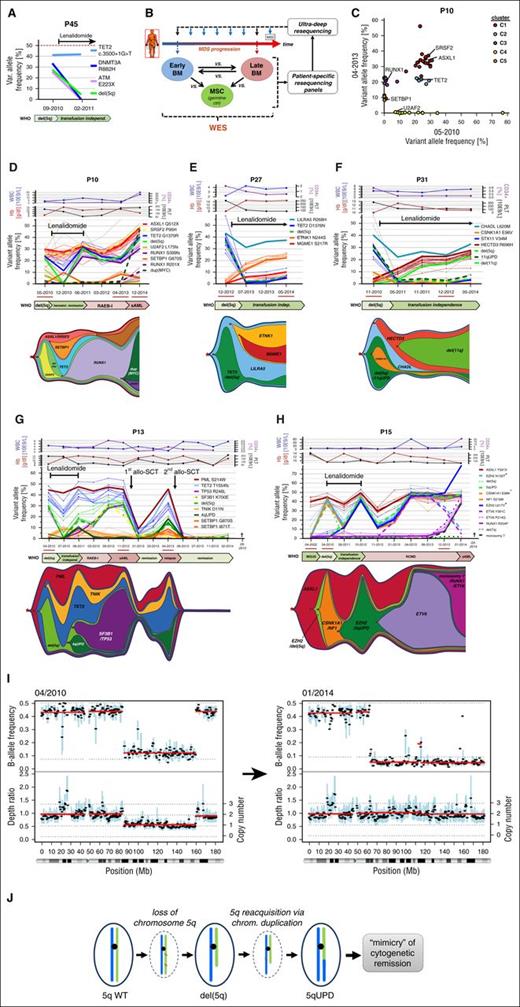
In a first case of a patient (P45), where 2 chronological samples were available, WES of the first sample revealed mutations in TET2, DNMT3A, and ATM as well as the presence of del(5q) (Figure 4A) with VAFs ranging between 25% and 41% (corresponding to 50% and 82% of affected diploid cells). Molecular analysis after 5 months of treatment with LEN revealed a substantial reduction of the later 3 molecular lesions but almost complete persistence of the candidate TET2 founding mutation (Figure 4A).
Branched evolution is a frequent route of clonal progression following treatment with LEN. (A) Mutational profiling of primary BM from P45 before and during LEN treatment reveals loss of subclonal lesions (DNMT3A/ATM/del(5q)) but stable maintenance of a mutation in TET2. (B) Schematic outline of an integrative NGS screening approach for unbiased evaluation of molecular disease evolution. (C) For 2 samples from P10 obtained before and after LEN treatment, validated mutations that have been requantified via UDS were bio-informatically separated into significant mutational subclusters with distinct MDS-specific driver lesions using the sciClone tool. (D-H) UDS-based profiling of somatically acquired mutations in P10 under LEN treatment and subsequent disease progression (D), and during long-term response under LEN therapy for P27 (E) and P31 (F) revealed complex branching evolution as illustrated in the graphical sketches. (G) Characterization of serial follow up BM aspirations from P13 during and after LEN treatment demonstrated linear acquisition of subclonal del(5q) and SF3B1 lesions during disease progression and emergence of an early PML/TET2-only “founder” clone after relapse from allo-SCT. (H) During 12 years of follow up, BM of P15 exhibited sequential acquisition of subclonal lesions including del(5q), which originated from an ASXL1-mutated “founder” clone that eventually gave rise to an independent branched subclone followed by clinical transformation into sAML and death of the patient. (Please note that loss of chromosome 7 carrying the EZH2 N130T-mutated allele results in decreasing VAF of this particular mutation and should not be mistaken with the disappearance of an early EZH2 N130T-mutated clone.) Mutations significantly associated with distinct clusters are equally colored. Depicted clinical parameters are WBC, Hb, PLT count, and proportion of CD34+ cells in BM quantified by FACS analysis. Samples that have been subjected to WES are underlined in red. Cytogenetic lesions throughout (D-H) were quantified via UDS-based SNP-skewing analysis. (I-J) Copy number and B-allele frequency profiling from WES data of P15 before (2010) and after (2014) LEN treatment (I), illustrates the transition of del(5q) into a copy number neutral UPD by duplication of the remaining nondeleted long arm of chromosome 5 (J). “+” indicates that mutational VAF was corrected for copy number bias. allo-SCT, allogeneic stem cell transplantation; chrom, chromosome; Hb, hemoglobin; LILRA5, leukocyte immunoglobulin-like receptor A5; PLT, platelet; vs, versus; WBC, white blood cell; WT, wild-type.
