Key Points
Two distinct regions of transmembrane somatic mutations in type I cytokine receptors IL7R and CRLF2 exist in acute lymphoblastic leukemias.
Noncysteine transmembrane mutations cause functional receptor dimerization and activation transforming pro-B cells.
Abstract
Gain-of-function somatic mutations introducing cysteines to either the extracellular or to the transmembrane domain (TMD) in interleukin-7 receptor α (IL7R) or cytokine receptor-like factor 2 (CRLF2) have been described in acute lymphoblastic leukemias. Here we report noncysteine in-frame mutations in IL7R and CRLF2 located in a region of the TMD closer to the cytosolic domain. Biochemical and functional assays showed that these are activating mutations conferring cytokine-independent growth of progenitor lymphoid cells in vitro and are transforming in vivo. Protein fragment complementation assays suggest that despite the absence of cysteines, the mechanism of activation is through ligand-independent dimerization. Mutagenesis experiments and ConSurf calculations suggest that the mutations stabilize the homodimeric conformation, positioning the cytosolic kinases in predefined orientation to each other, thereby inducing spontaneous receptor activation independently of external signals. Hence, type I cytokine receptors may be activated in leukemia through 2 types of transmembrane somatic dimerizing mutations.
Introduction
Interleukin-7 receptor α (IL7R) dimerizes with cytokine receptor-like factor 2 (CRLF2) to form the receptor for thymic stromal lymphopoietin and with interleukin-2 receptor γ to form the receptor for IL-7.1,2 We and others described acquired activating mutations in acute lymphoblastic leukemias (ALL) that insert cysteines into the juxtamembrane domains of IL7R or CRLF2 causing ligand independent dimerization via disulfide bonds.3-5 The creation of disulfide bonds is critical for the activation of the receptors because elimination of the cysteines abrogated the cytokine independent growth.4,6,7
Here we report and analyze a novel class of noncysteine mutations in cytokine type I receptors in ALL leading to ligand independent activation.
Study design
Patient samples
All specimens were collected with an informed consent and the approval of ethics committees.4 Samples were anonymized for the study. The study was approved by the Israeli Health Ministry Ethic committee, approval # 920070771.
Molecular studies
Mutation detection and analysis using appropriate primers (supplemental Table 1; see supplemental Data available at the Blood Web site). was performed as described.4,8 The human IL7R cloned into the MSCV-IRES-GFP and the human CRLF2 cloned into pMX-Puro retroviral vectors were used as templates for the generation of mutations by site-directed mutagenesis (QuikChange; Stratagene).
BaF3 cells were transduced with retroviruses and were sorted by flow cytometry (FACSAria; BD Biosciences) 2 to 4 days later using appropriate antibodies. BaF3 growth assays and immunoprecipitation and western analyses were performed as described previously.9
Protein fragment complementation assay
Mutated and wild-type (WT) cDNAs of human IL7R, CRLF2, and interleukin-2 receptor γ were inserted in pcDNA3.1/Zeo vector upstream of either the hGluc1 or hGluc2 fragments of Gaussia princeps luciferase10 and were cotransfected into HEK293 cells (see supplemental Methods). Signal intensities were read on an Infinite 200 reader (TECAN).
In vivo experiments
Six-week-old female Balb/c mice were injected intravenously with 1*106 BaF3 cells transduced with either WT or mutated CRLF2 or IL7R. Survival of the mice was monitored, and tissues from the sick mice were subjected to flow cytometry analysis (Galios; Coulter Inc.).
