Key Points
VAV1 rearrangements are recurrent in PTCLs, drive tumor cell growth, and can be targeted by clinically available RAC1 inhibitors.
Integrated mate-pair/RNA sequencing of PTCLs identifies expressed fusion transcripts that represent candidate therapeutic targets.
Abstract
Peripheral T-cell lymphomas (PTCLs) represent a heterogeneous group of T-cell malignancies that generally demonstrate aggressive clinical behavior, often are refractory to standard therapy, and remain significantly understudied. The most common World Health Organization subtype is PTCL, not otherwise specified (NOS), essentially a “wastebasket” category because of inadequate understanding to assign cases to a more specific diagnostic entity. Identification of novel fusion genes has contributed significantly to improving the classification, biologic understanding, and therapeutic targeting of PTCLs. Here, we integrated mate-pair DNA and RNA next-generation sequencing to identify chromosomal rearrangements encoding expressed fusion transcripts in PTCL, NOS. Two of 11 cases had novel fusions involving VAV1, encoding a truncated form of the VAV1 guanine nucleotide exchange factor important in T-cell receptor signaling. Fluorescence in situ hybridization studies identified VAV1 rearrangements in 10 of 148 PTCLs (7%). These were observed exclusively in PTCL, NOS (11%) and anaplastic large cell lymphoma (11%). In vitro, ectopic expression of a VAV1 fusion promoted cell growth and migration in a RAC1-dependent manner. This growth was inhibited by azathioprine, a clinically available RAC1 inhibitor. We also identified novel kinase gene fusions, ITK-FER and IKZF2-ERBB4, as candidate therapeutic targets that show similarities to known recurrent oncogenic ITK-SYK fusions and ERBB4 transcript variants in PTCLs, respectively. Additional novel and potentially clinically relevant fusions also were discovered. Together, these findings identify VAV1 fusions as recurrent and targetable events in PTCLs and highlight the potential for clinical sequencing to guide individualized therapy approaches for this group of aggressive malignancies.
Introduction
Peripheral T-cell lymphomas (PTCLs) represent a heterogeneous group of non-Hodgkin lymphomas (NHLs) of mature T-cell origin with poor prognosis.1 The most common subtype of PTCL in Western countries is PTCL, not otherwise specified (NOS), which has 5-year overall and failure-free survival rates of 32% and 20%, respectively.2 Although less common than B-cell NHLs, the incidence of PTCL, NOS and PTCLs in general has been increasing steadily in the United States in recent years.3 By definition, PTCL, NOS includes cases failing to meet criteria for a more specific PTCL subtype and has been referred to as a “wastebasket” category.4,5 Therefore, PTCL, NOS is markedly heterogeneous, and a major goal of PTCL research is to identify molecular abnormalities that improve classification and identify candidate therapeutic targets.6
Recurrent chromosomal rearrangements giving rise to expressed fusion transcripts play a key role in the pathogenesis and clinical management of hematologic malignancies.7 Among PTCLs, rearrangements of the tyrosine kinase gene, ALK, have diagnostic, prognostic, and therapeutic significance in ALK-positive anaplastic large cell lymphoma (ALCL).8-10 In addition, rearrangements producing fusion genes involving TP63, ROS1, and TYK2 recently have been identified in ALCLs that are negative for ALK.11-15 However, beyond rare ITK-SYK fusions,16 the contribution of gene fusions to the pathogenesis of PTCL, NOS remains poorly understood.
We previously have shown the efficacy of mate-pair DNA sequencing (MPseq) to identify chromosomal rearrangements in PTCLs and other human cancers.12,17,18 MPseq utilizes ligation of end-labeled DNA fragments followed by refragmentation and affinity purification to generate libraries of fragments containing 2 DNA sequences originally separated by a genomic distance of several kilobases. This technique has superb sensitivity for detecting rearrangements across the entire genome at a fraction of the cost of whole-genome sequencing; however, not all chromosomal rearrangements identified by MPseq involve named genes or give rise to expressed fusion transcripts. Conversely, RNA sequencing (RNAseq) preferentially identifies expressed fusion transcripts, but false positives remain a challenge due to difficulties mapping RNA reads to the genome.19 Taking advantage of the strengths of both approaches and simultaneously providing orthogonal validation of the results, we integrated MPseq and RNAseq data to identify expressed fusion transcripts in 11 cases of PTCL, NOS. We then performed further validation, functional studies, and assessment in additional PTCL tissue samples to explore potential clinical implications of selected findings.
Materials and methods
Patients and clinical samples
Eleven cases of PTCL, NOS were analyzed by integrating data from MPseq (including 2 patients reported previously12 ) and RNAseq. These cases were selected based on availability of sufficient frozen material for sequencing studies as outlined below. Data on biopsy site and treatment status at time of biopsy are presented in supplemental Table 1, available on the Blood Web site. Additional PTCLs (n = 137) were evaluated by fluorescence in situ hybridization (FISH) for VAV1 rearrangements. The study was approved by Mayo Clinic and University of Iowa Institutional Review Boards.
Sequencing and bioinformatics
MP libraries were prepared from genomic DNA extracted from frozen PTCL samples using the Mate Pair Library Prep Kit (Illumina) and sequenced on a HiSeq2000 (Illumina) as previously published.12,17 Reads were mapped to the human genome (GRCh38/hg38) using BIMA-V3 as published.18 Candidate events were eliminated if they had mapping scores that did not meet filtering criteria; were intrachromosomal events spanning <30 000 bp; were supported by ≤3 fragments; or were contained within a mask table including events occurring in noncancerous samples. RNAseq was performed on RNA extracted from frozen samples and chimeric transcripts were identified using SnowShoes-FTD as published.12,19
FISH
A breakapart FISH probe flanking the VAV1 locus on 19p13.3 was developed by labeling bacterial artificial chromosomes with SpectrumOrange (telomeric/5′: RP11-134L9, RP11-479K19, RP11-828J24, RP11-114A7) or SpectrumGreen (centromeric/3′: CTD-2596O14, CTD-2564J11, RP11-876D1), and interphase FISH was performed and scored on paraffin tissue sections of PTCLs as previously described.12,17,20 A normal intact VAV1 gene region is indicated by a yellow signal. Disruption of the VAV1 gene region is indicated by separation of the yellow signal into one red (5′) signal and 1 green (3′) signal. Two-sided χ2 tests were used to evaluate subtype specificity.