Branched evolution is a frequent route of clonal progression following treatment with LEN. (A) Mutational profiling of primary BM from P45 before and during LEN treatment reveals loss of subclonal lesions (DNMT3A/ATM/del(5q)) but stable maintenance of a mutation in TET2. (B) Schematic outline of an integrative NGS screening approach for unbiased evaluation of molecular disease evolution. (C) For 2 samples from P10 obtained before and after LEN treatment, validated mutations that have been requantified via UDS were bio-informatically separated into significant mutational subclusters with distinct MDS-specific driver lesions using the sciClone tool. (D-H) UDS-based profiling of somatically acquired mutations in P10 under LEN treatment and subsequent disease progression (D), and during long-term response under LEN therapy for P27 (E) and P31 (F) revealed complex branching evolution as illustrated in the graphical sketches. (G) Characterization of serial follow up BM aspirations from P13 during and after LEN treatment demonstrated linear acquisition of subclonal del(5q) and SF3B1 lesions during disease progression and emergence of an early PML/TET2-only “founder” clone after relapse from allo-SCT. (H) During 12 years of follow up, BM of P15 exhibited sequential acquisition of subclonal lesions including del(5q), which originated from an ASXL1-mutated “founder” clone that eventually gave rise to an independent branched subclone followed by clinical transformation into sAML and death of the patient. (Please note that loss of chromosome 7 carrying the EZH2 N130T-mutated allele results in decreasing VAF of this particular mutation and should not be mistaken with the disappearance of an early EZH2 N130T-mutated clone.) Mutations significantly associated with distinct clusters are equally colored. Depicted clinical parameters are WBC, Hb, PLT count, and proportion of CD34+ cells in BM quantified by FACS analysis. Samples that have been subjected to WES are underlined in red. Cytogenetic lesions throughout (D-H) were quantified via UDS-based SNP-skewing analysis. (I-J) Copy number and B-allele frequency profiling from WES data of P15 before (2010) and after (2014) LEN treatment (I), illustrates the transition of del(5q) into a copy number neutral UPD by duplication of the remaining nondeleted long arm of chromosome 5 (J). “+” indicates that mutational VAF was corrected for copy number bias. allo-SCT, allogeneic stem cell transplantation; chrom, chromosome; Hb, hemoglobin; LILRA5, leukocyte immunoglobulin-like receptor A5; PLT, platelet; vs, versus; WBC, white blood cell; WT, wild-type.
In order to profile clonal heterogeneity and its dynamic evolution over multiple interspaced chronological samples, we screened for candidate somatic lesions by performing at least 2 runs of WES on BM samples from an earliest possible vs a latest possible time point (Figure 4B) in 13 patients. Based on the somatically acquired mutations identified by WES, patient-individual UDS panels were designed to quantify allelic burdens of candidate mutations. Using this approach, we validated a median of 37 (range, 12-53) acquired mutations per patient (supplemental Figure 5C). Patient-specific UDS panels were then applied to interspaced chronologically acquired samples from up to 17 years of observation time (Figure 4B). Integrative bio-informatical cluster analysis using the sciClone tool21 revealed an average of 3.5 (range, 2-6) mutational clusters per patient, which were composed of both recurrent MDS driver and (putative) passenger mutations (Figure 4C).
