Key Points
Efforts to understand mechanisms of disease initiation in human adult pre-B ALL are hampered by lack of appropriate animal models.
Optimized xenotransplant assays show that niche-based SDF-1/CXCR4 interaction is crucial for adult non-t(4;11) pre-B ALL leukemia initiation.
Abstract
The distinct nature of acute lymphoblastic leukemia (ALL) in adults, evidenced by inferior treatment outcome and different genetic landscape, mandates specific studies of disease-initiating mechanisms. In this study, we used NOD/LtSz-scid IL2Rγ nullc (NSG) mouse xenotransplantation approaches to elucidate leukemia-initiating cell (LIC) biology in primary adult precursor B (pre-B) ALL to optimize disease modeling. In contrast with xenografting studies of pediatric ALL, we found that modification of the NSG host environment using preconditioning total body irradiation (TBI) was indispensable for efficient engraftment of adult non-t(4;11) pre-B ALL, whereas t(4;11) pre-B ALL was successfully reconstituted without this adaptation. Furthermore, TBI-based xenotransplantation of non-t(4;11) pre-B ALL enabled detection of a high frequency of LICs (<1:6900) and permitted frank leukemic engraftment from a remission sample containing drug-resistant minimal residual disease. Investigation of TBI-sensitive stromal-derived factor-1/chemokine receptor type 4 signaling revealed greater functional dependence of non-t(4;11) pre-B ALL on this niche-based interaction, providing a possible basis for the differential engraftment behavior. Thus, our studies establish the optimal conditions for experimental modeling of human adult pre-B ALL and demonstrate the critical protumorogenic role of microenvironment-derived SDF-1 in regulating adult pre-B LIC activity that may present a therapeutic opportunity.
Introduction
In contrast to children, outcomes for adults with acute lymphoblastic leukemia (ALL) are poor. Currently <50% of adults with ALL survive long term1,2 compared with a 5-year overall survival of >80% in children.3,4 Although treatment differences (intensity, tolerability, and compliance) do in part contribute to this differential, tumor biology itself may also play a significant role.
In vitro, adult ALL tumors exhibit greater cellular drug resistance compared with counterpart pediatric disease.5,6 Further support for an age-related ALL phenotype comes from molecular profiling of submicroscopic aberrations where the different distributions of cooperating mutations between adult7 and childhood ALL8 suggest divergent oncogenic pathways may operate. In addition to cell intrinsic genetic/epigenetic influence on tumor behavior, cell extrinsic microenvironmental interaction is also likely to play a vital role. Understanding requisite niche–tumor cell interactions for initiating and maintaining malignant growth, as well as the contribution of genetic determinism, requires optimized disease-specific animal models. However, such experimental systems of adult ALL are lacking. Available humanized models of ALL are almost entirely based on pediatric disease.9-14 We hypothesize that these systems may not recapitulate the mechanisms driving tumor initiation and progression in adult ALL and that conditions for adult leukemia-initiating cell (LIC) discovery have not been defined.
Functional LIC potential is measured by ability of tumor cells to initiate and sustain growth of the leukemic clone in vivo, with the immunodeficient mouse xenotransplantation assay considered the gold standard. However, assignment of tumorigenic potential as read out by this assay are affected by a number of variables including recipient mouse immune background,10,14,15 tumor cell manipulation,16 and nonhost factors such as method of tumor cell administration12 and use of preconditioning treatments such as total body irradiation (TBI).
Although studies of tumor initiation in human adult ALL are described, these reports focus on less permissive severe combined immunodeficient (SCID)17 or nonobese diabetic (NOD)/SCID xenotransplantation models,18,19 which can underestimate leukemogenic activity14 or have involved assessment of limited adult ALL subtypes.20 A wider assessment of primary adult ALL by more sensitive models for studying human cell engraftment, NOD SCID mice lacking a functional interleukin-2 receptor γ-chain, NOD/LtSz-scid/IL2Rγ nullc (NSG),21,22 has not been performed. Such studies are of fundamental importance to interrogating cancer biology in adult ALL, specifically for characterizing cellular drivers of malignant growth including self-renewing leukemia stem cell populations.
In this investigation, we use the tractable NSG mouse xenotransplantation assay to establish the optimal modeling system for adult precursor B-cell (pre-B) ALL. Our engraftment studies define the conditions for sensitive assaying of adult pre-B ALL LICs and low-frequency minimal residual disease (MRD) clones and reveal mechanistic insights into the functional parameters of the adult pre-B ALL tumor niche.
Materials and methods
Patient samples
All human cells were obtained after written informed consent in accordance with the Declaration of Helsinki and used according to protocols approved by the United Kingdom (UK) main research ethics committee. Mononuclear cell preparations were isolated by the Ficoll-Paque method from either bone marrow (BM) or peripheral blood (PB) of patients with newly diagnosed/relapsed ALL and cryopreserved in fetal bovine serum and 10% dimethylsulfoxide prior to use. Mobilized PB stem cells (MPBSCs) were obtained from fresh blood samples taken from both granulocyte colony-stimulating factor–mobilized and nonmobilized healthy donors.
Xenotransplantation of primary leukemia cells
All animal experiments were performed according to UK Home Office-approved protocols and institutional guidelines. NSG mice were obtained from The Jackson Laboratory and Charles Rivers, bred, and maintained in barrier accommodation. Baytril (2.5%)-supplemented water was additionally administered to mice for 7 days prior to receiving TBI preconditioning. Characteristics of the patients investigated for in vivo study are provided in Table 1. Leukemic cells resuspended in phosphate-buffered saline/2% fetal bovine serum were administered either intravenously (IV) into the tail vein or by intratibial BM (IBM) injection into 6- to 10-week-old mice. In mice receiving preconditioning, injection occurred +24 hours after TBI. Nonirradiated mice were additionally treated with anti-CD122 antibody (BioXCell) to abrogate host immune-mediated engraftment resistance. For chemokine receptor type 4 (CXCR4) blocking experiments, cells were preincubated for 30 minutes with 10 μg/mL of anti-CXCR4 monoclonal antibody (clone 12G5) prior to transplantation. Animals were regularly monitored and euthanized when signs of disease-related symptoms developed or electively at the experimental end point.