Results and discussion
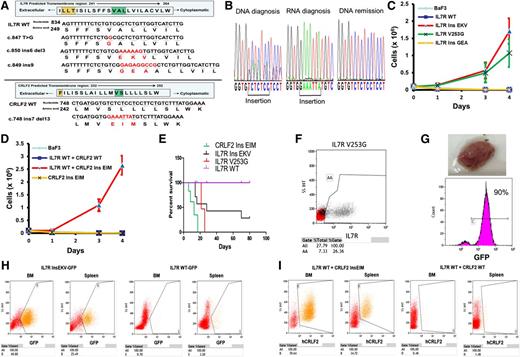
During our previously reported screens of childhood ALLs4,8 we identified mutations in IL7R and in CRLF2 that did not introduce cysteines (Figure 1A). All mutations were heterozygous, somatic, and the mutated mRNA was expressed (Figure 1B). Examination of published data showed additional similar mutations in IL7R (supplemental Table 2). These mutations cluster within a transmembranous region internal to the area afflicted by the cysteine mutations (Figure 1A; supplemental Figure 1). To test if the somatic mutations in IL7R and CRLF2 are gain-of-function mutations, BaF3 lines expressing either WT or mutated IL7R or CRLF2 were created. All proteins were expressed at the cell membrane (supplemental Figure 2). Mutated IL7RinsEKV, IL7RV253G, and CRLF2ins EIM enabled cytokine-independent growth of BaF3 cells. CRLF2insEIM required the formation of the thymic stromal lymphopoietin receptor by coexpression of IL7RWT (Figure 1C,D). One construct IL7RinsGEA did not transform BaF3 (Figure 1C). The transforming activity was confirmed in vivo in syngeneic Balb/c mice that developed fatal leukemia after intravenous injections of BaF3 cells expressing IL7RinsEKV, IL7RV253G, or CRLF2insEIMIL7RWT (Figure 1E), manifested by infiltration of the blood (Figure 1F), bone marrow, and spleen (Figure 1H-I) with the transformed cells. Similarly, subcutaneous injection caused lymphoma (Figure 1G). Thus noncysteine mutational activation of IL7R and CRLF2 is leukemogenic.
Noncysteine mutations in IL7R and CRLF2 are transforming leukemogenic mutations. (A) Predicted transmembrane domain (TMD) of IL7R and CRLF2 with location of cysteine mutations (orange box) and noncysteine mutations (green box) and alignment of WT and mutated TMD sequences. Numbers show the positions of nucleotides and corresponding amino acids. The inserted nucleotides and amino acids are shown in red. (B) Expression of CRLF2InsEIM mutation. The mutated allele is expressed in the RNA from diagnosis whereas only the normal allele is found in the remission DNA sample because CRLF2 is expressed only in leukemic B-cell as a result of a chromosomal rearrangement.8 (C,D) Cytokine withdrawal assay of BaF3 cells transduced with mutated or WT IL7R or CRLF2. (E) Overall survival of mice (7 mice in each group, 3 experiments) injected intravenously with 1-2*106 BaF3 cells expressing IL7R, WT or mutant, or IL7RWT and CRLF2insEIM compared by Kaplan-Meier analysis P ≤ .01. (F) Representative flow cytometric analysis of blood from mice injected intravenously with 1-2*106 BaF3 cells expressing IL7RV253G. (G) Lymphoma tumor (top) observed in a mouse injected subcutaneously with 1-2*106 BaF3 cells expressing IL7RIns EKV-GFP. Representative GFP expression from lymphoma cells (bottom). (H) Representative analysis of cells from spleen and bone marrow from mice injected intravenously with 1-2*106 BaF3 cells expressing IL7RIns EKV and sacrificed at day 14. (I) Representative analysis of cells from spleen and bone marrow from mice injected intravenously with 1-2*106 BaF3 cells coexpressing the IL7RWT and CRLF2WT or CRLF2insEIM mutant, and sacrificed at day 16. Del, deletion; Ins, insertion; WT, wild type.
Noncysteine mutations in IL7R and CRLF2 are transforming leukemogenic mutations. (A) Predicted transmembrane domain (TMD) of IL7R and CRLF2 with location of cysteine mutations (orange box) and noncysteine mutations (green box) and alignment of WT and mutated TMD sequences. Numbers show the positions of nucleotides and corresponding amino acids. The inserted nucleotides and amino acids are shown in red. (B) Expression of CRLF2InsEIM mutation. The mutated allele is expressed in the RNA from diagnosis whereas only the normal allele is found in the remission DNA sample because CRLF2 is expressed only in leukemic B-cell as a result of a chromosomal rearrangement.8 (C,D) Cytokine withdrawal assay of BaF3 cells transduced with mutated or WT IL7R or CRLF2. (E) Overall survival of mice (7 mice in each group, 3 experiments) injected intravenously with 1-2*106 BaF3 cells expressing IL7R, WT or mutant, or IL7RWT and CRLF2insEIM compared by Kaplan-Meier analysis P ≤ .01. (F) Representative flow cytometric analysis of blood from mice injected intravenously with 1-2*106 BaF3 cells expressing IL7RV253G. (G) Lymphoma tumor (top) observed in a mouse injected subcutaneously with 1-2*106 BaF3 cells expressing IL7RIns EKV-GFP. Representative GFP expression from lymphoma cells (bottom). (H) Representative analysis of cells from spleen and bone marrow from mice injected intravenously with 1-2*106 BaF3 cells expressing IL7RIns EKV and sacrificed at day 14. (I) Representative analysis of cells from spleen and bone marrow from mice injected intravenously with 1-2*106 BaF3 cells coexpressing the IL7RWT and CRLF2WT or CRLF2insEIM mutant, and sacrificed at day 16. Del, deletion; Ins, insertion; WT, wild type.