By integrating mutational clusters of the individual patients with serial sampling, we detected a dynamic evolution of branching or independent clones in 9/13 (69%) patients (Figures 4 and 5; supplemental Figure 6). Upon LEN treatment, P10, P27, and P31 demonstrated a loss of initially dominant subclones carrying del(5q), as well as mutations in U2AF2 (P10), TET2 (P27), or CSNK1A1 and 11qUPD (P31) (Figure 4D-F). Despite significant hematologic improvement such as increased Hb levels and reduction of blast counts, these patients displayed rapid emergence of previously undetectable branching subclones with new aberrations subsequent to LEN treatment. In all cases, the new subclones originated from persistent hierarchically earlier clones. Of note, the “founder” clone of P31 (Figure 4F) was defined by an acquired mutation in the gene chondroadherin-like, which was predominant throughout all BM samples and expected to be highly damaging for the encoded protein, according to the prediction from PolyPhen-2 and SIFT software. Similarly, P27 (Figure 4E) was characterized by a founding mutation affecting a regulatory arginine residue in the transmembrane domain of LILRA5, which is strongly implicated in proinflammatory cytokine secretion and innate immune regulation. In P10 (Figure 4D), following LEN treatment, a new SETBP1 branched clone dynamically expanded and acquired additional molecular alterations that likely improved its competitiveness against the TET2/del(5q)/RUNX1-mutated branching subclone. The additionally acquired lesions in the SETBP1 clone were not known MDS typical candidates. However, in silico prediction indicated them as highly damaging to the function of the corresponding proteins (supplemental Table 4) and as such, they likely had a biological impact on the emergence of this previously minor subclone. Finally, a duplication of the MYC oncogene in the TET2/del(5q)/RUNX1-mutated clone eventually led to the re-emergence of this branched clone and clinically resulted in progression to secondary AML (sAML) (Figure 4D; supplemental Figure 7).
Changes in clonal composition in BM are reflected in clinical alterations and course of the disease. UDS-based VAF quantification of mutational evolution during clinical follow up: (A) For P03, LEN treatment induced hemotoxicity without a profound reduction in MDS clonal burden resulting in transfusion dependence after treatment discontinuation and outgrowth of a novel EZH2-mutated subclone shortly after temsirolimus therapy. (B) P20 acquires 3 distinct subclonal mutation clusters during a follow-up of 11 years, prior to treatment with LEN. This induces a massive molecular remission, accompanied by a hematotoxic reaction, which is reversed after withdrawal of LEN and followed by rapid reconstitution of the pretreatment clone. (C) P37 received 5-aza in a high-risk disease stage and achieved transfusion independence, which was accompanied by overall clonal reduction. However, upon treatment discontinuation, his MDS clone acquired a novel subclonal LZTR1 mutation coinciding with prolonged transfusion dependence. Reinitiating the 5-aza treatment resulted only in a minor molecular response of the new subclone and was quickly followed by the patient’s death. (D) P19 shows a stable disease with fully clonal BM and novel appearance of a branched del(ETV6) clone following treatment with panobinostat, which remains stable on a subclonal level. Without any specific treatment applied, a rapid, diverse subclonal expansion can be observed in P29 (E) and P30 (F) leading to disease progression and rapid death. Completely independent clones were found in P02 (G) and P40 (H). (G) P02 showed a highly stable coexistence of a CHRM2/del(5q) clone and a DNMT3A-mutated clone for 16 years. Upon LEN treatment, the CHRM2/del(5q) clone completely disappeared, whereas the DNMT3A-mutant clones immediately fully clonal expanded, which changed the clinical phenotype of the patient, currently receiving phlebotomy. (H) In P40, a SF3B1/TET2/DNMT3A/del(5q) clone was likewise completely outcompeted by an independent DNMT3A-mutant clone after LEN treatment, ultimately resulting in a fatal outcome. Cytogenetic lesions throughout (A-H) were quantified via UDS-based SNP-skewing analysis unless stated otherwise. “+” indicates that mutational VAF was corrected for copy number bias.