Cell lines, transfection, and inhibitor treatment
Jurkat cells (ATCC) were maintained in RPMI, and NIH-3T3 and HEK-293T (ATCC) were maintained in Dulbecco’s modified Eagle medium, all containing 10% fetal bovine serum (Clontech) and 1% penicillin/streptomycin (Gibco). They were transfected using electroporation (Jurkat) or Lipofectamine 3000 (Life Technologies; NIH-3T3 and HEK-293T). Functional experiments were performed after 24 to 72 hours. RAC1 (Dharmacon) or control (AllStars; Qiagen) small interfering RNAs were transfected by electroporation, and cell growth was assessed at 72 hours. Cells were treated with azathioprine (Selleck) for 72 hours.
Expression plasmids
VAV1-GSS was cloned from TCL26 cDNA into TOPO-TA (Invitrogen) using primers 5′-GGCGACAGTTACAGGCAAAGAAG-3′ and 5′-TAGGAGAGGAATGACAAATACAGAGG-3′, sequenced verified, and cloned into pCI (Promega) using primers 5′-CTGTATGACTGCGTGGAGAATG-3′ and 5′-TAGGAGAGGAATGACAAATACAGAGG-3′. ITK-FER was cloned into TOPO-TA from TCL72 cDNA using primers 5′-ATGAACAACTTTATCCTCCTGG-3′ and 5′-AATAATGTTACTCTGCTGGAGG-3′ and then shuttled into pcDNA3.1(+) (Invitrogen) using BamHI and EcoRV restriction enzyme digestion. Human pCI2-VAV1 was from D.D.B.
Immunofluorescence and immunoblotting
NIH-3T3 cells were transfected and replated on acid-washed coverslips after 24 hours. At 48 hours after transfection, cells were fixed, permeabilized, and stained for actin (tetramethylrhodamine-phalloidin) and VAV1 (Abcam ab97574 and goat anti-rabbit Alexa-Fluor-488). Cells were mounted using Vectashield with 4′,6-diamidino-2-phenylindole (Vector Laboratories). Fluorescence micrographs (40×) were captured using iVision (BioVision) and processed using Adobe Photoshop. Circularity was calculated using ImageJ (National Institutes of Health) with a value of 1 indicating a perfect circle.
Immunoblotting was performed as previously described21 using antibodies for pVAV1(Y174) (Abcam; EP510Y), β-actin (Novus; AC-15), VAV1 (Cell Signaling; #2502), and RAC1 (Millipore; clone 23A8). Proteins were detected using IRDye 800CW and 680RD antibodies (LI-COR) on the LI-COR Odyssey CLx. Rac activity was assessed using GST-PAK1-PBD (p21-binding domain) pulldown as described previously.22
Functional assays
Cell migration was quantified by fluorometric assay 48 hours after transfection. Jurkat cells were labeled with 2 µM Calcein AM (Molecular Probes) for 45 minutes at 37°C in serum-free RPMI. Cells were then washed, resuspended in serum-free RPMI, and added to 5-µm inserts (Costar) equilibrated with RPMI containing 10% serum. Cells were allowed to migrate for 3 hurs at 37°C and 5% CO2. Migrated cells then were harvested and fluorescence was measured using a CytoFluor 4000 plate reader (Applied Biosystems). Percent of cells migrated was calculated by dividing the fluorescence of the migrated cell suspension by that of the starting cell suspension.
Colony formation was performed by seeding 200 cells per well of a 6-well plate at 24 hours after transfection and incubating for 8 days. Plates were imaged and counted with AlphaImager HP (ProteinSimple). Cell growth was measured by plating at 10 000 cells per well in a 96-well plate, treating for 48 to 72 hours, and incubating with 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS; Promega) for 2 to 4 hours. Absorbance was measured at 490 nm. Significance was evaluated using a Student t test.
Results
Integrated MPseq/RNAseq identifies novel expressed fusion transcripts in PTCL, NOS
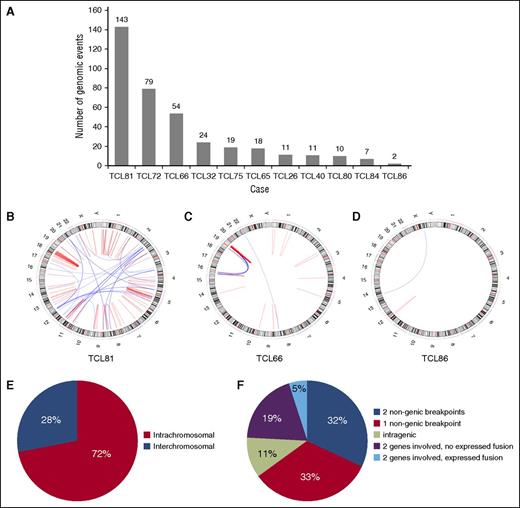
Eleven cases of PTCL, NOS were analyzed by integrated MP/RNA-sequation (9 men and 2 women; median age, 62 years; range, 48-75 years; Table 1). The number of genomic events identified in each case by MPseq varied from 2 to 143 (378 total events; mean/case ± standard deviation, 34 ± 43; Figures 1A-D). Seventy-two percent of events were intrachromosomal (range/case, 42-100%; Figure 1E). One case (TCL66) showed evidence of catastrophic chromosomal rearrangement, or chromothripsis,23 involving chromosomes 16 and 19 (Figure 1C). Sixty-five percent of events involved 1 to 2 nongenic regions (Figure 1F). An additional 11% of events were intragenic. The remaining 24% of events (n = 90) each involved 2 named genes.