Characteristics of the 22 pre-B ALL patients/leukemia samples investigated in engraftment studies
| UPN . | Age (years) . | Gender . | Disease status . | Sample type . | Cytogenetics . | Major cytogenetic group/risk status . |
|---|---|---|---|---|---|---|
| 1 | 59.4 | F | Diagnosis | PB | Failed* | t(9;22)/high |
| 2 | 44.5 | F | Diagnosis | PB | 45,XX,-7,t(9;22)(q34;q11)[4] | t(9;22)/high |
| 3 | 19.5 | M | Diagnosis | BM | Normal | Normal/standard |
| 4 | 26.1 | M | Diagnosis | BM | 55-56, XY,+X,+4,+6,+11,+12,+14,+17,+18,+21,+21[cp4]/46,XY[4] | High hyperdiploidy/ standard |
| 5 | 46.4 | F | Diagnosis | BM | 46,XX,t(9;22)(q34;q11)[1]/46,idem,der(10)t(10;20)(q22;p12)?del(10)(q22q34),der(20)t(10;20)[7]/46,XX[3] | t(9;22)/high |
| 6 | 33.6 | M | Diagnosis | PB | 46,XY,+1,dic(1;9)(p1;p1)[8]/46,XY[2] | Other/standard |
| 7 | 42.6 | M | Diagnosis | PB | 46,XY,t(4;11)(q21;q23)[10] | t(4;11)/high |
| 8 | 39.7 | M | Diagnosis | BM | 46,XY,der(9)add(9)(p11)add(9)(q34)[16]/46,XY[4] | Other/standard |
| 9 | 47.7 | M | Diagnosis | BM | 46,XY[20] | Normal/standard |
| 10 | 42.8 | F | Diagnosis | PB | 46,XX,t(2;11)(p11;p11),t(4;11)(q21;q23),i(7)(q10),add(22)(q13)[18]/46,XX[2] | t(4;11)/high |
| 11 | 39.8 | M | Diagnosis | PB | 59-62,XXYY,-1,-2,-3,-7,+8,add(9)(p1),der(9)t(9;22)(q34;q11)x2,-10,-12,-14,-15,-19,-20,+21,-22,-22,+2mar[cp6] | t(9;22)/high |
| 12 | 22.7 | M | Diagnosis | BM | 47,XY,+21c[20] | t(12;21)/standard |
| 13 | 50.4 | M | Diagnosis | BM | 46,XY,t(9;22)(q34;q11)[3]/46,XY[1] | t(9;22)/high |
| 14 | 48.2 | M | Diagnosis | PB | 46,XY,t(4;11)(q21;q23)[19]/46,XY[1] | t(4;11)/high |
| 15 | 49.3 | F | Diagnosis | PB | 46,XX,del(5)(q11q1?3),add(9)(p2?4)x2,der(19)t(1;19)(q23;p13)[35]/46,XX[5] | t(1;19)/standard |
| 16 | 42.7 | M | Diagnosis | PB | 46,XY,der(15)t(1;15)(q2;p1),der(19)t(1;19)(q23;p13)[11]† | t(1;19)/standard |
| 17 | 50.2 | F | Diagnosis | BM | Normal | Normal/standard |
| 18 | 52.4 | M | Diagnosis | BM | 45∼46,XX,inv(1)(p2q2),-4,add(4)(q2),del(6)(q1q2),add(8)(q24),add(9)(p2),add(9)(q2),-11,+2mar[cp10] | Complex/high |
| 19 | 48.7 | M | Diagnosis | BM | 46,XX,t(9;22)(q34;q11.2)[16]/46,XX[4]. | t(9;22)/high |
| 20 | 32.0 | M | Diagnosis | PB | 45,XY,der(1)inv(1)(p3q4)t(1;5)(?;q?),der(5)t(1;5),-7,t(9;22)(q34;q11)[10] | t(9;22)/high |
| 21 | 68.0 | F | 1st Relapse | BM | Failed‡ | NA |
| 22 | 27.0 | M | Diagnosis | BM | 46,XY,t(4;11)(q21;q23)[6]/46,XY[1] | t(4;11)/high |
| UPN . | Age (years) . | Gender . | Disease status . | Sample type . | Cytogenetics . | Major cytogenetic group/risk status . |
|---|---|---|---|---|---|---|
| 1 | 59.4 | F | Diagnosis | PB | Failed* | t(9;22)/high |
| 2 | 44.5 | F | Diagnosis | PB | 45,XX,-7,t(9;22)(q34;q11)[4] | t(9;22)/high |
| 3 | 19.5 | M | Diagnosis | BM | Normal | Normal/standard |
| 4 | 26.1 | M | Diagnosis | BM | 55-56, XY,+X,+4,+6,+11,+12,+14,+17,+18,+21,+21[cp4]/46,XY[4] | High hyperdiploidy/ standard |
| 5 | 46.4 | F | Diagnosis | BM | 46,XX,t(9;22)(q34;q11)[1]/46,idem,der(10)t(10;20)(q22;p12)?del(10)(q22q34),der(20)t(10;20)[7]/46,XX[3] | t(9;22)/high |
| 6 | 33.6 | M | Diagnosis | PB | 46,XY,+1,dic(1;9)(p1;p1)[8]/46,XY[2] | Other/standard |
| 7 | 42.6 | M | Diagnosis | PB | 46,XY,t(4;11)(q21;q23)[10] | t(4;11)/high |
| 8 | 39.7 | M | Diagnosis | BM | 46,XY,der(9)add(9)(p11)add(9)(q34)[16]/46,XY[4] | Other/standard |
| 9 | 47.7 | M | Diagnosis | BM | 46,XY[20] | Normal/standard |
| 10 | 42.8 | F | Diagnosis | PB | 46,XX,t(2;11)(p11;p11),t(4;11)(q21;q23),i(7)(q10),add(22)(q13)[18]/46,XX[2] | t(4;11)/high |
| 11 | 39.8 | M | Diagnosis | PB | 59-62,XXYY,-1,-2,-3,-7,+8,add(9)(p1),der(9)t(9;22)(q34;q11)x2,-10,-12,-14,-15,-19,-20,+21,-22,-22,+2mar[cp6] | t(9;22)/high |
| 12 | 22.7 | M | Diagnosis | BM | 47,XY,+21c[20] | t(12;21)/standard |
| 13 | 50.4 | M | Diagnosis | BM | 46,XY,t(9;22)(q34;q11)[3]/46,XY[1] | t(9;22)/high |
| 14 | 48.2 | M | Diagnosis | PB | 46,XY,t(4;11)(q21;q23)[19]/46,XY[1] | t(4;11)/high |
| 15 | 49.3 | F | Diagnosis | PB | 46,XX,del(5)(q11q1?3),add(9)(p2?4)x2,der(19)t(1;19)(q23;p13)[35]/46,XX[5] | t(1;19)/standard |
| 16 | 42.7 | M | Diagnosis | PB | 46,XY,der(15)t(1;15)(q2;p1),der(19)t(1;19)(q23;p13)[11]† | t(1;19)/standard |
| 17 | 50.2 | F | Diagnosis | BM | Normal | Normal/standard |
| 18 | 52.4 | M | Diagnosis | BM | 45∼46,XX,inv(1)(p2q2),-4,add(4)(q2),del(6)(q1q2),add(8)(q24),add(9)(p2),add(9)(q2),-11,+2mar[cp10] | Complex/high |
| 19 | 48.7 | M | Diagnosis | BM | 46,XX,t(9;22)(q34;q11.2)[16]/46,XX[4]. | t(9;22)/high |
| 20 | 32.0 | M | Diagnosis | PB | 45,XY,der(1)inv(1)(p3q4)t(1;5)(?;q?),der(5)t(1;5),-7,t(9;22)(q34;q11)[10] | t(9;22)/high |
| 21 | 68.0 | F | 1st Relapse | BM | Failed‡ | NA |
| 22 | 27.0 | M | Diagnosis | BM | 46,XY,t(4;11)(q21;q23)[6]/46,XY[1] | t(4;11)/high |
Cytogenetic profiles were determined by local laboratories using standard methods. NA, not applicable; UPN, unique patient number.
FISH positive for BCR/ABL gene rearrangement.
FISH positive for TCF3(E2A)-PBX1 gene fusion.
This case had low hypodiploidy (30-39 chromosomes) at diagnosis. FISH analysis at relapse detected loss of the ABL1 and BCR signals consistent with the low hypodiploid clone seen at diagnosis.
Flow cytometric assessment of human leukemia engraftment
Mononuclear preparations harvested from femoral/tibial BM of injected mice were stained with CD45-fluorescein isothiocyanate and CD19-phycoerythrin (PE) (Becton Dickinson San Jose, CA) to determine leukemia engraftment. Antibody stained cells were washed in Hanks balanced salt solution and stained with a viability dye before analysis. Gates were set up to exclude nonviable cells and isotype-stained populations. For certain experiments, extended immunophenotype analysis of the engrafted leukemia was performed (supplemental Methods A, available on the Blood Web site).
CXCR4 expression analysis
A total of 1 × 106 cells were stained with either CXCR4-PE or CXCR4-allophycocyanin (APC) (Becton Dickinson) or isotype control to assess cell surface receptor expression. Intracellular CXCR4 was investigated by first blocking CXCR4 at the cell surface with nonconjugated antihuman CXCR4 antibody (clone 12G5), 10 μg/mL, for 30 minutes at 4°C. Cells were then prepared for intracellular staining using Intrastain Fix and Perm (Dako, Cambridgeshire, UK) prior to labeling with Anti CXCR4-PE for 30 minutes at 4°C. We confirmed that fixation did not affect estimates of CXCR4-positive populations (supplemental Methods B).
Chemotaxis assays
Assays were performed as previously described23 (supplemental Methods C) with the exception that cell migration was assessed by enumerating the absolute number of CD19+ migrating cells against CountBright (Molecular Probes, Eugene, OR) precalibrated microspheres. For in vitro blocking experiments, cells were pretreated with AMD3100 100 μM/mL, or phosphate-buffered saline control for 1 hour at room temperature, washed, and assayed for transwell migration.
Fluorescence in situ hybridization
Interphase fluorescence in situ hybridization (FISH) studies were carried out on cytospin slides fixed in Carnoy’s fixative (3:1 methanol: acetic acid). The IGH and TCF3 Breakapart probes (Cytocell) were used to detect translocations involving 14q32 and 19p13, respectively. Hybridization was performed according to manufacturer’s instructions, and 100 nuclei were scored. FISH imaging was performed using an Olympus BX-61 camera equipped with an X-cite 120 mercury lamp (Lumen Dynamics) and Cytovision software (Leica Microsystems).
Clonal immunoglobulin T-cell receptor analysis
DNA was extracted from mononuclear cell preparations of BM from leukemic mice using the QIAmp DNA purification kit (Qiagen, Hilden, Germany).
Clonal immunoglobulin (Ig)/T-cell receptor (TCR) gene rearrangements were determined using published primer sets.24 Monoclonal polymerase chain reaction products were confirmed by heteroduplex gel assessment as previously described25 prior to gene sequencing. Sequence analysis and junctional region mapping was performed using DNASTAR. Leukemia xenograft Ig/TCR profiles were compared with the clonality profile of the original sample that had been determined by a reference laboratory using identical methodology.
Statistics
The significance of the differences between groups (±TBI) was performed using the Fisher’s exact test. All other tests unless otherwise indicated was performed using the Student t test. Limiting dilutions analysis was performed according to methods published by Hu et al26 using the publicly available Extreme Limiting Dilution Analysis (ELDA) web-based software (http://bioinf.wehi.edu.au/software/elda/).
Results
Differential requirements for TBI preconditioning therapy in human adult pre-B ALL engraftment
Several studies have described the successful engraftment of human ALL, primarily pediatric pre-B ALL in unconditioned immunodeficient hosts,9,12-14,27 suggesting that basal cellular/structural and soluble states within the xenogeneic environment are sufficient to support survival and proliferation of these tumors. To determine the potential for adult pre-B ALL tumors to proliferate under unmodified host conditions, we investigated leukemia repopulation potential of 17 consecutive diagnostic samples (UPN 1-17; Table 1) in either nonirradiated (n = 13) or TBI (2.5Gy) preconditioned (n = 12) NSG mice following IV injection of 5 to 10 million viable cells. ALL engraftment capability was assigned at disease manifestation or after prolonged (>5 months) modeling.
Engraftment outcomes of the 17 individual samples are shown in supplemental Table 1. A summary of these data is shown in Table 2. Overall, 13 of 17 samples engrafted. The addition of TBI preconditioning was associated with a higher proportion of engrafting samples compared with no TBI (11/12 vs 2/13, respectively; P = .0394). This difference was apparent in most cytogenetic subtypes, with the exception of t(4;11) pre-B ALL, where 2 of 3 samples efficiently engrafted unconditioned NSG recipients.
Comparison of adult pre-B ALL engraftment by either unconditioned (−TBI) or 2.5-Gy preconditioned (+TBI) xenotransplantation methods
| Major cytogenetic group . | No. of engrafting samples . | P . | |
|---|---|---|---|
| +TBI . | −TBI . | ||
| Normal/other | 7/7 | 0/6 | .0006 |
| t(9;22) | 3/3 | 0/4 | .0286 |
| t(4;11) | 1/2 | 2/3 | NS |
| Proportion of engrafting samples: % leukemic engraftment Mean no. of weeks to disease manifestation (range)* | |||
| 11/12 | 2/13 | .0394 | |
| 81.9 ± 5.4 | 96.4 ± 1.4 | NS | |
| 15.9 (5.9-29.7) | 27.7 (26.9-29.3) | .0231 | |
| Major cytogenetic group . | No. of engrafting samples . | P . | |
|---|---|---|---|
| +TBI . | −TBI . | ||
| Normal/other | 7/7 | 0/6 | .0006 |
| t(9;22) | 3/3 | 0/4 | .0286 |
| t(4;11) | 1/2 | 2/3 | NS |
| Proportion of engrafting samples: % leukemic engraftment Mean no. of weeks to disease manifestation (range)* | |||
| 11/12 | 2/13 | .0394 | |
| 81.9 ± 5.4 | 96.4 ± 1.4 | NS | |
| 15.9 (5.9-29.7) | 27.7 (26.9-29.3) | .0231 | |
NS, nonsignificant.
The number of samples with leukemic engraftment (CD45+/CD19+ > 0.01%) over the total number injected is shown.
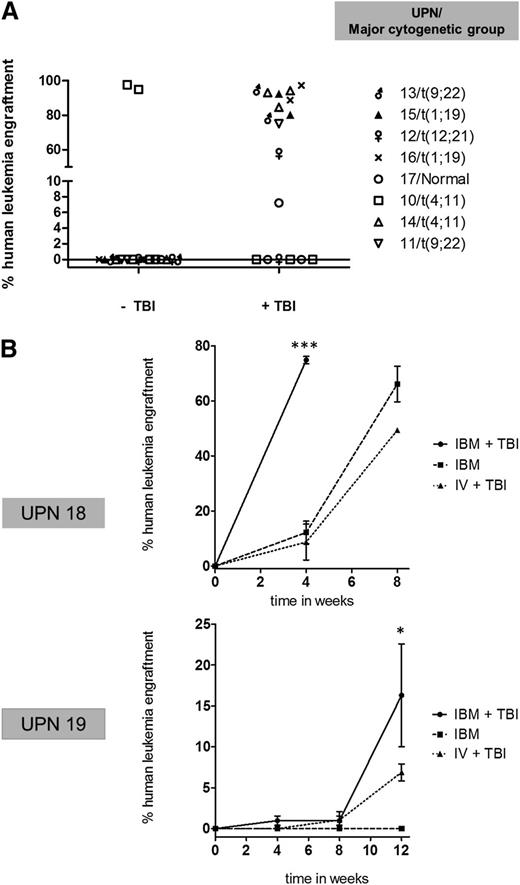
A paired analysis of the 8 samples that could be assessed by both models (with and without TBI preconditioning) is shown in Figure 1A and confirms the requirement of TBI preconditioning to facilitate non-t(4;11) pre-B ALL xenotransplantation, as the majority of samples (7/8) that failed to engraft unconditioned hosts gave rise to high levels of leukemic engraftment when TBI preconditioning was added.
Comparison of adult pre-B ALL engraftment by either unconditioned (−TBI) or 2.5-Gy TBI preconditioned (+TBI) xenotransplant assaying. (A) Comparison of percentage of engraftment in 8 samples assayed by ±TBI following IV transplantation. Symbols correspond to a single mouse. (B) Engraftment kinetics of 2 non-t(4;11) pre-B ALL samples by IBM alone, IBM injection + TBI, and IV injection + TBI. Mean leukemia engraftment (CD45+/CD19+) ± standard error of the mean (SEM) are shown at the indicated time points as ascertained by IBM sampling (first time point sampled from noninjected bone). More than 2 mice injected per condition. ***P < .0001 compared with IBM and IV + TBI. *P < .05 compared with IBM and IV + TBI.
Comparison of adult pre-B ALL engraftment by either unconditioned (−TBI) or 2.5-Gy TBI preconditioned (+TBI) xenotransplant assaying. (A) Comparison of percentage of engraftment in 8 samples assayed by ±TBI following IV transplantation. Symbols correspond to a single mouse. (B) Engraftment kinetics of 2 non-t(4;11) pre-B ALL samples by IBM alone, IBM injection + TBI, and IV injection + TBI. Mean leukemia engraftment (CD45+/CD19+) ± standard error of the mean (SEM) are shown at the indicated time points as ascertained by IBM sampling (first time point sampled from noninjected bone). More than 2 mice injected per condition. ***P < .0001 compared with IBM and IV + TBI. *P < .05 compared with IBM and IV + TBI.
Next we assessed whether this TBI-based differential engraftment behavior of non-t(4;11) pre-B ALL was also evident when cells were delivered by more efficient methods of tumor transplantation, using direct IBM injection, which bypasses the need for cells to home to the BM, the first part of the engraftment process. Figure 1B shows a comparison of the percentage leukemia engraftment (CD45+/CD19+) over time after injection of 1 × 106 cells from non-t(4;11) UPN 18 and 19. Leukemic engraftment following IBM transplantation into TBI (2.5 Gy)-irradiated vs nonirradiated recipients vs control IV injection plus TBI (2.5 Gy) were directly compared. In both cases, IBM transplantation with TBI preconditioning resulted in accelerated leukemia reconstitution, supporting significantly higher levels of CD45+/CD19+ engraftment compared with both IBM injection alone and IV injection with TBI. Notably, the absence of TBI preconditioning was the only factor associated with complete failure of engraftment of UPN 19.
Overall, these data indicate that in vivo clonal expansion of adult non-t(4;11) pre-B ALL is highly dependent on TBI-sensitive pathways that can operate independently of BM homing.
TBI preconditioning of NSG mice supports sensitive detection of non-t(4;11) pre-B ALL LICs and drug-resistant clones
To formally assess whether TBI-based xenotransplantation affects quantitative estimation of tumorigenic capacity, we determined LIC activity of 2 non-t(4;11) pre-B ALL tumors by limiting dilution transplantation using the optimized IBM injection route together with TBI preconditioning at 2.5 (UPN 20 and 21) and 2.0 Gy (UPN 21). Mice not exhibiting disease related symptoms were monitored until 6 months before assessment of engraftment. Results are shown in Table 3 and supplemental Table 2. Both non-t(4;11) pre-B ALL cases yielded a high LIC frequency (<1:6800) using the standard 2.5-Gy TBI assay conditions, and both engrafted at the lowest (103) cell dose tested. The lower dose (2.0 Gy) of TBI impaired leukemogenicity as indicated by a lower frequency of tumorigenic cells detected (UPN 21), confirming that non-t(4;11) pre-B ALL LIC activity is dependent on TBI.
LIC determination in non-t(4;11) pre-B ALL with TBI preconditioning based xenotransplantation
| UPN . | TBI dose (Gy) . | . | LIC frequency (95% confidence interval) . | |||
|---|---|---|---|---|---|---|
| Cells per injection | ||||||
| 20 | 2.5 | 1.8 × 106 | 1.8 × 105 | 1.8 × 104 | 1.8 × 103 | |
| No. of mice engrafting | <1/3900 (1/880-1/17 000) | |||||
| 3/3 | 2/2 | 3/3 | 1/3 | |||
| Cells per injection | ||||||
| 21 | 2.5 | 5.6 × 106 | — | — | 2.78 × 103 | |
| No. of mice engrafting | <1/6800 (1/960-1/49 000) | |||||
| 2/2 | ND | ND | 1/3 | |||
| Cells per injection | ||||||
| 21 | 2 | ND | 4.5 × 105 | 4.5 × 104 | 4.5 × 103 | |
| No. of mice engrafting | <1/20 000 (1/7000-1/59 000) | |||||
| ND | 4/4 | 4/4 | 0/5 | |||
| UPN . | TBI dose (Gy) . | . | LIC frequency (95% confidence interval) . | |||
|---|---|---|---|---|---|---|
| Cells per injection | ||||||
| 20 | 2.5 | 1.8 × 106 | 1.8 × 105 | 1.8 × 104 | 1.8 × 103 | |
| No. of mice engrafting | <1/3900 (1/880-1/17 000) | |||||
| 3/3 | 2/2 | 3/3 | 1/3 | |||
| Cells per injection | ||||||
| 21 | 2.5 | 5.6 × 106 | — | — | 2.78 × 103 | |
| No. of mice engrafting | <1/6800 (1/960-1/49 000) | |||||
| 2/2 | ND | ND | 1/3 | |||
| Cells per injection | ||||||
| 21 | 2 | ND | 4.5 × 105 | 4.5 × 104 | 4.5 × 103 | |
| No. of mice engrafting | <1/20 000 (1/7000-1/59 000) | |||||
| ND | 4/4 | 4/4 | 0/5 | |||
The number of mice engrafting over the total number injected is shown. LIC frequencies were calculated using the open access program ELDA (http://bioinf.wehi.edu.au/software/elda/). ND, not determined.
We additionally postulated that conditions that supported xenografting of non-t(4;11) pre-B ALL would enable proliferation of low-frequency drug-resistant clones—so-called MRD populations. Hence, we transplanted 6 polymerase chain reaction-determined MRD-positive (range, 8.33 × 10−2-3.6 × 10−4) non-t(4;11) pre-B ALL BM samples (supplemental Table 3) by IBM into TBI (2.0 Gy) preconditioned NSG mice. To prevent any attrition arising during cell sorting, we injected unpurified MRD-positive BM samples. MRD-induced leukemic engraftment (CD45+/CD19+; 83.6%) was achieved in 1 of 6 samples (UPN 25) after extended modeling of 7 months and, importantly, gave rise to overt leukemia-associated symptoms in the recipient mouse, demonstrating that MRD populations have functional disease-mediating ability. The majority of features of the original leukemia at diagnosis were reconstituted in the MRD-ALL xenograft (Figure 2A-D). Importantly, the patient-specific Ig/TCR receptor gene rearrangements that had characterized the original MRD population in the patient were also fully recapitulated in the MRD-xenografted leukemia (Figure 2E).