Changes in clonal composition in BM are reflected in clinical alterations and course of the disease. UDS-based VAF quantification of mutational evolution during clinical follow up: (A) For P03, LEN treatment induced hemotoxicity without a profound reduction in MDS clonal burden resulting in transfusion dependence after treatment discontinuation and outgrowth of a novel EZH2-mutated subclone shortly after temsirolimus therapy. (B) P20 acquires 3 distinct subclonal mutation clusters during a follow-up of 11 years, prior to treatment with LEN. This induces a massive molecular remission, accompanied by a hematotoxic reaction, which is reversed after withdrawal of LEN and followed by rapid reconstitution of the pretreatment clone. (C) P37 received 5-aza in a high-risk disease stage and achieved transfusion independence, which was accompanied by overall clonal reduction. However, upon treatment discontinuation, his MDS clone acquired a novel subclonal LZTR1 mutation coinciding with prolonged transfusion dependence. Reinitiating the 5-aza treatment resulted only in a minor molecular response of the new subclone and was quickly followed by the patient’s death. (D) P19 shows a stable disease with fully clonal BM and novel appearance of a branched del(ETV6) clone following treatment with panobinostat, which remains stable on a subclonal level. Without any specific treatment applied, a rapid, diverse subclonal expansion can be observed in P29 (E) and P30 (F) leading to disease progression and rapid death. Completely independent clones were found in P02 (G) and P40 (H). (G) P02 showed a highly stable coexistence of a CHRM2/del(5q) clone and a DNMT3A-mutated clone for 16 years. Upon LEN treatment, the CHRM2/del(5q) clone completely disappeared, whereas the DNMT3A-mutant clones immediately fully clonal expanded, which changed the clinical phenotype of the patient, currently receiving phlebotomy. (H) In P40, a SF3B1/TET2/DNMT3A/del(5q) clone was likewise completely outcompeted by an independent DNMT3A-mutant clone after LEN treatment, ultimately resulting in a fatal outcome. Cytogenetic lesions throughout (A-H) were quantified via UDS-based SNP-skewing analysis unless stated otherwise. “+” indicates that mutational VAF was corrected for copy number bias.
Rapid oligoclonal turnover could also be observed for 2 further patients undergoing LEN treatment. Although cases P13 and P15 (Figure 4G-H) exhibited cytogenetic remissions of their del(5q) bearing subclones, earlier founding clones carrying PML and TET2 (P13) or ASXL1-only (P15) strongly expanded in the BM. However, P13 lost drug response upon reappearance of the initial del(5q) subclone, with additionally acquired mutations in SF3B1 and TP53 that eventually dominated until disease progression to sAML (Figure 4G). Subsequently, the patient was subjected to an allogeneic stem cell transplantation from which he rapidly relapsed with a third branching clone carrying 4qUPD but not the previously described sAML clone (Figure 4G). Comparable observations were made in the case of P15 (Figure 4H). During the first 11 years, this patient exhibited sequential linear acquisition of mutations in genes such as EZH2, ETV6, and del(5q) (Figure 4H). Upon progression to sAML, the dominant ETV6-mutated subclone was fully substituted by an independent branching subclone carrying a monosomy 7, and novel mutations in ETV6 and RUNX1 (Figure 4H). In response to LEN treatment, the patient initially appeared to be in complete cytogenetic remission by classical cytogenetic analysis of his del(5q) aberration. However, WES-based allelokaryotyping of the BM before and after LEN treatment revealed that the remaining long-arm of chromosome 5 (carrying a mutated CSNK1A1 allele) was duplicated during therapy, which resulted in an acquired UPD (Figure 4I) mimicking cytogenetic remission (Figure 4J).
Although the above described patients showed good clinical responses in association with strong oligoclonal fluctuations under LEN therapy, 2 other cases lacked any hematologic improvement. In P03 (Figure 5A), LEN induced only a minor reduction of the overall MDS mutational burden. In the other case, LEN treatment of P20 (Figure 5B) resulted in a strong reduction of all MDS clones even though this patient did not carry the putative target lesion of LEN, del(5q). In both cases, the treatment had to be stopped due to hemotoxicity and in P20 the initial clone re-emerged quickly. Further follow up of P03 revealed that temsirolimus induced a molecular response of an early del(5q)/ASXL1-mutated clone, but simultaneously led to the emergence of an EZH2-mutated subclone that subsequently expanded after cessation of temsirolimus (Figure 5A).
In addition to patients treated with LEN, we characterized longitudinal samples from patients subjected to other drug regimens. For P37, we observed an overall reduction of the dominant clone’s burden during treatment with 5-azacytidine (5-aza) (Figure 5C). However, after a short treatment discontinuation, a novel clone carrying an LZTR1 mutation appeared (Figure 5C). Although not typical in MDS, LZTR1 was previously reported to be mutated in glioblastoma.22 Despite this increasingly complex clonal BM composition, the patient’s clinical symptomatology initially improved, which coincided with an overall reduction of the MDS clonal burden. However, this was later reversed by re-expansion of the earlier clone with subsequent fatal outcome. P19 was a case of MDS RARS, observed for almost 4 years, and in part treated with panobinostat and an experimental anti-CD95 antibody (APG-101) within a phase 1 clinical trial (Figure 5D). Although this patient remained clinically and molecularly relatively stable during the observation time, we observed the emergence of a minor branching subclone with an ETV6 deletion following treatment with panobinostat (Figure 5D).
Despite seemingly elevated clonal fluctuations induced by drug treatment, rapid molecular evolution accompanied by clinical progression could also be observed in cases without any therapeutic intervention. P29 initially presented with a highly advanced clone carrying 4 different recurrent MDS mutations, all of which originated from our previously defined “founder” gene category (Figure 5E). This clone rapidly evolved and acquired additional mutations in BCOR, STAG2, and RUNX1, directly followed by disease progression from RCMD to RAEB-1 and subsequent death. Similarly, P30 was a case observed over 8 months with rapid progression to sAML without any drug treatment. The molecular follow-up of this patient revealed the outgrowth of a novel branching del(5q) subclone that outcompeted the preexisting NF1-mutated subclone (Figure 5F).
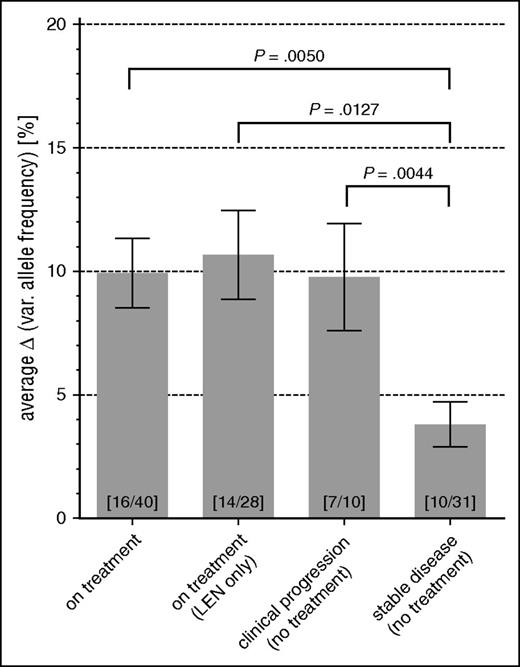
The strongest shifts of clonal composition under therapy were detectable in the cases P40 and P02: although P40 initially presented with a dominant clone carrying del(5q), SF3B1, TET2, and DNMT3A, BM biopsies of P02 collected over a period of 15 years displayed stable VAFs of several lesions such as del(5q) (Figure 5G-H). These were almost undetectable in the posttreatment biopsy after the beginning of LEN treatment. However, new and completely independent clones carrying DNMT3A mutations simultaneously expanded in these patients (Figure 5G-H) and an additional case (P52) that was screened using UDS (supplemental Figure 8F). In these cases, we could not identify a single potential “founder” lesion shared between the new arising DNMT3A-positive clones and the initially discovered del(5q)-bearing clones. Despite such severe clonal fluctuations and persistent clonal BM involvement, all 3 patients achieved significant hematologic responses after LEN treatment. Additional patients’ follow-up samples, which were investigated by UDS screening for every time point available, further confirmed the observed patterns of clonal fluctuations (supplemental Figure 8). Importantly, by integrating the degree of VAF changes with the clinical status between any two time points for all patients, we could readily demonstrate a significant increase of VAF fluctuations during treatment (2.6-fold, P = .005) or clinical progression (2.6-fold, P = .0044) as compared with clinically stable periods (Figure 6).
Changes in VAF are significantly increased during treatment and clinical change as compared with stable disease. For any 2 consecutive aspirations in patient follow ups, their median ΔVAF was calculated from all identified somatic mutations. Bars depict mean and standard error of calculated median ΔVAFs, and are grouped according to the clinical status (eg, clinical change in WHO status, LEN treatment) during the period between the 2 aspirations. The total numbers of patients and data points contributing to each specific group are indicated at the bottom of each bar (patients/data points).
Changes in VAF are significantly increased during treatment and clinical change as compared with stable disease. For any 2 consecutive aspirations in patient follow ups, their median ΔVAF was calculated from all identified somatic mutations. Bars depict mean and standard error of calculated median ΔVAFs, and are grouped according to the clinical status (eg, clinical change in WHO status, LEN treatment) during the period between the 2 aspirations. The total numbers of patients and data points contributing to each specific group are indicated at the bottom of each bar (patients/data points).
To our knowledge, this study represents the first systematic comprehensive data set providing high throughput molecular monitoring of long-term serial follow-ups in a significant (n = 22) cohort of patients. Importantly, this cohort includes 14 LEN-treated MDS patients. In almost all cases, LEN induced an effective reduction of cell clones carrying the controlling feature del(5q). However, it frequently did not induce complete molecular remission of all clones carrying MDS typical mutations. After LEN treatment, the BM of these patients was still infiltrated or substituted by clones carrying MDS typical founder lesions, such as TET2, ASXL1, and DNMT3A, or even gene mutations that were not previously implicated in MDS pathogenesis. Retention of such early “founder” or independent clones may represent a reservoir of “primed cells” that can further evolve to give rise to pathogenic clones that could ultimately drive relapse, underlining the importance of their early detection for clinical stratification.
Finally, we analyzed the influence of branching vs linear evolutionary histories on overall survival. At this stage, this analysis did not yield significant differences, which is likely due to the sample size necessary for this type of analysis in particular, in the context of a tremendously heterogeneous disease such as MDS (data not shown). However, novel mutations after drug therapy frequently affected genes that we previously defined as hierarchically late in MDS, underlining the importance of their early detection for prediction of disease progression.
Discussion
Cross-sectional studies investigating single time point BM specimens from MDS patients had indicated mutations in genes involved in the splicing machinery and epigenetic modification as relatively early events in MDS6,7 but did not succeed in isolating single “founder” events on an MDS patient-specific level. Here, we demonstrate on a patient-individual basis that mutations affecting epigenetic modifiers (eg, TET2, ASXL1) and RNA splicing factors (eg, SF3B1, SRSF2) are predominantly “founder” events in MDS. Likewise, mutations of, for example, DNMT3A and TET2, have recently been suggested as preleukemic events in the development of AML23-27 and homozygous knock out of DNMT3A in a mouse model has been shown to predispose to a wide range of hematologic malignancies.28 In line with this, mice with conditional inactivation of ASXL1,29 TET2,30 or knock-in of mutated SRSF2,31 closely resembled features of human MDS.
Our data further demonstrated that genes involved in signaling cascades (eg, JAK2 and CBL), transcription factors (eg, RUNX1 and ETV6), and cytogenetic lesions (eg, monosomy 7, trisomy 8, and del(5q)) were almost exclusively acquired as late events in MDS, emphasizing their potential use as indicators of disease progression. Among patients carrying a cytogenetic lesion, our cohort contained a total of 28 del(5q)-bearing cases, of which 21 were classified as isolated del(5q) according to the World Health Organization (WHO). Our thorough molecular analysis demonstrated that del(5q) was acquired as an unequivocal secondary lesion, or constituted a minor independent clone in 54% (n = 15/28) of all del(5q)-bearing cases and 62% (n = 13/21) of patients classified as “isolated del(5q).” This is in stark contrast to a recent report proposing del(5q) as the initiating lesion in isolated del(5q) patients.32 In our cohort, del(5q) appears to be a potential “founder” event in only a minority (21.4%) of the cases (n = 6/28), whereas its VAF is comparable to other key MDS “founder” lesions in 28.5% (n = 7/28) of the del(5q) cases, making it impossible to decipher their order of acquisition. These discrepancies could possibly result from the smaller number of del(5q)-bearing cases analyzed in the Woll study32 (n = 15), the use of limited NGS gene panels, or most likely due to the lack of appropriate germ line controls for the majority of patients in the former study. Most importantly, both studies clearly demonstrate that a “founder” lesion in 1 patient can appear as a secondary hit in another one, thereby highlighting the importance of “patient-specific” molecular characterization to guide therapeutic decision.
Recent studies revealed that ASXL1, TET2, and DNMT3A but not RNA splicing gene mutations were common in hematopoietic cells from healthy elderly people12-15 and proposed the existence of a pre-disease state characterized by clonal hematopoiesis of indeterminate potential (CHIP).33 Importantly, our data also suggest that MDS likely develop following the acquisition of “founder” genetic events in patients diagnosed with CHIP. Overall, the striking overlap of “founder” events identified in MDS and other hematologic neoplasia34 might suggest a common CHIP-like phase of preleukemic initiation followed by more confined diversification through the acquisition of disease-specific lesions such as del(5q) in MDS.
Our ability to identify individual mutational hierarchies is a necessary step toward personalized therapy. Such knowledge could guide patient-specific drug targeting of particular clones with the aim to either: (1) eradicate MDS by targeting the “founder” lesion thereby affecting all subclones, or (2) eradicate advanced clones allowing for hematopoietic reconstitution and hematologic improvement through the establishment of a CHIP phenotype driven by the founder clones. Therefore, it is necessary to evaluate the likelihood that the patient marrow still contains residual lesion-free HSCs to ensure hematopoietic reconstitution posttreatment. Alternatively, the maintenance of a CHIP phenotype in an MDS patient might represent the best clinical course for a subset of patients with high clonal involvement. Our finding that “founder” mutations are often observed in lymphoid cells of MDS patients argues in favor of maintenance of multilineage potential in these “primed” founder clones, at least in a subset of patients.
Conceptually, the eradication of “founder” clones has already been shown to be an effective strategy for disease control in an AML mouse model for IDH2,35 and clinical trials evaluating the efficacy of IDH and SF3B1 inhibitors are currently underway.36 However, this “founder” targeting approach might be compromised by the emergence of subclonal mutations maintaining independent self-renewal, which would require combinatorial therapeutic intervention.
Previous studies investigating del(5q) and TET2 mutations identified rare cases of lymphocytic involvement.37-39 Our exhaustive analysis of a large cohort of 31 MDS patients in combination with our detection of mutated B cells in long-term xenoengrafted MDS hematopoiesis now provides definitive proof that genetically defined MDS HSC clones frequently contribute to lymphopoiesis. These findings raise important questions about the potential influence of intrinsically aberrant lymphopoiesis on the pathogenesis of MDS. Importantly, our identification of somatically mutated lymphocytes was only made possible thanks to the use of nonhematopoietic MSCs as germ line controls. Critically, the usage of lymphocytes as a germ line control, particularly in the context of NGS studies, likely hampers the ability to detect important MDS-associated lesions, thus providing an incomplete overview of the mutational make-up.
In recent years, our understanding of mutational landscapes in MDS greatly improved6,7,39-41 with studies suggesting linear subclonal evolution accompanied by continuous acquisition of molecular lesions as the putative main driver of disease progression.41,42 However, the influence of drug treatment on the molecular plasticity of oligoclonal hierarchies remained poorly understood. WES and targeted deep-sequencing at multiple time points of MDS evolution allowed us to precisely resolve the molecular dynamics during drug treatment and progression (Figure 4B). Our data confirmed the existence of a linear path of clonal evolution, but importantly, also uncovered previously unrecognized branching evolution leading to the emergence of genetically advanced MDS subclones during disease progression that are capable of mediating drug resistance. Although recently reported in the context of single MDS/CMML cases and other tumor entities,32-39,43-49 the observed frequency of this phenomenon was largely unrecognized in MDS so far. Moreover, we observed that treatment with drugs such as LEN, 5-aza, or temsirolimus was immediately followed by significantly elevated fluctuations in both the oligoclonal BM composition and clinical parameters. Although this may suggest a higher vulnerability for transformation, the initial clinical consequences of such therapies were either positive or at worst neutral, ie, patients became transfusion independent and cell counts improved. Nevertheless, in most cases, the BM remained largely clonal due to the rapid outgrowth of founder- or initially minor subclones (eg, P10 and P31) or the emergence of resistant clones carrying additional mutations that significantly expanded under active therapy (eg, P03 and P15).