Expressed fusion transcripts identified by integrated MPseq/RNAseq in PTCL, NOS
| Case . | Age (years) . | Sex . | Diagnosis . | DNA breakpoints . | Fusion transcript(s) . |
|---|---|---|---|---|---|
| TCL26 | 70 | Male | PTCL, NOS | 19p13.3, 20q11.22 | VAV1 ↔ GSS |
| TCL32 | 72 | Male | PTCL, NOS | None | None |
| TCL40 | 50 | Female | PTCL, NOS | None | None |
| TCL65 | 61 | Male | PTCL, NOS | 3q28, 6p22.3 | ATXN1 → TP63 |
| 1p36.23, 11q24.1 | CRTAM ↔ TNFRSF9 | ||||
| TCL66 | 59 | Male | PTCL, NOS | 16p13.3, 19q13.33 | BAX → TCEB2 |
| 19q13.2, 19q13.33 | GSK3A → ZNF541 | ||||
| 19q13.12, 19q13.43 | LOC100128398 → THAP8 | ||||
| TCL72 | 48 | Male | PTCL, NOS | 5q33.1, 5q33.1 | ANXA6 → RBM22 |
| 19p13.2, 19p13.2 | CARM1 → ZNF627 | ||||
| 1p33, 1p35.3 | PPP1R8 → EFCAB14 | ||||
| 5q21.3, 5q33.3 | ITK → FER | ||||
| 1p35.2, 16q22.3 | PUM1 → ZFHX3 | ||||
| 4p16.3, 4p16.3 | STX18 → ZNF718 | ||||
| TCL75 | 67 | Male | PTCL, NOS | 2q34, 2q34 | IKZF2 → ERBB4 |
| 2q12.3, 2q13 | RANBP2 → LIMS1 | ||||
| TCL80 | 56 | Male | PTCL, NOS | 19p13.2, 19p13.3 | VAV1 ↔ MYO1F |
| TCL81 | 64 | Female | PTCL, NOS | 14q32.13, 14q32.2 | ATG2B → CLMN |
| 1p13.1, 1q23.2 | COPA → CD58 | ||||
| 1p33, 1p34.2 | SZT2 → EFCAB14 | ||||
| TCL84 | 62 | Male | PTCL, NOS | None | None |
| TCL86 | 75 | Male | PTCL, NOS | None | None |
| Case . | Age (years) . | Sex . | Diagnosis . | DNA breakpoints . | Fusion transcript(s) . |
|---|---|---|---|---|---|
| TCL26 | 70 | Male | PTCL, NOS | 19p13.3, 20q11.22 | VAV1 ↔ GSS |
| TCL32 | 72 | Male | PTCL, NOS | None | None |
| TCL40 | 50 | Female | PTCL, NOS | None | None |
| TCL65 | 61 | Male | PTCL, NOS | 3q28, 6p22.3 | ATXN1 → TP63 |
| 1p36.23, 11q24.1 | CRTAM ↔ TNFRSF9 | ||||
| TCL66 | 59 | Male | PTCL, NOS | 16p13.3, 19q13.33 | BAX → TCEB2 |
| 19q13.2, 19q13.33 | GSK3A → ZNF541 | ||||
| 19q13.12, 19q13.43 | LOC100128398 → THAP8 | ||||
| TCL72 | 48 | Male | PTCL, NOS | 5q33.1, 5q33.1 | ANXA6 → RBM22 |
| 19p13.2, 19p13.2 | CARM1 → ZNF627 | ||||
| 1p33, 1p35.3 | PPP1R8 → EFCAB14 | ||||
| 5q21.3, 5q33.3 | ITK → FER | ||||
| 1p35.2, 16q22.3 | PUM1 → ZFHX3 | ||||
| 4p16.3, 4p16.3 | STX18 → ZNF718 | ||||
| TCL75 | 67 | Male | PTCL, NOS | 2q34, 2q34 | IKZF2 → ERBB4 |
| 2q12.3, 2q13 | RANBP2 → LIMS1 | ||||
| TCL80 | 56 | Male | PTCL, NOS | 19p13.2, 19p13.3 | VAV1 ↔ MYO1F |
| TCL81 | 64 | Female | PTCL, NOS | 14q32.13, 14q32.2 | ATG2B → CLMN |
| 1p13.1, 1q23.2 | COPA → CD58 | ||||
| 1p33, 1p34.2 | SZT2 → EFCAB14 | ||||
| TCL84 | 62 | Male | PTCL, NOS | None | None |
| TCL86 | 75 | Male | PTCL, NOS | None | None |
→, gene direction of fusion transcript; ↔, reciprocal fusion transcript also present.
Distribution of 378 genomic events identified by mate-pair sequencing in 11 cases of PTCL, NOS. (A) Number of genomic events per case. (B-D) Representative Circos diagrams illustrating heterogeneity in the degree of complexity of genomic events among PTCLs: (B) high complexity; (C) focally high complexity with multiple rearrangements involving chromosomes 16 and 19 (chromothripsis); and (D) low complexity. (E) Distribution of intrachromosomal and interchromosomal events. (F) Distribution of events based on involvement of genes or nongenic regions and identification of expression fusion transcripts by RNA sequencing.
Distribution of 378 genomic events identified by mate-pair sequencing in 11 cases of PTCL, NOS. (A) Number of genomic events per case. (B-D) Representative Circos diagrams illustrating heterogeneity in the degree of complexity of genomic events among PTCLs: (B) high complexity; (C) focally high complexity with multiple rearrangements involving chromosomes 16 and 19 (chromothripsis); and (D) low complexity. (E) Distribution of intrachromosomal and interchromosomal events. (F) Distribution of events based on involvement of genes or nongenic regions and identification of expression fusion transcripts by RNA sequencing.
