Key Points
Accelerated telomere attrition precedes chromosomal loss and malignant transformation to MDS/AML arising from aplastic anemia.
Abstract
The pathophysiology of severe aplastic anemia (SAA) is immune-mediated destruction of hematopoietic stem and progenitor cells (HSPCs). Most patients respond to immunosuppressive therapies, but a minority transform to myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML), frequently associated with monosomy 7 (–7). Thirteen SAA patients were analyzed for acquired mutations in myeloid cells at the time of evolution to −7, and all had a dominant HSPC clone bearing specific acquired mutations. However, mutations in genes associated with MDS/AML were present in only 4 cases. Patients who evolved to MDS and AML showed marked progressive telomere attrition before the emergence of −7. Single telomere length analysis confirmed accumulation of short telomere fragments of individual chromosomes. Our results indicate that accelerated telomere attrition in the setting of a decreased HSPC pool is characteristic of early myeloid oncogenesis, specifically chromosome 7 loss, in MDS/AML after SAA, and provides a possible mechanism for development of aneuploidy.
Introduction
Human aplastic anemia (AA) is characterized by reduced peripheral blood counts caused by destruction of hematopoietic stem and progenitor cells (HSPCs). Most patients with severe aplastic anemia (SAA) respond to immunosuppressive therapy (IST), implicating an immune pathophysiology. However, malignant “clonal evolution” to myelodysplastic syndrome (MDS) or acute myeloid leukemia (AML) occurs in ∼15% of cases,1-3 and its pathophysiology is not well understood.4 The application of next-generation sequencing to de novo MDS/AML has led to definition of recurrently mutated genes as possible drivers of leukemogenesis.5-7 In the current study, 13 SAA patients who developed −7 MDS after IST were analyzed for acquired mutations by deep sequencing and for telomere attrition, because short telomeres may predict clonal evolution in SAA.8
Methods
Patients and blood samples
Peripheral blood and bone marrow cells were collected after informed consent according to institutional review board–approved protocols and in accordance with the Declaration of Helsinki. All SAA patients treated between 2001 and 2010 at our institution who had developed −7 MDS and had vailable serial blood and marrow samples were analyzed. Diagnosis, response to IST, clonal evolution, and relapse criteria have been described.1
Telomere length measurements
Mean telomere content of peripheral blood leukocytes by quantitative polymerase chain reaction (qPCR) and TERT/TERC sequencing were performed as previously described.9 Single telomere length assay (STELA) uses single-molecule PCR to amplify chromosome-specific telomeres based on the sequence specificity of subtelomeric regions, allowing for direct visualization and quantification of very short telomeres.10
Whole-exome sequencing and targeted sequencing
DNA was extracted from peripheral blood and bone marrow mononuclear cells using a Maxwell DNA purification kit (Promega, Madison, WI). DNA extracted from MicroBeads (Miltenyi, Germany)-sorted CD3 T cells was used as “germline” control DNA. A detailed protocol of whole-exome sequencing (WES) and analysis algorithm is available in the supplemental Methods, available on the Blood Web site. Targeted sequencing for genes associated with de novo MDS/AML (supplemental Table 1) was done using SureSelect custom kit from Agilent Technology, and sequencing was done using HiSequation 2000 instrument from Illumina with the 100-bp paired-end (PE) read option, according to the manufacturer’s protocol. Acquired mutations were called as previously shown.5 Deep sequencing of PCR-amplified sequences was performed for validation.
Results and discussion
Acquired mutations at clonal evolution to −7 MDS
We hypothesized that clonal evolution would be accompanied by mutations in genes previously identified in de novo MDS and AML.5-7 We used a similar strategy to identify and quantify mutations, isolating DNA from marrow mononuclear cells obtained at the time of progression to −7 MDS from a cohort of 13 SAA patients, followed by WES. We identified 209 somatic mutations in myeloid cells, both single-nucleotide variants (SNVs) and small insertions/deletions (indels), with variant allele frequency (VAF) >0.1 (supplemental Table 2). All 13 patients had a dominant HSPC clone, as inferred from SNVs and indels, at VAF 0.25 to 0.5 (Figure 1A). Within the dominant HSPC clone, 4 patients from this cohort (supplemental Table 2) had somatic mutations in genes known to be recurrently mutated in de novo MDS/AML (supplemental Table 1). Sanger sequencing of serial blood and bone marrow samples confirmed these recurring mutations. No other novel, recurrently mutated genes were identified, and mutated genes did not cluster in any pathway associated with cancer.
Oligoclonal hematopoiesis at the time of clonal evolution to –7 MDS in SAA patients. (A) Dominant hematopoietic clone VAF in 13 SAA patients at the time of clonal evolution to MDS with −7. Each patient’s mutations with VAF >0.25 are shown. (B-C) VAF of acquired mutations in UPN#1 and UPN#2, with neutrophil count. Direct Sanger sequencing confirmed the presence of mutations and VAF. The upper part of the graph shows representative hematoxylin-eosin staining of the core biopsy and CD34 antigen staining at the time of clonal evolution. The images were taken on an Olympus BX41 microscope with an Olympus DP72 camera, using a 4× UPlanFL N Olympus objective (original magnification ×20). Neutrophil count (left y-axis) is shown by blue bars. VAF of acquired mutations (right y-axis) at various time points from diagnosis of SAA until diagnosis of MDS are shown in red and green line graphs. Arrows on the x-axis mark ISTs and time to progression to MDS.
Oligoclonal hematopoiesis at the time of clonal evolution to –7 MDS in SAA patients. (A) Dominant hematopoietic clone VAF in 13 SAA patients at the time of clonal evolution to MDS with −7. Each patient’s mutations with VAF >0.25 are shown. (B-C) VAF of acquired mutations in UPN#1 and UPN#2, with neutrophil count. Direct Sanger sequencing confirmed the presence of mutations and VAF. The upper part of the graph shows representative hematoxylin-eosin staining of the core biopsy and CD34 antigen staining at the time of clonal evolution. The images were taken on an Olympus BX41 microscope with an Olympus DP72 camera, using a 4× UPlanFL N Olympus objective (original magnification ×20). Neutrophil count (left y-axis) is shown by blue bars. VAF of acquired mutations (right y-axis) at various time points from diagnosis of SAA until diagnosis of MDS are shown in red and green line graphs. Arrows on the x-axis mark ISTs and time to progression to MDS.
Among the 4 patients with candidate gene mutations, one (UPN#1) had completely responded to IST, later had relapse, and then responded to cyclosporine (Figure 1B); 2 years later she developed −7 and blood counts were stable despite 5% to 10% bone marrow blasts for the next 3 years. At 6 months after initial IST, a clone was characterized by mutations in ASXL1 and DNMT3A, which was unchanged for 3 years. A subclone emergent at relapse had mutations in SETBP1, DOT1L, and STAT3; it increased over 30 months until it was present in 60% of myeloid cells at diagnosis of −7. UPN#2 was refractory to IST before developing −7. He died 6 months later from infection, and the last marrow showed 7% myeloblasts (Figure 1C). In 2 patients (UPN#9, UPN#10), acquired mutations were detected in RUNX1 and CSF3R, respectively, only at the time of clonal evolution and also present at a lower VAF in the lymphoid lineage.
Targeted sequencing of genes associated with de novo MDS and AML (supplemental Table 1) confirmed all mutations identified by WES and identified small subclones at low VAF mutations in 3 additional patients (supplemental Table 3).
WES allowed characterization of the oligoclonal architecture of hematopoiesis at development of −7 MDS. Somatic mutations in genes associated with de novo MDS and AML were present in a subset of patients whose disease had evolved, but candidate gene abnormalities were not prevalent in the group as a whole.
Accelerated telomere attrition and accumulation of very short telomeres in SAA evolving to MDS
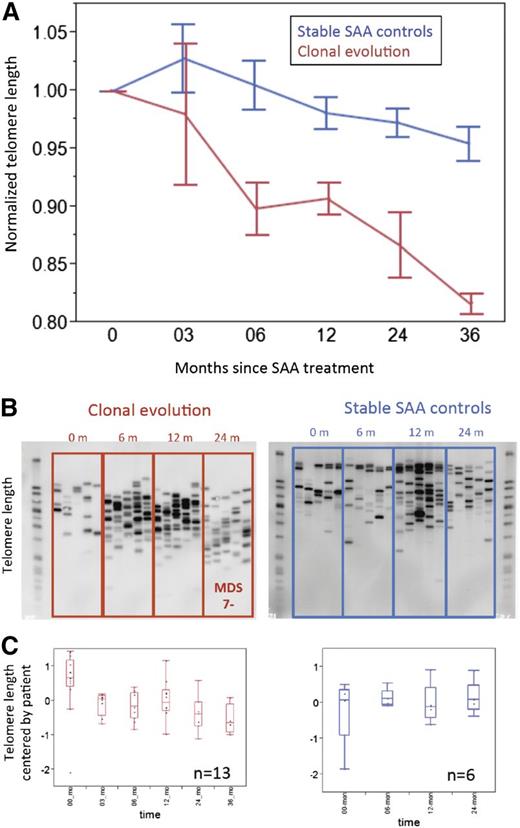
Accelerated telomere attrition causes increased chromosomal instability, aneuploidy, and progression to malignancy in mouse models.11 We have linked short telomeres at SAA diagnosis with later MDS and AML.8 We postulated that accelerated telomere attrition might be pathogenic in the development of myeloid neoplasms after marrow failure. First, we estimated mean telomere content loss from the time of SAA diagnosis until clonal evolution. In the 13 patients who progressed to −7 MDS, the average attrition rate was 419 bp per year (Figure 2A); among controls, the rate was much lower, 65 bp per year (Figure 2A; P < .01). (The control group of 30 stable SAA patients was treated in the same time period and matched by age, sex, and IST response to −7 cases; supplemental Table 4). The attrition rate was significantly higher in the clonal evolution group compared with the control SAA group, starting at 6 months after IST (Figure 2A). STELA confirmed increased attrition and revealed that chromosome-specific, very short telomeres accumulated in SAA patients with clonal evolution and were present before development of a cytogenetic abnormality and diagnosis of MDS. In the stable SAA group, XpYp profiles showed no increase in shorter telomeres to 48 months after IST (Figure 2B-C). A similar shift in telomere XpYp STELA profiles was observed with cultured bone marrow cells; accumulation of shorter telomeres was observed in samples from SAA patients who developed MDS (supplemental Figure 1A) but not in cells from stable SAA patients (supplemental Figure 1B).
Accumulation of very short telomeres precedes development of –7 MDS in SAA patients. (A) Mean telomere content for clonal evolution group (red line, n = 13) and SAA stable controls (blue line, n = 30) measured by qPCR. For each patient in both groups, mean telomere content at each available time point was normalized to mean telomere content at presentation. (B) Examples of XpYp STELA profile of serial peripheral blood leukocytes for patients from the time of diagnosis of SAA until progression to −7 MDS (left) or SAA controls (right). (C) Quantification of telomere length measured by XpYp STELA. For each of the 2 groups, all available samples at various time points after IST were assayed by both qPCR and STELA. For the clonal evolution group, samples up to the time of −7 diagnosis were used; thus for each time point, at least 4 patients samples were used. Similarly for the SAA control group, 30 patients’ samples were available to 24 months after initial IST; each time point included more than 15 patient samples. For serial blood samples’ telomere length measured by XpYp STELA, average lengths were mean-centered by patient. Discrete bands lengths were quantified using ImageQuant TL software (GE Healthcare Life Sciences, Piscataway, NJ). Bands were quantified based on a standard DNA ladder. A 1-way Wilcoxon test on equal length of the telomeres of the patients over all time points was statistically significant (P = .0027) for the clonal evolution group but not for controls (P = .62).
Accumulation of very short telomeres precedes development of –7 MDS in SAA patients. (A) Mean telomere content for clonal evolution group (red line, n = 13) and SAA stable controls (blue line, n = 30) measured by qPCR. For each patient in both groups, mean telomere content at each available time point was normalized to mean telomere content at presentation. (B) Examples of XpYp STELA profile of serial peripheral blood leukocytes for patients from the time of diagnosis of SAA until progression to −7 MDS (left) or SAA controls (right). (C) Quantification of telomere length measured by XpYp STELA. For each of the 2 groups, all available samples at various time points after IST were assayed by both qPCR and STELA. For the clonal evolution group, samples up to the time of −7 diagnosis were used; thus for each time point, at least 4 patients samples were used. Similarly for the SAA control group, 30 patients’ samples were available to 24 months after initial IST; each time point included more than 15 patient samples. For serial blood samples’ telomere length measured by XpYp STELA, average lengths were mean-centered by patient. Discrete bands lengths were quantified using ImageQuant TL software (GE Healthcare Life Sciences, Piscataway, NJ). Bands were quantified based on a standard DNA ladder. A 1-way Wilcoxon test on equal length of the telomeres of the patients over all time points was statistically significant (P = .0027) for the clonal evolution group but not for controls (P = .62).
In this report, we show that SAA patients who progress to −7 MDS have oligoclonal hematopoiesis and increased telomere attrition caused by accumulation of very short telomeres before development of aneuploidy. Rapid telomere attrition did not appear to be genetic, because these patients did not have clinical features of a telomeropathy; rather they had telomeres within the normal range at diagnosis and they lacked mutations in TERT and TERC. Accelerated telomere loss likely is secondary to restricted clonal hematopoiesis and proliferative stress, leading to chromosomal instability. The increased average number of acquired mutations identified by WES in this cohort (16/case) compared with 5 to 10 mutations that were previously reported in healthy volunteers of similar ages6 also would result from a long replicative history.
In the 13 patients who had clonal evolution, no novel recurrently mutated genes were identified, and acquired mutations in genes postulated to be drivers of myeloid malignancies5-7 were present in only 4 patients in the dominant HSPC clone. Acquired low VAF mutations in these genes were identified in 3 additional patients by targeted sequencing. However, because of their low VAF, these subclones were unlikely to be drivers of progression to aneuploidy. We cannot exclude novel unique gene mutations for the remaining patients. Oncogenesis might be driven by aneuploidy through haploinsufficiency of genes on chromosome 7.12,13 Moreover, acquired mutations in the same gene set were recently reported by targeted sequencing in 10% to 19% of AA patients, and not all had progressed to overt MDS.14-16 In the largest reported cohort of 150 AA patients,14 17 had clonal evolution, but only 4 patients had −7 MDS.
We previously reported that mean telomere content of leukocytes at diagnosis of SAA was the only known predictor of malignant progression.9 Aneuploidy would be expected if chromosomal instability were responsible, and indeed cytogenetic clonal evolution in SAA is almost invariably accompanied by whole or partial loss of chromosome 7, and less frequently by gain of chromosome 8, deletions of 13q and 20q, and similar aberrations.2,3 Moreover, telomerase-deficient bone marrow cells in culture show increased −7, linking this aneuploidy to telomere pathology.17
Although our study is limited by sample size and, as with most investigations of human biology, shows association rather than causation, it allows some cautious inferences. Accelerated telomere attrition preceded aneuploidy and malignant transformation at an early stage of oncogenesis, arising from bone marrow failure. Chromosome instability, as the molecular mechanism of aneuploidy, has been suggested by many cell culture experiments and animal models.18,19 Identification of critically short telomeres before development of clinical progression might allow for timely management of patients at risk of clonal evolution. Therapeutic upregulation of telomerase, for example by sex hormones20 or other pharmacologic agents, might reduce the risk of clonal evolution in the setting of a reduced stem cell pool, in bone marrow failure syndromes or after intensive chemotherapy.
Presented orally at the 55th Annual Meeting of the American Society of Hematology, New Orleans, LA, December 10, 2013.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Cynthia Dunbar, Andre LaRochelle, and Stephen Chanock for their careful reading of the manuscript, Dr Delong Liu for support with statistical analysis, and Drs Mark Hill, Duncan Baird, and Peter Lansdorp for help developing the STELA method.
This research was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute, National Institutes of Health.
Authorship
Contribution: B.D., X.F., D.M.T., R.T.C., and N.S.Y. designed experiments; B.D., X.F., Y.U., T.Y., Y.Y., Y.W., and S.K. performed experiments; B.D., X.F., T.Y., Y.Y., S.O., J.Z., and N.S.Y. analyzed the data; and B.D. and N.S.Y. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bogdan Dumitriu, 10 Center Dr (MSC 1475), Building 10, CRC 3-5216, Bethesda, MD 20892; e-mail: dumitriub@nhlbi.nih.gov.
References
Author notes
B.D. and X.F. contributed equally to this study.