In conclusion, the emergence and disappearance of specific clones in the BM frequently correlated with changes of clinical features in PB, such as Hb and platelet levels, highlighting their distinct and pervasive functional properties. Similarly, increasingly complex mutational hierarchies were frequently associated with phenotypic disease progression, such as transition into worse WHO MDS categories. Importantly, patient death was often conjunct with the rapid emergence of genetically highly progressive clones carrying additional genetic lesions. Therefore, whereas the increasing repertoire of new therapeutic options has led to a tremendous improvement for MDS patients, it is also highly warranted to identify those patients who would benefit from a “watch and wait” strategy that maintains a clonal equilibrium with residual functionality, as previously suggested for chronic lymphocytic leukemia.47
In most patients, subclones appeared to originate from preexisting earlier clones, which were largely insensitive to treatment. In contrast, for P02, P40, and P52, an unequivocal “founder” lesion could not be identified, suggesting that LEN treatment resulted in the outgrowth of independent and novel MDS clones in these patients. Moreover, only WES-based mutation screening allowed the identification of several nonrecurrent lesions that uniquely characterized initiating but also subclones that lacked any known MDS typical mutations. In summary, our results strongly emphasize the critical need for unbiased mutational monitoring as deep and broad as possible during regular staging. This will undoubtedly enhance the ability of clinicians to precisely distinguish the disappearance of advanced subclones (eg, carrying del(5q) and RUNX1) from true molecular remission in which all clones (including early “founder” clones) are below detection threshold, and most importantly, allow for the early detection of more advanced subclones to anticipate disease progression and guide clinical decisions.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Verena Haselmann and Romy Eichner (Institute of Clinical Chemistry, Mannheim, Germany) for providing their pyrosequencing device.
This work was supported by funds from the National Center for Tumor Diseases (Heidelberg, Germany), the Deutsche Krebshilfe within a German-wide MDS collaborative initiative (Deutschlandweites MDS Verbundprojekt, Teilprojekt I), the Gutermuth Foundation, the ZOBEL initiative within the Innovation Fund for Medicine Baden-Wuerttemberg, the H. W. and J. Hector fund (Baden-Wuerttemberg), the Dietmar Hopp Foundation, the SFB 873 funded by the Deutsche Forschungsgemeinschaft, the European Research Council (#639795) (H.M.), and a research grant from the Celgene Corporation. Fellowships were provided by the Studienstiftung des Deutschen Volkes (J.-C.J.) and by the Deutsche José Carreras Leukämie Stiftung (F 11/05–Young Investigator Fellowship [M. Mossner] and A 14/01–Career Development Award [H.M.]).
Authorship
Contribution: M. Mossner, J.-C.J., W.-K.H., H.M., and D.N. designed the study, analyzed data, and wrote the manuscript; M. Mossner and J.-C.J. conducted the experimental design, executed bio-informatic analyses and most of the experiments; H.M. performed mouse experiments and FACS sorting; A.T. and W.-K.H. supervised the whole study and provided research infrastructure; C.H. provided FISH analyses; S.H.W., H.D., H.B., and M. Meggendorfer provided NGS resources and data; J.W., S.F., V.N., J.O., J.P., I.P., C.X., and C.K. provided assistance for molecular analyses, primary MSC expansion, and mouse procedures; S.S. performed FACS sorting; A.F. performed cytogenetic analyses; P.G. contributed mathematical calculations of NGS-derived copy number data; F.N., T.B., G.M., U.P., E.B., T.J.S., H.R., and D.N. provided patient material and clinical data.
Conflict-of-interest disclosure: M. Meggendorfer is employed by the MLL Munich Leukemia Laboratory GmbH. C.H. declares part ownership of the MLL Munich Leukemia Laboratory GmbH. The remaining authors declare no competing financial interests.
Correspondence: Daniel Nowak, Department of Hematology and Oncology, Medical Faculty Mannheim, University of Heidelberg, Pettenkoferstr 22, 68169 Mannheim, Germany; e-mail: daniel.nowak@medma.uni-heidelberg.de; and Hind Medyouf, Georg-Speyer-Haus Institute for Tumor Biology and Experimental Therapy, Paul-Ehrlich-Str 42-44, 60596 Frankfurt, Germany; e-mail: medyouf@gsh.uni-frankfurt.de.
References
Author notes
M. Mossner and J.-C.J. contributed equally to this study.
H.M. and D.N. contributed equally to this study.