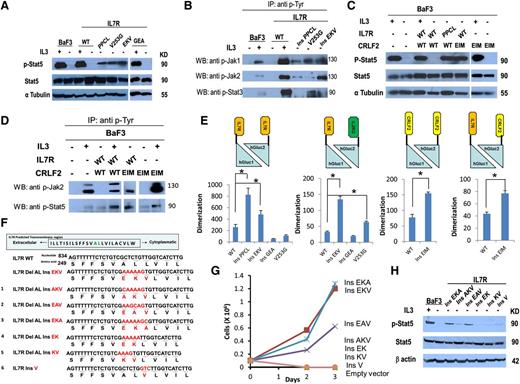
Biochemical analysis was consistent with the growth assays. Stat5 (Figure 2A), Jak1, Jak2, and Stat3 (Figure 2B) were phosphorylated in the absence of cytokine in BaF3 cells transduced with the IL7RinsPPCL (positive control that constitutively activates the JAK-STAT pathway),4 IL7RV253G, and IL7RinsEKV but not in cells expressing IL7RWT or IL7RinsGEA. In IL7RinsEKV we found higher expression of Jak2, which correlated with the higher activation capacity of this mutant in BaF3 cytokine withdrawal assays. Similarly, coexpression of IL7RWT with CRLF2insEIM, but not the expression of CRLF2insEIM by itself, caused constitutive phosphorylation of Stat5 and Jak2 (Figure 2C,D) consistent with the known binding of Jak2 to CRLF211 .
Noncysteine mutations in IL7R and CRLF2 cause constitutive activation of the JAK-STAT pathway and increase receptor dimerization. (A) Constitutive phosphorylation of Stat5 in BaF3 cells expressing IL7R mutants after 5 hours of cytokine deprivation. One mutant with insertion of cysteine (IL7RinsPPCL) was used as a positive control. IL-3+ indicates cells harvested after 5 hours of IL-3 deprivation followed by 20 minutes of IL-3 stimulation. (B) Identification of constitutive phosphorylation of Jak1, Jak2, and Stat3 in BaF3 cells expressing IL7R mutants after 5 hours of cytokine deprivation. IL-3+ indicates cells harvested after 5 hours of IL-3 deprivation followed by 20 minutes of IL-3 stimulation. Cells were subjected to lysis and immunoprecipitation with anti p-Tyr antibody (sc-508; Santa Cruz Biotechnology). The presence of Jak1, Jak2, and Stat3 was visualized by western blotting with anti-Jak1/Jak2/Stat3 antibodies. (C) Constitutive phosphorylation of Stat5 in BaF3 cells expressing CRLF2 mutant after 5 hours of cytokine deprivation. IL-3+ indicates cells harvested after 5 hours of IL-3 deprivation followed by 20 minutes of IL-3 stimulation. (D) Identification of constitutive phosphorylation of Jak2 and Stat5 in BaF3 cells expressing CRLF2 mutants after 5 hours of cytokine deprivation. IL-3+ indicates cells harvested after 5 hours of IL-3 deprivation followed by 20 minutes of IL-3 stimulation. Cells were subjected to lysis and immunoprecipitation with anti p-Tyr antibody (sc-508; Santa Cruz Biotechnology). The presence of Jak2 and Stat5 was visualized by western blotting with anti-Jak2 or Stat5 antibodies. (E) Relative dimerization level of IL7R and CRLF2 from HEK293 cells transiently transfected with the WT or mutant receptor. Dimerization was calculated by dividing luminescence by the mean fluorescence intensity of each treatment, normalizing the luminescence signal for experimental variability resulting from transfection efficiency. * P < .01, 1-way ANOVA, and Student t test. (F) Alignment of WT, natural mutant (EKV), and 6 experimental mutants (numbered 1 to 6) in the TMD of IL7R. Numbers show the positions of nucleotides and corresponding amino acids. The inserted nucleotides and amino acids are shown in red. The deleted AL amino acids are shown in green. (G) Cytokine withdrawal assay of BaF3 cells transduced with IL7R experimental mutants. (H) Constitutive phosphorylation of Stat5 in BaF3 cells expressing IL7R experimental mutants after 5 hours of cytokine deprivation. IL-3+ indicates cells harvested after 5 hours of IL-3 deprivation followed by 20 minutes of IL-3 stimulation. Del, deletion; Ins, insertion.