Among the 90 events involving 2 genes, RNAseq identified expressed fusion transcripts in 18 (mean, 1.6 fusions/case; range, 0–6 fusions/case; Table 1); 13 (72%) were derived from intrachromosomal events, identical to the proportion among all DNA events. Three DNA events yielded reciprocal fusion transcripts. All gene pairs were unique and novel except ATXN1→TP63 as previously reported.12 Two genes, VAV1 and EFCAB14, were involved in fusions in >1 case.
VAV1 rearrangements are recurrent events in PTCLs and show subtype specificity
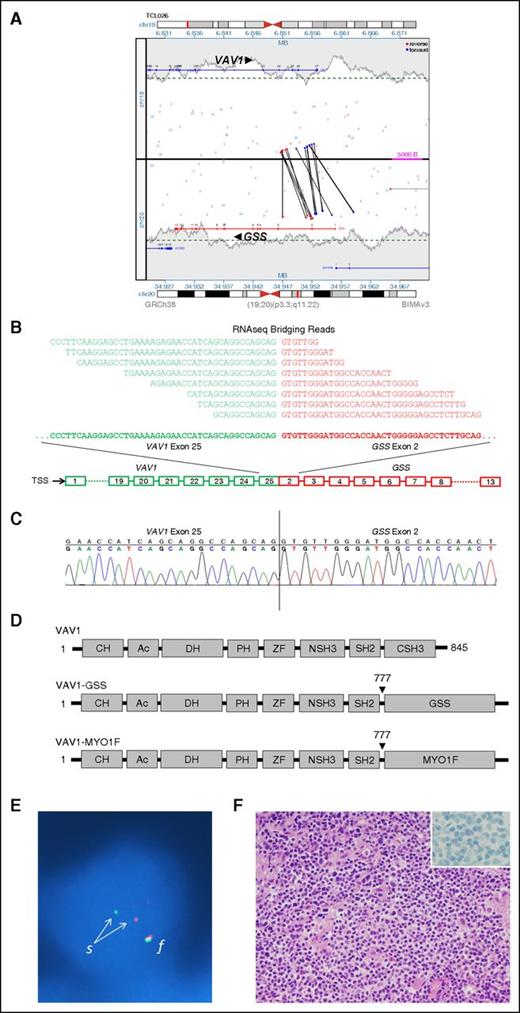
We examined VAV1 rearrangements in greater detail because they were recurrent and because VAV1 encodes a guanine nucleotide exchange factor (GEF) critical in T-cell receptor (TCR) signaling.24,25 The 2 cases showed rearrangements with GSS on 20q11.22 (Figure 2A) and MYO1F on 19p13.2, respectively. RNAseq identified reciprocal expressed fusion transcripts in both cases. VAV1-GSS is shown in Figure 2B and also was revalidated by Sanger sequencing (Figure 2C). We also confirmed the presence of the resultant VAV1-GSS fusion protein by western blot (supplemental Figure 1). VAV1-GSS (GSS exons 2-13) and VAV1-MYO1F (MYO1F exons 26-28) showed identical VAV1 fusion sites, resulting in loss of the VAV1 C-terminal SH3 (CSH3) domain (Figure 2D). The reciprocal fusions, GSS-VAV1 and MYO1F-VAV1, encoded only the CSH3 domain of VAV1.
Identification and validation of VAV1 fusions in PTCL, NOS. (A) Rearrangement of VAV1 and GSS genes as detected in genomic DNA by MPseq. Vertical and oblique black lines represent aberrant read-pairs; blue ends indicate mapping to (+) strand, whereas red ends indicate mapping to (−) strand. (B) VAV1-GSS fusion transcript as detected by RNAseq. Bridging reads spanning the fusion site are shown. TSS, transcription start site. (C) Sanger sequencing of the fusion site in VAV1-GSS fusion. (D) Schematic diagrams of VAV1 protein domains in wild-type VAV1 and resulting from VAV1 fusions. (E) FISH confirming a VAV1 rearrangement in a tumor cell nucleus (blue) from a PTCL, NOS specimen. The normal intact VAV1 allele is indicated by a red-green fusion (f) signal. Disruption of the VAV1 gene region on the other allele is indicated by separation (s) into one red (5′) signal and 1 green (3′) signal. (F) Pathologic features of PTCL, NOS with VAV1 fusion (hematoxylin and eosin stain; inset, CD30 immunostain). Original magnification ×400 (inset, ×1000).
Identification and validation of VAV1 fusions in PTCL, NOS. (A) Rearrangement of VAV1 and GSS genes as detected in genomic DNA by MPseq. Vertical and oblique black lines represent aberrant read-pairs; blue ends indicate mapping to (+) strand, whereas red ends indicate mapping to (−) strand. (B) VAV1-GSS fusion transcript as detected by RNAseq. Bridging reads spanning the fusion site are shown. TSS, transcription start site. (C) Sanger sequencing of the fusion site in VAV1-GSS fusion. (D) Schematic diagrams of VAV1 protein domains in wild-type VAV1 and resulting from VAV1 fusions. (E) FISH confirming a VAV1 rearrangement in a tumor cell nucleus (blue) from a PTCL, NOS specimen. The normal intact VAV1 allele is indicated by a red-green fusion (f) signal. Disruption of the VAV1 gene region on the other allele is indicated by separation (s) into one red (5′) signal and 1 green (3′) signal. (F) Pathologic features of PTCL, NOS with VAV1 fusion (hematoxylin and eosin stain; inset, CD30 immunostain). Original magnification ×400 (inset, ×1000).
We then investigated the frequency of VAV1 rearrangements by interphase FISH on 148 PTCLs (Table 2; Figure 2E). FISH results were 100% concordant with MPseq/RNAseq findings for the 11 cases analyzed by both methods. VAV1 rearrangements were present in 10 of 148 cases (7%). These rearrangements were restricted solely to PTCL, NOS (5 of 45; 11%; P = .0153) and ALCL (5 of 47; 11%; P = .0175). Because some PTCLs, NOS show similarities to ALCL and may represent a pathologic spectrum characterized by CD30 expression,26 we examined CD30 expression in the cases with VAV1 rearrangements. Although all ALCLs strongly expressed CD30, 5 of 5 cases of PTCL, NOS with VAV1 rearrangement were negative for CD30 (Figure 2F).
Frequency of VAV1 rearrangements in 148 PTCLs
| . | MP/RNA cases* . | Additional FISH cases . | Total . | P value† . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n positive . | n tested . | % . | n positive . | n tested . | % . | n positive . | n tested . | % . | ||
| PTCL, NOS | 2 | 11 | 18 | 3 | 34 | 9 | 5 | 45 | 11 | 0.0153 |
| AITL | 0 | 0 | — | 0 | 36 | 0 | 0 | 36 | 0 | — |
| ALCL | 0 | 0 | — | 5 | 47 | 11 | 5 | 47 | 11 | 0.0175 |
| ALK+ | 0 | 0 | — | 1 | 18 | 6 | 1 | 18 | 6 | 0.2685 |
| ALK− | 0 | 0 | — | 4 | 25 | 16 | 4 | 25 | 16 | 0.0060 |
| Cutaneous | 0 | 0 | — | 0 | 4 | 0 | 0 | 4 | 0 | — |
| EATL | 0 | 0 | — | 0 | 5 | 0 | 0 | 5 | 0 | — |
| NKTL | 0 | 0 | — | 0 | 7 | 0 | 0 | 7 | 0 | — |
| CTCL | 0 | 0 | — | 0 | 6 | 0 | 0 | 6 | 0 | — |
| T-LGL | 0 | 0 | — | 0 | 2 | 0 | 0 | 2 | 0 | — |
| Total | 2 | 11 | 18 | 8 | 137 | 6 | 10 | 148 | 7 | — |
| . | MP/RNA cases* . | Additional FISH cases . | Total . | P value† . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n positive . | n tested . | % . | n positive . | n tested . | % . | n positive . | n tested . | % . | ||
| PTCL, NOS | 2 | 11 | 18 | 3 | 34 | 9 | 5 | 45 | 11 | 0.0153 |
| AITL | 0 | 0 | — | 0 | 36 | 0 | 0 | 36 | 0 | — |
| ALCL | 0 | 0 | — | 5 | 47 | 11 | 5 | 47 | 11 | 0.0175 |
| ALK+ | 0 | 0 | — | 1 | 18 | 6 | 1 | 18 | 6 | 0.2685 |
| ALK− | 0 | 0 | — | 4 | 25 | 16 | 4 | 25 | 16 | 0.0060 |
| Cutaneous | 0 | 0 | — | 0 | 4 | 0 | 0 | 4 | 0 | — |
| EATL | 0 | 0 | — | 0 | 5 | 0 | 0 | 5 | 0 | — |
| NKTL | 0 | 0 | — | 0 | 7 | 0 | 0 | 7 | 0 | — |
| CTCL | 0 | 0 | — | 0 | 6 | 0 | 0 | 6 | 0 | — |
| T-LGL | 0 | 0 | — | 0 | 2 | 0 | 0 | 2 | 0 | — |
| Total | 2 | 11 | 18 | 8 | 137 | 6 | 10 | 148 | 7 | — |
Bold entries denote statistical significance, P < .05.
AITL, angioimmunoblastic T-cell lymphoma; ALK, anaplastic lymphoma kinase; CTCL, cutaneous T-cell lymphoma; EATL, enteropathy-associated T-cell lymphoma; NKTL, extranodal NK-/T-cell lymphoma, nasal type; T-LGL, T-cell large granular lymphocytic leukemia.
All cases also tested by FISH with 100% concordance.
Versus AITL, EATL, NKTL, CTCL, and T-LGL together.
VAV1-GSS promotes cell migration and growth
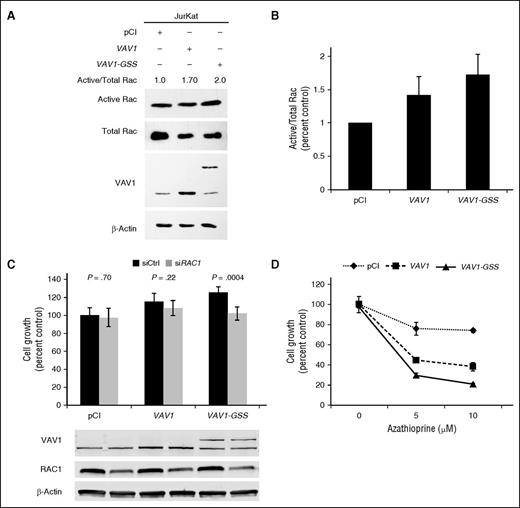
Because the VAV1 CSH3 domain mediates autoinhibition of VAV1 GEF activity,27 we hypothesized that VAV1 fusions, which lack this domain, promote VAV1 activation. Because VAV1 mediates cytoskeletal remodeling,28 we first performed immunofluorescence for actin in adherent NIH-3T3 fibroblasts transfected with VAV1-GSS, wild-type (wt)VAV1, or an empty control vector. We observed cytoskeletal rearrangement characterized by increased circularity in VAV1-GSS transfected cells (0.834 ± 0.033, where 1 indicates a perfect circle) compared with either wtVAV1 (0.765 ± 0.048; P = .045) or control (0.453 ± 0.089; P < .0001; Figure 3A-B).
VAV1-GSS fusion drives cytoskeletal reorganization, migration, and proliferation. (A) VAV1-GSS fusion promotes cytoskeletal reorganization and (B) increased cell circularity in adherent NIH-3T3 cells. A circularity value of 1 indicates a perfect circle. (C) The VAV1-GSS fusion protein is phosphorylated at VAV1 Y174 in Jurkat cells. In Jurkat, VAV1-GSS drives cell (D) migration and (E) growth relative to both wild-type VAV1 and an empty vector control.
VAV1-GSS fusion drives cytoskeletal reorganization, migration, and proliferation. (A) VAV1-GSS fusion promotes cytoskeletal reorganization and (B) increased cell circularity in adherent NIH-3T3 cells. A circularity value of 1 indicates a perfect circle. (C) The VAV1-GSS fusion protein is phosphorylated at VAV1 Y174 in Jurkat cells. In Jurkat, VAV1-GSS drives cell (D) migration and (E) growth relative to both wild-type VAV1 and an empty vector control.
We then examined the effect of the VAV1-GSS fusion protein on migration and growth in the neoplastic T-cell line, Jurkat. VAV1-GSS was phosphorylated at VAV1(Y174), indicative of GEF activity29 (Figure 3C). Cells expressing VAV1-GSS also showed increased migration (124% of empty vector control, P = .0016; P = .05 vs wtVAV1; Figure 3D). Finally, VAV1-GSS strongly promoted cell growth (159% of empty vector control; P < .0001 vs both control and wtVAV1; Figure 3E).
VAV1-GSS drives growth through RAC1 and can be targeted by Rac inhibition
Because VAV1 GEF activity predominantly targets the Rho family GTPase RAC1 (and to a lesser extent RHOA and CDC42),30,31 and RAC1 has been demonstrated to promote migration in an ALCL model,32 we measured Rac activation following VAV1-GSS transfection in Jurkat cells. VAV1-GSS overexpression increased active Rac (GTP-Rac; 173% of control; P = .054; Figure 4A-B). Furthermore, RAC1 knockdown abrogated the growth advantage conferred by VAV1-GSS, reducing growth from 125% to 102% of the baseline control value (P = .0004; Figure 4C). RAC1 knockdown had no effect on growth of control-transfected Jurkat cells. These data indicate that VAV1-GSS function is at least in part RAC1 dependent.
VAV1-GSS fusion induces targetable RAC1 dependence. (A) VAV1-GSS fusion induces Rac activation in Jurkat cells. (B) Rac activation data summarizing 3 independent experiments as shown in A. (C) RAC1 knock-down abrogates effect of VAV1-GSS fusion on cell growth. (D) Jurkat cells transfected with VAV1-GSS exhibit greater sensitivity to azathioprine than cells transfected with wild-type VAV1 or an empty vector control.
VAV1-GSS fusion induces targetable RAC1 dependence. (A) VAV1-GSS fusion induces Rac activation in Jurkat cells. (B) Rac activation data summarizing 3 independent experiments as shown in A. (C) RAC1 knock-down abrogates effect of VAV1-GSS fusion on cell growth. (D) Jurkat cells transfected with VAV1-GSS exhibit greater sensitivity to azathioprine than cells transfected with wild-type VAV1 or an empty vector control.
We then treated transfected Jurkat cells with azathioprine, a clinically available immunosuppressant and Rac inhibitor.33-35 At a dose of 10 µM, azathioprine inhibited growth of VAV1-GSS–transfected cells by 79.4 ± 1.4% compared with 26.0 ± 2.0% in control-transfected cells (P < .0001), and 62.1 ± 4.5% in wtVAV1-transfected cells (P = .0001; Figure 4D). Taken together, these findings demonstrate that VAV1-GSS promotes increased GEF activity and Rac activation that drives cell migration and growth and confers enhanced sensitivity to Rac inhibition.
MPseq/RNAseq identifies novel kinase fusions in PTCL, NOS
Among the other events identified by MPseq/RNAseq (Table 1) were fusions involving kinase genes known to be involved in oncogenic events in other cancers. Case TCL72 had an in-frame ITK-FER fusion (Figure 5A-C). Although ITK is expressed consistently in PTCL, NOS,36 read-count data showed FER expression only in TCL72 exons 11 to 20 encoded by ITK-FER (Figure 5D). Of note, ITK-SYK fusions are recurrent in PTCL, NOS and SYK inhibitors have been proposed therapeutically.16,37 ITK, SYK, and FER all encode SH2 domain-containing nonreceptor tyrosine kinases.38 Like ITK-SYK, ITK-FER retains the N-terminal plekstrin homology domain of ITK required for ITK-SYK function39 and contains the FER C-terminal kinase domain in place of the SYK kinase domain (Figure 5E).16 In line with this, expression of ITK-FER in HEK-293T cells resulted in increased colony formation (148% of control, P = .0001; Figure 5F). FER fusions have not been reported in PTCLs, although an SSBP2-FER fusion has been reported in T-cell acute lymphoblastic leukemia.40 FER is sensitive to kinase inhibitors under evaluation, including aminopyridine 8e, a drug developed to target mutant ALK.41
Novel ITK-FER kinase fusion in PTCL, NOS. (A) ITK-FER rearrangement as detected by MPseq. (B) ITK-FER fusion transcript as detected by RNAseq. (C) Sanger sequencing of the ITK-FER fusion site. (D) Exon-level RNAseq read plot for FER in TCL72 (“Case”) compared with the remaining 10 sequenced cases of PTCL, NOS. RPKM, reads per kilobase per million. (E) Schematic diagrams of protein domains in ITK, FER, and the ITK-FER fusion protein. (F) ITK-FER expression promotes colony formation in HEK-293 cells. PH, pleckstrin homology; TH, Tec homology; SH, Src homology; F-BAR, FER-CIP4 homology–Bin-amphiphysin-Rvs.
Novel ITK-FER kinase fusion in PTCL, NOS. (A) ITK-FER rearrangement as detected by MPseq. (B) ITK-FER fusion transcript as detected by RNAseq. (C) Sanger sequencing of the ITK-FER fusion site. (D) Exon-level RNAseq read plot for FER in TCL72 (“Case”) compared with the remaining 10 sequenced cases of PTCL, NOS. RPKM, reads per kilobase per million. (E) Schematic diagrams of protein domains in ITK, FER, and the ITK-FER fusion protein. (F) ITK-FER expression promotes colony formation in HEK-293 cells. PH, pleckstrin homology; TH, Tec homology; SH, Src homology; F-BAR, FER-CIP4 homology–Bin-amphiphysin-Rvs.
We also identified a novel IKZF2-ERBB4 fusion in case TCL75 (supplemental Figure 2A-C). IKZF2 encodes Helios, a T cell–restricted Ikaros-family transcription factor, and is expressed in various PTCLs.42-45 IKZF2 was expressed similarly in TCL75 and other cases, whereas ERBB4 was expressed only in TCL75 exons 2 to 28 encoded by IKZF2-ERBB4, which includes the ERBB4 kinase domain (supplemental Figure D-E). Thus, the fusion, which contains only exons 1 to 2 of IKZF2, co-opts the IKZF2 promoter for expression.46 These findings complement a recent report of truncated ERBB4 transcripts promoted by intronic long terminal repeats in ALCL, which retain ERBB4 kinase activity and are sensitive to ERBB family kinase inhibitors such as lapatinib.47
Examination of exon-level read plots for genes involved in the remaining fusions in Table 1 demonstrated 2 additional events with altered read counts in the case of interest: diminished CD58 expression in TCL81 (COPA-CD58 fusion; supplemental Figure 3A) and increased ZNF541 expression (exons 11-15) in TCL66 (GSK3A-ZNF541 fusion; supplemental Figure 3B).
Discussion
Here, we integrated MPseq and RNAseq data to identify expressed fusion transcripts in PTCL, NOS, an aggressive malignancy that is the most common form of T-cell NHL.2 Our findings highlight the marked genomic heterogeneity underlying this disorder, both in overall structural complexity and in the distribution of specific genetic events. We identified recurrent, novel, and targetable rearrangements of the VAV1 gene, which encodes a critical component of TCR signaling. We also discovered novel kinase fusions that represent candidate therapeutic targets. These data suggest that clinical sequencing approaches may help match PTCL patients to targeted therapeutics for personalized cancer therapy.
We identified VAV1 rearrangements in 11% of PTCL, NOS and ALCL, but not in other PTCL subtypes, suggesting that the biologic role of the resultant fusion proteins in T-cell lymphomagenesis may be highly context dependent. Of note, knockdown of VAV1 previously has been shown to lead to cell cycle arrest and apoptosis of ALCL cells.48 In addition, predicted gain-of-function hotspot mutations of VAV1 recently have been reported in adult T-cell leukemia/lymphoma (ATL).49 ATL was not evaluated in our study group as it is associated with human T-cell leukemia virus type-1 infection, which is nonendemic in the Midwestern United States.
We demonstrated that VAV1 fusions increased Rac activation and promoted cell growth. We focused on RAC1 because mutation of the VAV1 CSH3 domain strongly activates RAC1, whereas its effects on the ρ GTPases CDC42 and RHOA are more modest.27 RAC1 also has been shown to be activated by NPM-ALK in ALCL cells, and a Rac small molecule inhibitor blocked lymphoma development and dissemination.50,51 We showed that cells expressing VAV1-GSS were exquisitely sensitive to azathioprine, which inhibits activation of RAC1 but not of CDC42 or RHOA,33,34 Other clinically available drugs that inhibit RAC1 and have been proposed to be repurposed as anticancer agents include metformin, statins such as simvastatin, and R-ketorolac.52-54 RAC1 small molecule inhibitors are in ongoing development.55,56 Taken together, these data suggest RAC1 inhibition is a potential therapeutic strategy for PTCLs with VAV1 fusions, noting, however, that these fusions are present in the minority of PTCLs.
Our data add to a growing body of evidence supporting the importance of ρ-family GTPases in PTCL, including recently reported RHOA mutations in PTCL, NOS and angioimmunoblastic T-cell lymphomas57-59 and redundant and nonredundant roles for RAC1 and CDC42 in ALCL cell growth, migration, and dissemination.32,48 More broadly, these findings suggest the importance of TCR signaling as a key pathway involved in lymphomagenesis,60 as has been demonstrated in mice61 and is evidenced further by other genes altered by various mechanisms in PTCLs, including rearrangements of SYK, copy number gains of ITK, and point mutations of FYN.16,49,57,62 These potentially targetable molecular alterations show promise for advances analogous to those resulting from elucidation of dysregulated B-cell receptor signaling in B-cell NHLs.63,64
It should be noted that, in addition to the RAC1-mediated effects we demonstrated in VAV1-GSS-transfected cells, VAV1 has non–GEF-associated functions in T cells as well.65 For example, loss of the VAV1 CSH3 domain also might increase activity of nuclear factor of activated T cells through loss of nuclear factor of activated T cells–inhibitory residues.66 RAC1 also has multiple mechanisms of oncogenic activity, including its ability to enhance cytokine-mediated activation of signal transducer and activator of transcription 3 (STAT3),67 which plays a central pathogenetic role in many PTCLs.14,68-71 Finally, the role of the partner gene in VAV1 fusions, as well the potential function of the reciprocal expressed transcripts, may merit further study.