Functional modeling of MRD-positive ALL. (A) Schematic of the disease course and treatment from initial diagnosis of UPN 25. BM cells from the +7.8-month MRD-positive (pos) sample: 3.65 × 10−2 were transplanted into 3 individual recipient mice (8 × 10−4 nucleated cells/mouse). (B) Phenotypic characterization of BM from the single MRD engrafted recipient mouse shows CD34− xenografted leukemia; CD34+ disease was characterized at diagnosis. (C) (a) MGG staining of BM shows infiltration by monomorphic lymphoid blasts in the MRD engrafted recipient, ×60. (b) Hematoxylin and eosin–stained histological section of the liver parenchyma, ×20, and (c) human CD10 immunostaining of liver, ×20 in the MRD engrafted recipient mouse. Images were taken using Nikon Eclipse E600 microscope and Nikon DS-Fi1 camera. Black arrows indicate focal areas of leukemic infiltration. (D) FISH analysis (×100), showing presence of the original IgH@CRLF2 chromosomal rearrangement in cells harvested from BM as separated red (red arrow) and green signals (green arrow); the normal copy of the IgH gene is seen as a red-green fused signal in the same cell (yellow arrow). IgH rearrangement was detected in 97% of BM cells. (E) (Upper) Heteroduplex assessment of the original diagnostic sample from patient UPN 25 and (lower) murine BM that had engrafted with the patients MRD-ALL. MRD-ALL reproduced the identical Ig/TCR clonal rearrangements of the diagnostic sample as confirmed by sequence analysis.
Functional modeling of MRD-positive ALL. (A) Schematic of the disease course and treatment from initial diagnosis of UPN 25. BM cells from the +7.8-month MRD-positive (pos) sample: 3.65 × 10−2 were transplanted into 3 individual recipient mice (8 × 10−4 nucleated cells/mouse). (B) Phenotypic characterization of BM from the single MRD engrafted recipient mouse shows CD34− xenografted leukemia; CD34+ disease was characterized at diagnosis. (C) (a) MGG staining of BM shows infiltration by monomorphic lymphoid blasts in the MRD engrafted recipient, ×60. (b) Hematoxylin and eosin–stained histological section of the liver parenchyma, ×20, and (c) human CD10 immunostaining of liver, ×20 in the MRD engrafted recipient mouse. Images were taken using Nikon Eclipse E600 microscope and Nikon DS-Fi1 camera. Black arrows indicate focal areas of leukemic infiltration. (D) FISH analysis (×100), showing presence of the original IgH@CRLF2 chromosomal rearrangement in cells harvested from BM as separated red (red arrow) and green signals (green arrow); the normal copy of the IgH gene is seen as a red-green fused signal in the same cell (yellow arrow). IgH rearrangement was detected in 97% of BM cells. (E) (Upper) Heteroduplex assessment of the original diagnostic sample from patient UPN 25 and (lower) murine BM that had engrafted with the patients MRD-ALL. MRD-ALL reproduced the identical Ig/TCR clonal rearrangements of the diagnostic sample as confirmed by sequence analysis.
SDF-1/CXCR4 axis mediates adult non-t(4;11) pre-B ALL migration and in vivo repopulation
To understand the dependence of non-t(4;11) pre-B ALL engraftment on TBI preconditioning, we next examined TBI-sensitive stromal-derived factor-1 (SDF-1)/CXCR4 signaling and its functional role in adult pre-B ALL. Interaction of SDF-1 with receptor CXCR4 is known to mediate pre-B ALL BM homing and repopulation.23,28,29 Based on studies of pediatric disease, pre-B ALL cells are more sensitive than normal hematopoietic stem cell (HSC) progenitors to SDF-1 and respond maximally to low concentrations.23 This is taken as an explanation for why TBI preconditioning, which induces SDF-1 release by murine stromal cells,30,31 is dispensable for efficient engraftment of human (pediatric) pre-B ALL but not necessarily HSC populations.32
As TBI preconditioning significantly improved engraftment in the setting of adult non-t(4;11) pre-B ALL, this led us to hypothesize that, in this group, (1) SDF-1/CXCR4 had an important functional role and (2) sensitivity to SDF-1 would be more comparable to human HSC populations.
To investigate, we first determined expression of CXCR4 both at the cell surface and intracellularly in both non-t(4;11) (n = 7) and t(4;11) (n = 3) rearranged pre-B ALL and MPBSCs (n = 3). Figure 3A and Table 4 show expected patterns of CXCR4 expression.23 Pre-B ALL cells, irrespective of t(4;11) status, had higher levels of receptor expression at the surface of the cell compared to inside the cell (intracellularly), whereas the distribution was reversed in CD34+ MPBSC, with CXCR4 predominantly localized within the cells. Notably, we found differences in the level of receptor expression between adult ALL subtypes with t(4;11) pre-B ALL exhibiting a significantly lower percentage of cells expressing CXCR4 expression at the cell surface compared with non-t(4;11) pre-B ALL (8.7± 3.3% vs 59.7 ± 10.6%, P = .02).
Role of the SDF-1/CXCR4 axis in adult ALL migration and engraftment. (A) Representative histograms of CXCR4 receptor expression in t(4;11), non-t(4;11) pre-B ALL, and CD34+ MPBSCs. Illustrations correspond to cases 3, 7, and 1, respectively, in Table 4. CXCR4 at the cell surface (gray line), intracellularly (dotted line), and isotype control (black line) are shown. (B) Transwell migration toward a gradient of SDF-1 [0-1000 ng/mL in non-t(4;11), n = 7; t(4;11), n = 3; and CD34+ MPBSCs, n = 3]. Results show mean ± SEM of fold change in absolute numbers of migrating cells per microliter normalized to spontaneous migration without SDF-1. Results are from 3 to 5 independent experiments from each group.*P < .05 compared with respective control; ●P < .05; ▲P = .07. (C) Non-t(4;11) pre-B ALL cells from 2 patients were pretreated with vehicle or AMD1300 and then assayed for transwell migration toward SDF-1 (125 ng/mL). The number of migrating cells was determined after 4 hours in transwell culture. Data points show duplicates for each sample together with mean ± SEM. **P = .003 compared with vehicle. (D) Pre-B ALL cells from a single patient were treated with isotype control or anti-CXCR4 blocking antibody. Results show (lower) the percentage of CD45+/CD19+ cells in BM +14 days after injection in representative mice and (upper) the absolute number of leukemia cells in harvested BM 4 weeks following injection in individual recipients. Four mice were used for each treatment. *P = .03 compared with control.
Role of the SDF-1/CXCR4 axis in adult ALL migration and engraftment. (A) Representative histograms of CXCR4 receptor expression in t(4;11), non-t(4;11) pre-B ALL, and CD34+ MPBSCs. Illustrations correspond to cases 3, 7, and 1, respectively, in Table 4. CXCR4 at the cell surface (gray line), intracellularly (dotted line), and isotype control (black line) are shown. (B) Transwell migration toward a gradient of SDF-1 [0-1000 ng/mL in non-t(4;11), n = 7; t(4;11), n = 3; and CD34+ MPBSCs, n = 3]. Results show mean ± SEM of fold change in absolute numbers of migrating cells per microliter normalized to spontaneous migration without SDF-1. Results are from 3 to 5 independent experiments from each group.*P < .05 compared with respective control; ●P < .05; ▲P = .07. (C) Non-t(4;11) pre-B ALL cells from 2 patients were pretreated with vehicle or AMD1300 and then assayed for transwell migration toward SDF-1 (125 ng/mL). The number of migrating cells was determined after 4 hours in transwell culture. Data points show duplicates for each sample together with mean ± SEM. **P = .003 compared with vehicle. (D) Pre-B ALL cells from a single patient were treated with isotype control or anti-CXCR4 blocking antibody. Results show (lower) the percentage of CD45+/CD19+ cells in BM +14 days after injection in representative mice and (upper) the absolute number of leukemia cells in harvested BM 4 weeks following injection in individual recipients. Four mice were used for each treatment. *P = .03 compared with control.
CXCR4 receptor expression studies
| . | . | non t(4;11) . | . | t(4;11) . | . | MPBSC . | |||
|---|---|---|---|---|---|---|---|---|---|
| . | Case . | Surface . | Intra . | Case . | Surface . | Intra . | Case . | Surface . | Intra . |
| 1 | 17.1 | 0.8 | 1 | 2.19 | 1.4 | 1 | 67.2 | 82 | |
| 2 | 50.9 | 3.3 | 2 | 13.1 | 8.5 | 2 | 46.1 | 83.6 | |
| 3 | 50.3 | 38.8 | 3 | 10.9 | 8.2 | 3 | 82.3 | 84.4 | |
| 4 | 51.3 | 12.1 | |||||||
| 5 | 84.1 | 28.7 | |||||||
| 6 | 88.9 | 78 | |||||||
| 7 | 75.1 | 36.7 | |||||||
| Mean ± SE | 59.7 ± 9.4 | 28.4 ± 10.1 | 8.7 ± 3.3 | 6.0 ± 2.3 | 65.2 ± 10.5 | 83.3 ± 0.7 | |||
| . | . | non t(4;11) . | . | t(4;11) . | . | MPBSC . | |||
|---|---|---|---|---|---|---|---|---|---|
| . | Case . | Surface . | Intra . | Case . | Surface . | Intra . | Case . | Surface . | Intra . |
| 1 | 17.1 | 0.8 | 1 | 2.19 | 1.4 | 1 | 67.2 | 82 | |
| 2 | 50.9 | 3.3 | 2 | 13.1 | 8.5 | 2 | 46.1 | 83.6 | |
| 3 | 50.3 | 38.8 | 3 | 10.9 | 8.2 | 3 | 82.3 | 84.4 | |
| 4 | 51.3 | 12.1 | |||||||
| 5 | 84.1 | 28.7 | |||||||
| 6 | 88.9 | 78 | |||||||
| 7 | 75.1 | 36.7 | |||||||
| Mean ± SE | 59.7 ± 9.4 | 28.4 ± 10.1 | 8.7 ± 3.3 | 6.0 ± 2.3 | 65.2 ± 10.5 | 83.3 ± 0.7 | |||
Values represent the percentage of cells expressing CXCR4. Intra, intracellular; SE, standard error.
To determine sensitivity to SDF-1, we compared the capacity of 10 different primary adult pre-B ALL tumors to migrate toward increasing concentrations of SDF-1 (0, 10, 50, 125, and 1000 ng/mL) to that of CD34+ MPBSCs using a transwell system. Figure 3B shows the fold change compared with control (no SDF-1). Both non-t(4;11) pre-B ALL and CD34+ MPBSCs were significantly SDF-1 responsive, with the maximal (55.6 ± 27.1-fold) migratory response of non-t(4;11) pre-B ALL tumors occurring at 125 ng/mL of SDF-1, which is higher than that reported in pediatric ALL (50 ng/mL)23 but lower than that of CD34+ MPBSCs (1000 ng/mL). Migration was significantly (P = .016) inhibited in non-t(4;11) pre-B ALL (but not CD34+ MPBSCs) at the highest (1000 ng/mL) concentration of SDF-1, reflecting the expected aberrant cell surface receptor desensitization in malignant cells.23,29 Notably, t(4;11)-positive tumors were relatively less sensitive to SDF-1-induced migration (maximum fold change, 3.13 ± 1.4), which, together with the lower cell surface expression of CXCR4, may explain the lack of preferential NSG engraftment of this ALL subgroup with the addition of TBI preconditioning.
To further confirm the role of SDF-1/CXCR4 in non-t(4;11) pre-B ALL migration, leukemic blasts from 2 patients were pretreated with AMD3100, a small peptide antagonist of CXCR-4, and SDF-1 induced directional migration at the optimal, 125-ng/mL, dose assessed. Figure 3C shows that AMD3100 significantly blocks SDF-1 (125 ng/mL)-induced directional migration. In contrast, directional migration was not inhibited by AMD3100 in t(4;11) pre-B ALL (supplemental Figure 1).
In addition to migration, SDF-1 also promotes survival30 and proliferation33,34 of pre-B ALL cells. Hence, we investigated the role of SDF-1/CXCR4 on long-term engraftment of primary non-t(4;11) pre-B ALL. Leukemia cells from UPN 18 were pretreated with anti-CXCR4 antibody prior to injection in TBI (2.5 Gy)-preconditioned hosts. At +14 days, human CD45+/19+ engraftment was detected in all control mice (mean, 30.1 ± 12.9%), whereas no human cells could be detected in the BM of recipients that had received cells pretreated with CXCR4 blocking antibody (Figure 3D, lower). At 4 weeks, although all mice had some evidence of ALL engraftment, anti-CXCR4 pretreatment was significantly associated with reduced human leukemia cell chimerism in the BM (Figure 3D, upper).
Taken together, these data suggest that the SDF-1/CXCR4 interaction differentially regulates the migration of adult non-t(4;11) pre-B ALL and is also required for long-term in vivo development of this ALL subtype. Furthermore, aberrant patterns of CXCR4 receptor expression and SDF-1-induced migration (including receptor desensitization) can also be a feature of TBI responsive pre-B ALL.
Characterization of adult pre-B ALL xenografts
A major goal of this study was to establish robust in vivo tumor models that faithfully recapitulate human adult ALL. Thus, we determined the clinical, immunophenotypic, and genetic characteristics of ALL in mice that developed leukemia to assess the relevance of our models to human disease. Mice developing leukemia-related symptoms invariably exhibited splenomegaly (supplemental Figure 2) with BM and organs completely effaced by infiltrating monomorphic blasts (supplemental Figure 3), consistent with overt disseminated leukemia development.
To assess the stability of the immunophenotype after NSG passage, we performed detailed 6-color flow cytometric analysis of all leukemia xenografts derived from the 2 samples (UPN 20 and 21) that were subjected to limiting dilution transplantation (Table 3). Although there was overall preservation of the original immunophenotype, some changes in the repopulating leukemia were observed in select cases (supplemental Figure 4A-B). This manifested as either loss of antigen expression (Figure 4A; supplemental Figure 4A) or loss of immunophenotypically defined blast populations (supplemental Figure 4B).
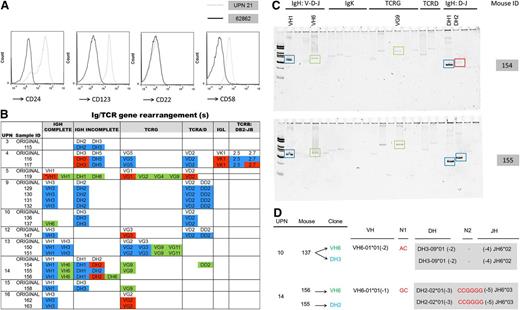
Characterization of adult ALL xenografts. (A) Comparison of flow cytometric analysis of original ALL from UPN 21 (gray dashed) and resulting leukemia xenograft from BM of mouse 62862 (black) showing down-regulation of CD24, CD123, and CD58 expression. CD19+/CD34+ gates are presented. (B) Schematic of clonal Ig/TCR rearrangements in the original sample (non-color highlighted text) compared with the xenografted leukemia. Ig/TCR rearrangements that are preserved (blue), lost (red), or newly emergent (green) in leukemia xenografts are color highlighted. *Newly emergent VH1 clone accompanied loss of the original VH1 rearrangement in this xenograft. (C) Heteroduplex gel images of PCR-amplified Ig/TCR rearrangements in BM from 2 leukemic mice (154 and 155), both transplanted with cells from UPN 14 showing differences in clonal composition between engrafted leukemias. Preservation (blue box) and loss (red box) of original rearrangements and newly emergent clones (green box) are indicated. (D) Newly emergent VH6 clones (highlighted green) in xenografts share identical DH-N-JH stems (gray box) with rearrangements that predominated in the original sample (highlighted blue), indicating that clonal evolution by variable (VH) gene addition to preexisting DH-JH rearrangements had occurred in the respective human tumor.
Characterization of adult ALL xenografts. (A) Comparison of flow cytometric analysis of original ALL from UPN 21 (gray dashed) and resulting leukemia xenograft from BM of mouse 62862 (black) showing down-regulation of CD24, CD123, and CD58 expression. CD19+/CD34+ gates are presented. (B) Schematic of clonal Ig/TCR rearrangements in the original sample (non-color highlighted text) compared with the xenografted leukemia. Ig/TCR rearrangements that are preserved (blue), lost (red), or newly emergent (green) in leukemia xenografts are color highlighted. *Newly emergent VH1 clone accompanied loss of the original VH1 rearrangement in this xenograft. (C) Heteroduplex gel images of PCR-amplified Ig/TCR rearrangements in BM from 2 leukemic mice (154 and 155), both transplanted with cells from UPN 14 showing differences in clonal composition between engrafted leukemias. Preservation (blue box) and loss (red box) of original rearrangements and newly emergent clones (green box) are indicated. (D) Newly emergent VH6 clones (highlighted green) in xenografts share identical DH-N-JH stems (gray box) with rearrangements that predominated in the original sample (highlighted blue), indicating that clonal evolution by variable (VH) gene addition to preexisting DH-JH rearrangements had occurred in the respective human tumor.
To assess the molecular relevance of the engrafted leukemia to that of the original sample, we screened a subset (n = 19) of xenografts derived from 10 patients (UPN 3-5, 9,10, and 12-16) for >70 Ig/TCR gene rearrangements. A schematic summarizing results for the 19 analyzed xenografts together with the Ig/TCR clonal profile of the original corresponding leukemia is shown in Figure 4B.
In all cases, the xenograft was closely related to the original leukemia as indicated by the preservation of some of the original Ig/TCR gene rearrangements. However, 12 of 19 xenografts derived from UPN 4, 5, 10, and 12 to 16 also exhibited a loss or gain from the original Ig/TCR repertoire, with 20 of 33 (60.6%) changes being a gain. Interestingly, changes in the Ig/TCR profile were evident between xenograft recipients injected with the same sample (Figure 4C). In certain cases, newly emergent Ig/TCR rearrangements in the xenograft could clearly be backtracked as descending from clones that had predominated in the original tumor (Figure 4D). Overall, propagation of the original genetic rearrangements together with the loss of genetic markers in leukemia xenografts strongly supports the existence of multiple subclones in the original ALL that vary in their ALL tumorigenic capability. These findings are consistent with intratumoral genetic and functional heterogeneity that has been described in ALL20,35 and with the xenotransplantation assay enabling segregation of subclonal ALL architecture.
Discussion
This report details the largest investigation of engraftment model systems for primary adult ALL. The experimental systems defined here help inform future studies looking into mechanisms of disease pathogenesis in adult ALL, specifically the cancer stem cell model,36 which critically relies on optimized xenotransplantation approaches for assigning serial leukemia propagating ability. Although it is clear that unconditioned xenotransplant assays reliably and reproducibly reinitiate a diverse range of pediatric ALL9,12,27 and enable elucidation of a broad spectrum of self-renewing LSC populations,9,10,13 our study suggests that conditions for optimally establishing in vivo leukemogenic capability and self-renewing LSC populations in adult ALL are distinct.
An unmodified murine host environment either did not support or was significantly less efficient in supporting tumor modeling of primary non-t(4;11) pre-B adult ALL in IV and IBM xenotransplant models, respectively. However, adaptation of murine hosts with TBI preconditioning enabled virtually all non-t(4;11) ALL tumors to engraft and proliferate. Furthermore, there was no evidence that TBI preconditioning was detrimental to pre-B ALL leukemic engraftment as suggested from pediatric cell line data.23
In contrast, t(4;11) pre-B ALL did not absolutely require TBI for successful engraftment and was less SDF-1 responsive. Furthermore, we found that TBI preconditioning may in fact interfere with adult t(4;11) engraftment (supplemental Figure 5). Together, these data suggest that adult t(4;11) ALL may harbor distinct engraftment requirements that is not inconsistent with the unique clinical,37 immunophenotypic,38 and genetic/epigenetic39 nature of this ALL subgroup. Further study is required into the conditions that best reveal the intrinsic oncogenic potential of primary adult t(4;11) ALL, not only for understanding the biology of this extremely poor disease group, but also, importantly, because engineered animal models that faithfully recapitulate this ALL subtype are lacking.40
Arguably, a principal means by which sublethal TBI augments human cell engraftment in an already heavily immunocompromised NSG environment is by induction of key growth promoting cross-reactive soluble factors.41 Human and murine SDF-1 are cross-reactive.42 In TBI-responsive adult non-t(4;11) pre-B ALL, the SDF-1/CXCR4 axis had an important functional role in cell migration and long-term in vivo repopulation. Hence, the improved engraftment of non-t(4;11) pre-B ALL with TBI preconditioning is likely due to induction of a tumor-promoting cytokine milieu that creates a supportive leukemia niche. This points to extrinsic (SDF-1) microenvironment-derived cues playing a crucial role in driving clonal expansion of non-t(4;11) adult pre-B ALL.
Because SDF-1/CXCR4 interaction is also required for in vivo tumor initiation of pediatric pre-B ALL,23,29 the seemingly distinct xenografting requirement of adult non-t(4;11) pre-B ALL compared with equivalent subtypes described in pediatric studies merits some discussion. Maximum migration was achieved with higher (125 ng/mL) SDF-1 levels than in pediatric pre-B ALL,23 raising the intriguing possibility that adult non-t(4;11) pre-B ALL cells are intrinsically less sensitive to SDF-1 stimulation. Lending support to such an age-related variation in SDF-1 response is the lower migration potential of adult CD34+ cells compared with cord blood CD34+ progenitors following SDF-1 stimuation.43 Of course, formal proof of a difference in this niche-based interaction between adult and pediatric ALL would require head-to-head comparison of SDF-1 sensitivity within a single study together with assessment of other determinants of repopulation potential (eg, cell cycle status and LIC frequency). Pending such an investigation, we might predict that, although a single phase 1 study using the CXCR4 antagonist, AMD1300, has reported disappointing results in pediatric ALL,44 adult non-t(4;11) pre-B ALL tumors may exhibit greater vulnerability to niche-based therapeutic strategies using CXCR4 blockade given their requirement for higher levels of SDF-1. Further study is required to determine whether age-related tumor microenvironment variation contributes to the significant differences observed in the clinical behavior of adult and pediatric ALL.
Importantly, many aspects of the original ALL were mirrored in our engraftment models, confirming the high potential for use of these systems as preclinical tools. A potentially exploitable aspect is the segregation of subclones on xenografting of the primary ALL tumor and exposure of minor populations not apparent in the original sample. Such a phenomenon is well described.20,36,45 Clonal selection, indicated by the loss of existing Ig/TCR rearrangements and unmasking of previously unidentified clones in leukemia xenografts, suggests that the xenotransplantation assay offers an opportunity to interrogate functional aspects of subclonal ALL including differential therapy sensitivity and its relevance to clonal selection. The importance of intraclonal assessment in ALL pathogenesis is underscored by multiple reports that point to subclonal origins of relapsed ALL.35,46-48
MRD populations are of considerable interest given the strong association with treatment failure both in children49,50 and adults.51-53 Due to the low frequency of these cells, little is known about the genetic or cellular basis of these chemo-resistant populations, and therefore strategies for their interrogation are of considerable interest. To the best of our knowledge, this is the first description of experimental modeling of primary MRD-ALL. However, despite optimized assay conditions (IBM transplantation plus TBI preconditioning), only 1 of 6 samples successfully engrafted. As MRD positivity was molecularly determined opposed to being determined by flow cytometric assessment, it cannot be certain the residual leukemic populations in each case were viable and therefore capable of functional in vivo repopulation. Further progress toward the development of a reproducible model of MRD-ALL may be gained through a cell sorting strategy in which live cells exhibiting a leukemia associated immunophenotype are directly transplanted under supportive xenotransplant assay conditions. Additionally, we applied a lower (2.0 Gy) TBI dose for modeling MRD with the rationale that this would provide a competitive advantage over HSC populations which may also engraft. A higher TBI dose (2.5 Gy) may yield further improvements in the rate at which MRD-positive populations engraft, which would then make this approach feasible. We are currently investigating this possibility.
In summary, our data establish the parameters of a functional system for in vivo study of adult pre-B ALL. We identified distinct in vivo growth requirements between adult ALL subtypes and potentially with that of pediatric ALL, underlining the importance of age/leukemia subtype-specific investigation. Our optimized system provides a powerful tool for characterizing serially transplantable adult ALL stem cells and potentially treatment-refractory MRD populations. Such studies should yield improvements in our understanding of disease biology in adult ALL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Mohammad Elham, Rachel Mitchell, and Krisztina Zuborne Alapi (UK Adult ALL Molecular Diagnostic Laboratory) for sample processing and Professor Tsvee Lapidot and Dr Orit Kollet for insightful discussions. Professor Stephen Mackinnon is gratefully acknowledged for supporting the research.
This work was supported by Leukaemia Lymphoma Research grants 07062 (B.P.), 09026 (A.Z.C. and A.K.F.), 10005, (T.E.), and 10007 (J.S.); Specialist Programme grant 11004 (A.V.M. and C.S.); Comprehensive Biomedical Research Centre National Institute of Health grant G502 (T.E.); Cancer Research UK grant CRUK/09/006 (A.K.F.); and Gabrielle’s Angels Foundation (B.P.).
Authorship
Contribution: B.P. designed the study, performed research, analyzed data, and wrote the manuscript; A.D., A.Z.C., C.S., E.S., J.S., B.B., N.Z., C.Y.Z., L.R. and A.K.F. performed research; A.K.F. and A.V.M. provided research support; and A.K.F. and T.E. critically assessed the data and contributed to writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bella Patel, University College London, Royal Free Campus, Rowland Hill St, London NW3 2PF, United Kingdom; e-mail b.patel@ucl.ac.uk.


![Figure 3. Role of the SDF-1/CXCR4 axis in adult ALL migration and engraftment. (A) Representative histograms of CXCR4 receptor expression in t(4;11), non-t(4;11) pre-B ALL, and CD34+ MPBSCs. Illustrations correspond to cases 3, 7, and 1, respectively, in Table 4. CXCR4 at the cell surface (gray line), intracellularly (dotted line), and isotype control (black line) are shown. (B) Transwell migration toward a gradient of SDF-1 [0-1000 ng/mL in non-t(4;11), n = 7; t(4;11), n = 3; and CD34+ MPBSCs, n = 3]. Results show mean ± SEM of fold change in absolute numbers of migrating cells per microliter normalized to spontaneous migration without SDF-1. Results are from 3 to 5 independent experiments from each group.*P < .05 compared with respective control; ●P < .05; ▲P = .07. (C) Non-t(4;11) pre-B ALL cells from 2 patients were pretreated with vehicle or AMD1300 and then assayed for transwell migration toward SDF-1 (125 ng/mL). The number of migrating cells was determined after 4 hours in transwell culture. Data points show duplicates for each sample together with mean ± SEM. **P = .003 compared with vehicle. (D) Pre-B ALL cells from a single patient were treated with isotype control or anti-CXCR4 blocking antibody. Results show (lower) the percentage of CD45+/CD19+ cells in BM +14 days after injection in representative mice and (upper) the absolute number of leukemia cells in harvested BM 4 weeks following injection in individual recipients. Four mice were used for each treatment. *P = .03 compared with control.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/1/10.1182_blood-2014-01-549352/4/m_96f3.jpeg?Expires=1765923031&Signature=qT8ir7FaFnSWqy5bCkzrtR4un4-dpwJXk7R~qJ7WJYZuWXOBClV7KoOT67pG3WBn2MRoix814LrTy99aeB6FxB2tcbuCsD6JwesA3EoyPIYX9unkERgjZG19okBucF1sWtiR~vjsd82udoZNW4a7daDVwbLGkG2-22R0mq-zEh1FxftN744Qp38Xk2QdE-Hw2eFGzE4swCgmWCRaO6T4cHF44PR21rrqS23PKpWLnsx6-3Y0d54S99y1sqr2AXn4M-ZZFHpYVpi-1UHtSUU9C-YJ1n3kRzRCmsUPadHfxS9WjK7ZZwMgeJ0f-Y64P9C47MVMNDANWJxliPjetbG6AQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)