Noncysteine mutations in IL7R and CRLF2 cause constitutive activation of the JAK-STAT pathway and increase receptor dimerization. (A) Constitutive phosphorylation of Stat5 in BaF3 cells expressing IL7R mutants after 5 hours of cytokine deprivation. One mutant with insertion of cysteine (IL7RinsPPCL) was used as a positive control. IL-3+ indicates cells harvested after 5 hours of IL-3 deprivation followed by 20 minutes of IL-3 stimulation. (B) Identification of constitutive phosphorylation of Jak1, Jak2, and Stat3 in BaF3 cells expressing IL7R mutants after 5 hours of cytokine deprivation. IL-3+ indicates cells harvested after 5 hours of IL-3 deprivation followed by 20 minutes of IL-3 stimulation. Cells were subjected to lysis and immunoprecipitation with anti p-Tyr antibody (sc-508; Santa Cruz Biotechnology). The presence of Jak1, Jak2, and Stat3 was visualized by western blotting with anti-Jak1/Jak2/Stat3 antibodies. (C) Constitutive phosphorylation of Stat5 in BaF3 cells expressing CRLF2 mutant after 5 hours of cytokine deprivation. IL-3+ indicates cells harvested after 5 hours of IL-3 deprivation followed by 20 minutes of IL-3 stimulation. (D) Identification of constitutive phosphorylation of Jak2 and Stat5 in BaF3 cells expressing CRLF2 mutants after 5 hours of cytokine deprivation. IL-3+ indicates cells harvested after 5 hours of IL-3 deprivation followed by 20 minutes of IL-3 stimulation. Cells were subjected to lysis and immunoprecipitation with anti p-Tyr antibody (sc-508; Santa Cruz Biotechnology). The presence of Jak2 and Stat5 was visualized by western blotting with anti-Jak2 or Stat5 antibodies. (E) Relative dimerization level of IL7R and CRLF2 from HEK293 cells transiently transfected with the WT or mutant receptor. Dimerization was calculated by dividing luminescence by the mean fluorescence intensity of each treatment, normalizing the luminescence signal for experimental variability resulting from transfection efficiency. * P < .01, 1-way ANOVA, and Student t test. (F) Alignment of WT, natural mutant (EKV), and 6 experimental mutants (numbered 1 to 6) in the TMD of IL7R. Numbers show the positions of nucleotides and corresponding amino acids. The inserted nucleotides and amino acids are shown in red. The deleted AL amino acids are shown in green. (G) Cytokine withdrawal assay of BaF3 cells transduced with IL7R experimental mutants. (H) Constitutive phosphorylation of Stat5 in BaF3 cells expressing IL7R experimental mutants after 5 hours of cytokine deprivation. IL-3+ indicates cells harvested after 5 hours of IL-3 deprivation followed by 20 minutes of IL-3 stimulation. Del, deletion; Ins, insertion.
To assess whether the functional and biochemical evidence of receptor activation is associated with the oligomerization of IL7R and CRLF2 mutated proteins in cell membranes, we used the luciferase protein fragment complementation assay10 (supplemental Figure 3A). HEK293 cells were transduced with homodimeric or heterodimeric combinations of IL7R and CRLF2 fused to hGluc1 or hGluc2 (supplemental Figure 3B). After transduction, luciferase expression and receptor expression were analyzed in live cells (supplemental Figure 4). Dimerization was calculated by dividing luminescence by the mean fluorescence intensity of each receptor thereby enabling normalization of the luminescence signal for experimental variability resulting from transfection efficiency.