Our institution and others have developed clinical infrastructure to facilitate a personalized medicine approach to malignant diseases, in which genomic testing might inform therapeutic strategies specific for each individual.72 Although this concept is still evolving, data on PTCLs from our group and others, as well as our institutional experience with an Individualized Medicine Clinic, suggest PTCL patients might be particularly suitable for this approach.13,73-75 PTCLs are relatively uncommon and generally are clinically aggressive. They are markedly heterogeneous, both at the genetic–biologic level and at the clinicopathologic level, with >15 subtypes recognized by the World Health Organization.76 Current standard therapy is acknowledged to be suboptimal, but a number of targeted agents have shown clinical efficacy in subsets of patients.1,74 With this in mind, we examined the events identified by MPseq/RNAseq for their potential to suggest therapeutic options.
In addition to VAV1 rearrangements, we also discovered 2 novel kinase gene fusions, ITK-FER and IKZF2-ERBB4. As previously exemplified among PTCLs by ALK fusions, both fusions resulted in overexpression of a kinase not normally expressed in the cell-of-origin under the control of the partner gene promoter. In keeping with its predicted oncogenic function, ITK-FER promoted colony formation in vitro. ERBB4 encodes a member of the ERBB family, a group of targetable receptor tyrosine kinases that also includes epidermal growth factor receptor (ERBB1), HER2 (ERBB2), and ERBB3, and historically has been associated with the pathogenesis of epithelial and other solid tumors.77 Recent data, however, support the relevance of this family in PTCLs, including aberrant ERBB4 transcripts, elevated circulating epidermal growth factor levels, and enrichment for somatic mutations targeting the ERBB signaling pathway.47,78,79
We also rediscovered a previously reported ATXN1-TP63 fusion. TP63 rearrangements are recurrent in PTCL, NOS and ALK-negative ALCL and are characterized by high proliferation indices and poor prognosis.12,13 In fact, this patient (TCL65) had aggressive disease refractory to multiple regimens, did not achieve a response sufficient to be transplant eligible, and died 13 months after diagnosis. Poor outcomes with standard therapy suggest that patients with TP63-rearranged PTCLs might be considered for more intensive or experimental upfront treatment approaches.12,13
We identified 2 other genes, CD58 and ZNF541, whose expression differed between rearranged and nonrearranged cases. TCL81 carried a COPA-CD58 rearrangement associated with decreased CD58 expression. CD58 encodes a member of the immunoglobulin superfamily, CD58/LFA-3, an adhesion molecule known to be genetically inactivated by deletions and mutations in B-cell NHLs as a mechanism of immune escape.80 Deletions and mutations, but not rearrangements, of CD58 recently have been reported in PTCLs as well.57,58,81 Rearrangements are a less common structural cause of gene inactivation than deletions but have been reported. For example, we have shown that rearrangements disrupting the DUSP22 phosphatase gene are associated with decreased expression of the putative tumor suppressor gene, DUSP22, in ALCL.17 We also identified marked expression of ZNF541 only in TCL66, which carried a GSK3A-ZNF541 fusion. GSK3A, which was ubiquitously expressed in all sequenced samples (data not shown), encodes glycogen synthase kinase-3a, which itself has been proposed as a therapeutic target in cancer.82 ZNF541 normally is expressed in germ cells, where it mediates chromatin remodeling and is associated with histone hypoacetylation.83 Notably, treatment with histone deacetylase inhibitors, a class of drugs US Food and Drug Administration approved for PTCL,74 dramatically reduced ZNF541 protein levels.
EFCAB14 was involved in fusions in 2 of 11 sequenced cases. This gene encodes a member of the EF-hand superfamily of calcium-binding proteins, which play a role in TCR signaling.84 In 1 of these cases (TCL72), EFCAB14 was fused to PPP1R8, which encodes a phosphatase inhibitor critical for the activity of EZH2, a promising therapeutic target in PTCL.85,86 We also identified a reciprocal fusion of TNFRSF9 (TCL65) encoding CD137/4-1BB, a costimulatory molecule proposed as a therapeutic target in PTCL.87 Of note, fusions and mutations affecting the costimulatory molecules CD28 and ICOS recently have been observed in PTCL.49,88 We also identified a fusion involving the proapoptotic gene, BAX (TCL66), which has been reported to undergo loss-of-function mutations, but not rearrangements, in hematologic malignancies.89 Sequencing of additional PTCLs is likely to help prioritize these events for further study and identify additional candidates.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by awards R01 CA177734 (A.L.F.), P30 CA15083 (Mayo Clinic Cancer Center), and P50 CA97274 (University of Iowa/Mayo Clinic Lymphoma Specialized Program of Research Excellence) from the National Institutes of Health, National Cancer Institute, the Fraternal Order of Eagles Cancer Research Fund, Mayo Clinic Cancer Center (R.L.B. and G.L.R.), Clinical and Translational Science Award grant UL1 TR000135 from the National Center for Advancing Translational Science, award CI-48-09 from the Damon Runyon Cancer Research Foundation (A.L.F.), a scholarship award from the China Scholarship Council (Y.Z.), the Center for Individualized Medicine and the Department of Laboratory Medicine and Pathology, Mayo Clinic; and the Predolin Foundation.
Authorship
Contribution: R.L.B. and A.L.F. designed the study and wrote the manuscript; D.D.B., M.A.M., and G.V. contributed to study design; R.L.B., G.L.R., Y.Z., G.H., J.C.P., and B.W.E. conducted experiments; R.L.B., S.D., J.I.D., S.H.J., J.B.S., Y.W.A., G.V., and A.L.F. analyzed data; R.A.K. and P.T.G. conducted cytogenetic studies; and P.J.K., B.K.L., S.M.A., and J.R.C. contributed clinical specimens.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andrew L. Feldman, Department of Laboratory Medicine and Pathology, Mayo Clinic, 200 First Street SW, Rochester, MN 55905; e-mail: feldman.andrew@mayo.edu; or George Vasmatzis, Center for Individualized Medicine, Mayo Clinic, 200 First St SW, Rochester, MN 55905; e-mail: vasmatzis.george@mayo.edu.