Consistent with the requirement of complementation for luciferase expression, none of the single-receptor hGluc1 or hGluc2 constructs that were individually transfected in HEK293 cells generated a signal (supplemental Figure 3C). Yet when the IL7RWT homodimer or heterodimer was expressed, a signal was obtained reflecting a basal level of receptor dimerization, as recently reported.12 We then analyzed the influence of the somatic mutations of IL7R and CRLF2 on receptor dimerization. As shown in Figure 2E, IL7Rins EKV, IL7RV253G, and CRLF2insEIM increased receptors dimerization (P < .05) whereas the IL7RinsGEA mutant that failed to provide cytokine-independent survival of BaF3 cells, or to cause Stat5 phosphorylation, also did not increase receptor dimerization (Figure 2E). Thinking that it may be a “passenger” mutation, we looked for additional activating mutations but found none (supplemental Table 3).
As CRLF2insEIM alone increased receptor dimerization but did not induce downstream constitutive signaling or transformed BaF3 cells, we concluded that there is no absolute correlation between dimerization and activation.
We next proceeded to decipher the amino acids that are important for the highly activating IL7RinsEKV mutation. We designed 6 variants of the EKV mutation: AKV, EAV, EKA, EK, KV (including DelAL like the natural mutation), and ins V (Figure 2F). BaF3 lines expressing each variant were created (supplemental Figure 5). The variants IL7RinsEAV and IL7RinsEKA enabled cytokine-independent growth of BaF3 cells, whereas IL7RinsAKV, IL7RinsEK, IL7RinsKV, and IL7RinsV did not transform BaF3 (Figure 2G). Biochemical analysis was consistent with the growth assays. Stat5 was phosphorylated in the absence of cytokine in BaF3 cells transduced with the IL7RinsEAV and IL7RinsEKA but much less in cells expressing IL7RinsAKV, IL7RinsEK, IL7RinsKV, and IL7RinsV (Figure 2H).
ConSurf13 calculations with IL7R showed that the TMD segment manifests a unique evolutionary conservation pattern with i/i+4 periodicity, equivalent to that of a perfect α-helix (supplemental Figure 6). The conserved positions would reside in the same helix face, providing an interface for IL7R homodimerization. All of the natural and experimental mutations can be interpreted assuming that activation requires IL7R dimerization with the cytosolic kinases in predefined orientation with respect to each other. The conserved face of the TMD helix in WT IL7R marks the orientation. Dimerization along a different TMD helix face would not activate because the kinases will not be facing each other. Molecular detailed interpretations of the mutations’ effect are provided in supplemental Figures 7 and 8 and supplemental Table 4.
Taken together, these data indicate that the functional and biochemical evidence for receptor activation by mutations lacking cysteine correlates with ligand independent dimerization. There are several examples of TMD mutations in receptors that cause constitutive activation without introducing cysteine: V664E in the Neu receptor tyrosine kinase14-16 and the dimerization inducing mutations in MPL17,18 that was identified in rare familial thrombocytosis patients.19 Our observations of noncysteine transmembrane mutations in type I cytokine receptors in patients with ALL demonstrate for the first time that such somatic mutations activate these receptors through ligand-independent dimerization and are leukemogenic.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Israel Science Foundation Legacy and iCORE programs, the Israel Cancer Research Foundation, Swiss Bridge Foundation, USA Israel Binational Science Foundation, Waxman Cancer Research Foundation, and William Lawrence and Blanche Hughes Foundation. C.S. and N.T. are PhD candidates at Tel Aviv University and this work is submitted in partial fulfillment of the requirement for a PhD.
Authorship
Contribution: S.I. designed the study and wrote the paper; C.S. performed research, analyzed data, and wrote the paper; N.T., N.G., Y.B., and N.B.-T. performed research and analyzed data; V.G., S.N.C., M.R.M., J.-C.T., and D.B. provided reagents and scientific expertise; and O.R.B., G.C., A.E.K., A.B., and M.U.M. provided reagents and data.
Conflict of interest disclosure: The authors declare no competing financial interests.
Correspondence: Shai Izraeli, Sheba Medical Center, Tel Hashomer Ramat Gan, 52621, Israel; e-mail: shai.izraeli@sheba.health.gov.il.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal