Key Points
The most abundant miRNA in CLL, miR-150, is expressed at lower levels in cases with unfavorable clinicobiological markers and worse prognosis.
miR-150 regulates expression of genes encoding proteins that modulate BCR signaling in CLL.
Abstract
We examined the microRNAs (miRNAs) expressed in chronic lymphocytic leukemia (CLL) and identified miR-150 as the most abundant, but with leukemia cell expression levels that varied among patients. CLL cells that expressed ζ-chain–associated protein of 70 kDa (ZAP-70) or that used unmutated immunoglobulin heavy chain variable (IGHV) genes, each had a median expression level of miR-150 that was significantly lower than that of ZAP-70–negative CLL cells or those that used mutated IGHV genes. In samples stratified for expression of miR-150, CLL cells with low-level miR-150 expressed relatively higher levels of forkhead box P1 (FOXP1) and GRB2-associated binding protein 1 (GAB1), genes with 3′ untranslated regions having evolutionary-conserved binding sites for miR-150. High-level expression of miR-150 could repress expression of these genes, which encode proteins that enhance B-cell receptor signaling, a putative CLL-growth/survival signal. Also, high-level expression of miR-150 was a significant independent predictor of longer treatment-free survival or overall survival, whereas an inverse association was observed for high-level expression of GAB1 or FOXP1 for overall survival. This study demonstrates that expression of miR-150 can influence the relative expression of GAB1 and FOXP1 and the signaling potential of the B-cell receptor, thereby possibly accounting for the noted association of expression of miR-150 and disease outcome.
Introduction
Chronic lymphocytic leukemia (CLL) is the most common leukemia among adults in the Western world. The clinical course of CLL patients is heterogeneous, ranging from indolent to highly aggressive. Several prognostic markers have been described in CLL that can reliably segregate patients into subgroups that differ in treatment-free survival (TFS) or overall survival (OS).1-3 Some of these markers, such as the immunoglobulin heavy chain variable (IGHV) gene mutation status or expression of ζ-chain–associated protein of 70 kDa (ZAP-70) or CD38, are associated with the B-cell receptor (BCR) signaling pathway.4-6 This suggests that BCR signaling may be involved in the pathogenesis and/or progression of CLL.
The intensity of BCR signaling varies between CLL cells of different patients, which in turn might account for some of the heterogeneity observed in the proclivity for disease progression (reviewed in Kipps7 ). Some CLL cells are more responsive to ligation of surface immunoglobulin, particularly CLL cells that express ZAP-70, the expression of which is associated with more aggressive disease.1,7 Similarly, there might be differences in other BCR-associated kinases, phosphatases, and their adaptor molecules between the CLL cells of different patients that also could modulate BCR signaling and potentially contribute to differences in the tendency for disease progression.8 As such, understanding the factors that modulate BCR signaling intensity in CLL cells may identify other features that are associated with prognosis and/or response to newly defined inhibitors of BCR signaling, which are found to have clinical activity in patients with this disease.9
Factors that might regulate expression of genes encoding proteins involved in BCR-signaling are microRNAs (miRNAs).10 These short noncoding RNAs each can regulate expression of a variety of different genes at the posttranscriptional level. miRNAs can regulate the stability and translation of a large number of target messenger RNAs (mRNAs) and thus “fine tune” essential cell functions.11-14 In lymphoid cells, such gene-dose regulation is needed for survival and proper maturation of B and T cells, immunoglobulin production by B cells, and relative proficiency of T-cell receptor signaling in T lymphocytes.10,12,15-19 The miRNAs that regulate essential pathways in immune cells generally are abundantly expressed and evolutionarily conserved.12,20-23 Aberrations in such miRNA-mediated regulation were directly implicated in cancer pathogenesis (reviewed in O’Connell and Baltimore12 ). This is particularly the case for CLL, the first human disease in which deregulation of miRNAs was linked to pathogenesis.20,24
In CLL deletion of miR-15a/miR-16-1, located at 13q14, is implicated in the pathogenesis of about 50% of all cases.20,24 Aberrant expression of miRNAs also is observed in other B-cell malignancies.25 Comparison of miRNA expression in CLL cases with favorable vs unfavorable prognosis have revealed other miRNAs that are associated with more aggressive disease or unfavorable cytogenetics (reviewed in Mraz et al26 and Mraz and Pospisilova27 ).
We and others have observed differential expression of approximately 20 miRNAs between CLL cells of patients with progressive disease vs those from patients with indolent disease.17,28-33 Several of these miRNAs contribute to the deregulation of anti-apoptotic molecules, such as bcl-2 (miR-15a/16-1),34 mcl-1 (miR-29),35 or tcl-1 (miR-29, miR-181),36 molecules that play important roles in the resistance to apoptosis or growth of neoplastic B cells. Lack of functional p53 also can affect expression of miRNAs that might influence apoptosis (eg, miR-34a).32 Additionally, the levels of certain miRNAs, such as miR-181b, can decrease with disease progression, whereas others, such as miR-125b, can influence the metabolic adaptation of B cells to their malignant phenotype.33,37 However, the pathways regulated by most of the miRNAs expressed in CLL cells and their contributions to CLL-cell biology remain unknown.
We hypothesized that miRNAs that are abundantly expressed in CLL may regulate expression of genes encoding proteins involved in key molecular pathways, such as those involved in BCR signaling. We identified the most abundant miRNA in CLL to be miR-150, which we found expressed at different levels in CLL cells of different patients. We examined for genes that are differentially expressed between CLL cells that have relatively high vs low levels of miR-150, allowing us to discover the regulatory activity of miR-150 on 2 genes encoding proteins that can modulate the intensity of BCR signaling and potentially contribute to the heterogeneity noted in disease progression of patients with CLL.
Methods
CLL cohort
Blood samples were collected from patients (n = 168) at the University of California-San Diego Moores Cancer Center who satisfied diagnostic and immunophenotypic criteria for common CLL after providing written informed consent in compliance with the Declaration of Helsinki and the institutional review board of University of California-San Diego. Peripheral blood mononuclear cells were isolated from CLL patients using density centrifugation with Ficoll-Hypaque (GE Healthcare; obtained purity of ≥95% of CD5+19+ cells). The basic clinicobiological characteristics of this patient cohort are summarized in Table 1.
Cohort characteristics (n = 168)
| . | N . | % . |
|---|---|---|
| Median age at diagnosis (range), y | 56.5 (34-79) | |
| Median follow up (y) | 8.3 | |
| Median survival (y) | 19.8 | |
| Median time from diagnosis to blood collection (y) | 2.6 | |
| Treated during follow-up* | 105 | 63.0 |
| Gender | ||
| Male | 107 | 63.7 |
| Female | 61 | 36.3 |
| Rai stage | ||
| 0 | 54 | 42.6 |
| I | 43 | 32.4 |
| II | 25 | 18.4 |
| III-IV | 9 | 6.6 |
| Not determined | 37 | |
| IGHV/ZAP-70/CD38 status | ||
| unmut IGHV/mut IGHV | 72/96 | 42.9/57.1 |
| ZAP-70+/ZAP-70− | 73/95 | 43.5/56.5 |
| ZAP-70− AND mut IGHV | 83 | 49.4 |
| ZAP-70+ AND unmut IGHV | 60 | 35.7 |
| CD38+/CD38− | 49/119 | 29.2/70.8 |
| Hierarchical cytogenetics (FISH) | ||
| del 17p13 | 12 | 10.4 |
| del 11q23 | 12 | 10.4 |
| del 13q14 | 47 | 40.8 |
| Trisomy 12 | 15 | 13.0 |
| Normal karyotype | 29 | 25.2 |
| Not determined | 53 | |
| . | N . | % . |
|---|---|---|
| Median age at diagnosis (range), y | 56.5 (34-79) | |
| Median follow up (y) | 8.3 | |
| Median survival (y) | 19.8 | |
| Median time from diagnosis to blood collection (y) | 2.6 | |
| Treated during follow-up* | 105 | 63.0 |
| Gender | ||
| Male | 107 | 63.7 |
| Female | 61 | 36.3 |
| Rai stage | ||
| 0 | 54 | 42.6 |
| I | 43 | 32.4 |
| II | 25 | 18.4 |
| III-IV | 9 | 6.6 |
| Not determined | 37 | |
| IGHV/ZAP-70/CD38 status | ||
| unmut IGHV/mut IGHV | 72/96 | 42.9/57.1 |
| ZAP-70+/ZAP-70− | 73/95 | 43.5/56.5 |
| ZAP-70− AND mut IGHV | 83 | 49.4 |
| ZAP-70+ AND unmut IGHV | 60 | 35.7 |
| CD38+/CD38− | 49/119 | 29.2/70.8 |
| Hierarchical cytogenetics (FISH) | ||
| del 17p13 | 12 | 10.4 |
| del 11q23 | 12 | 10.4 |
| del 13q14 | 47 | 40.8 |
| Trisomy 12 | 15 | 13.0 |
| Normal karyotype | 29 | 25.2 |
| Not determined | 53 | |
mut, mutated; unmut, unmutated.
Details provided in supplemental Table 3.
Gene expression microarray analysis and quantitative real-time polymerase chain reaction
Total RNA was isolated, labeled, and hybridized to Affymetrix HG-U133+2 GeneChips according to the manufacturer‘s protocol, as described previously.38 Expression of individual protein-coding genes/miR-150 (TaqMan Assays; Applied Biosystems) and miRNA expression data (TaqMan Array MicroRNA Cards; Applied Biosystems) were obtained and normalized according to the manufacturer‘s protocol, as described previously39 (see supplemental Methods on the Blood Web site).
Cell transfection
B-cell lines MEC-1 and Raji were obtained from American Type Culture Collection and cultured in RPMI-1640 supplemented with 10% fetal bovine serum in 5% CO2 at 37°C. Cell lines or CLL cells were respectively suspended at 2 × 106 per mL or 1 × 107 per mL in transfection medium for transfection using the DharmaFECT Duo Transfection Reagent (Dharmacon; Thermo Scientific) with a short artificial miR-150 (MISSION microRNA Mimic, 100 nM; Sigma-Aldrich), control RNA (MISSION microRNA Mimic Negative Control, 100 nM), short interfering RNA (siRNA) (ON-TARGET plus siRNA-SMARTpool, 100 nM; Thermo Scientific), or fluorochrome-labeled short RNA (siGLO; Thermo Scientific).17 Raji and MEC-1 cell lines were used for the transfection experiments because of their relatively low-level expression of miR-150, resilience to transfection with high cell viability (viability > 90%), and, in the case of Raji cells, high transfection efficiency (>85%; supplemental Figure 1A-B). Raji and MEC-1 cell lines both express miR-150 at ∼100-fold lower levels than the average level expressed by CLL cells. The immunoblot analysis of transfected cells is described in supplemental Methods.
Luciferase assay
Luciferase reporter assay was performed using the LightSwitch Luciferase Assay System (SwitchGear Genomics), according to the manufacturer's protocol, as described previously40 (see supplemental Methods).
BCR crosslinking and measurement of intracellular calcium flux
BCR signaling induced by BCR ligation was evaluated by flow cytometry to measure changes in intracellular calcium flux induced by treatment with goat F(ab′)2 anti-human immunoglobulin M (IgM; anti-μ, Southern Biotechnology; final concentration 10 µg/mL), as described elsewhere4,5,41 (see supplemental Methods).
Methylation arrays
We obtained data on the methylation of the miR-150 promoter region using an Illumina Human Methylation 450 BeadChip, following the manufacturer’s protocol (see supplemental Methods).
Statistical analysis
Univariate and multivariate analysis of OS and TFS were computed using R package (see supplemental Methods). All other statistical analyses were performed with GraphPad Prism Software, v. 5.0 (GraphPad Software). All statistical tests were 2-sided and P values <.05 were considered significant.
Results
Expression of miR-150 and its relationship with clinicobiological features of CLL
We examined for the expression of 754 human miRNAs in CLL cells isolated from 10 different patients (cohort characteristics are listed in supplemental Table 1). Altogether, we detected 271 different miRNAs (threshold cycle [Ct] value <38) in at least 5 CLL samples, and another 171 different miRNAs in 1 to 4 samples. We did not detect expression of 312 different miRNAs in any of the samples. Of the 15 most abundantly expressed miRNAs, 11 had previously been found to vary between patients with a low risk vs a high risk for early disease progression (Figure 1A).26,27 All 15 of these abundantly expressed miRNAs were evolutionarily conserved,42 including miR-150, which was the most abundantly expressed miRNA in CLL (Figure 1A; supplemental Figure 2). CLL cells expressed higher median levels of miR-150 than normal blood B cells or tonsillar B cells (3.5-fold and 6.5-fold, respectively, P < .05) (supplemental Figure 3A).
Expression of miR-150 and its relationship with clinicobiological features. (A) The expression of 754 human miRNAs (TaqMan Array MicroRNA Cards; ABI) was screened in 10 purified CLL samples (>95% of CD5+19+ cells purified using RosseteSep Human B Cell Enrichment Cocktail, StemCell Technologies; RNA Integrity number >8). Results are visualized as geometric mean (black bars) and standard deviation (gray error bars). The lower Ct value corresponds to higher expression; Ct values >38 were considered as nondetectable miRNAs (n = 312). (B) The methylation levels in the region upstream of miR-150 in CLL cases stratified as low miR-150 expression (<median, n = 13) and high miR-150 (>median, n = 13). The prediction of transcription start sides for miR-150 in regions 1 and 2 is described in supplemental Figure 3. Analyzed region 1 contained 5 probes and region 2 contained 1 probe (region is defined by a size of ∼1000 nt). The methylation levels were compared using the nonparametric Mann-Whitney U test. The error bars represent standard deviation. (C-G) miR-150 expression was quantified in a cohort of 168 CLL patients (cohort characteristics in Table 1) and correlated to the clinicobiological characteristics of CLL cells such as IGHV mutation status (C), ZAP-70 expression (D), Rai stage (E), CD38 expression (F), and hierarchical classification of FISH abnormalities (G). The differences in expression were compared using the nonparametric Mann-Whitney U test. U IgHV, unmutated IGHV; M IgHV, mutated IGHV.
Expression of miR-150 and its relationship with clinicobiological features. (A) The expression of 754 human miRNAs (TaqMan Array MicroRNA Cards; ABI) was screened in 10 purified CLL samples (>95% of CD5+19+ cells purified using RosseteSep Human B Cell Enrichment Cocktail, StemCell Technologies; RNA Integrity number >8). Results are visualized as geometric mean (black bars) and standard deviation (gray error bars). The lower Ct value corresponds to higher expression; Ct values >38 were considered as nondetectable miRNAs (n = 312). (B) The methylation levels in the region upstream of miR-150 in CLL cases stratified as low miR-150 expression (<median, n = 13) and high miR-150 (>median, n = 13). The prediction of transcription start sides for miR-150 in regions 1 and 2 is described in supplemental Figure 3. Analyzed region 1 contained 5 probes and region 2 contained 1 probe (region is defined by a size of ∼1000 nt). The methylation levels were compared using the nonparametric Mann-Whitney U test. The error bars represent standard deviation. (C-G) miR-150 expression was quantified in a cohort of 168 CLL patients (cohort characteristics in Table 1) and correlated to the clinicobiological characteristics of CLL cells such as IGHV mutation status (C), ZAP-70 expression (D), Rai stage (E), CD38 expression (F), and hierarchical classification of FISH abnormalities (G). The differences in expression were compared using the nonparametric Mann-Whitney U test. U IgHV, unmutated IGHV; M IgHV, mutated IGHV.
Despite expressing relatively high levels of miR-150, CLL cells of different patients (n = 168) varied in their relative expression of this miRNA (∼12-fold difference between cases in lower 10th percentile vs those in the 90th percentile). The distribution in expression of miR-150 among patients is provided in supplemental Figure 4. Two regions in the putative miR-150 promoter were significantly less methylated in cases with high-level miR-150 than in cases with low-level miR-150, suggesting epigenetic differences influence expression of this miRNA (Figure 1B; supplemental Figure 5). In our cohort of patients (Table 1), miR-150 levels were significantly lower in cases that used unmutated IGHV or that expressed ZAP-70 (Figure 1C-D). Discordant cases that did not express ZAP-70, but used unmutated IGHV, expressed lower levels of miR-150 than cases that lacked expression of ZAP-70 or that expressed mutated IGHV (supplemental Figure 6A). There were wide variations in the expression levels of miR-150 among samples collected from patients who had Rai stage 0-II disease. However, those patients in our cohort that had Rai stage III-IV disease had CLL cells that expressed relatively low levels of miR-150 (P < .005, Figure 1E). The expression levels of miR-150 did not have any discernible relationship with the expression levels of CD38, or with specific chromosomal abnormalities, as detected by fluorescence in situ hybridization (FISH) (Figure 1F-G; supplemental Figure 6B).
Identification of miR-150 targets by gene expression analysis
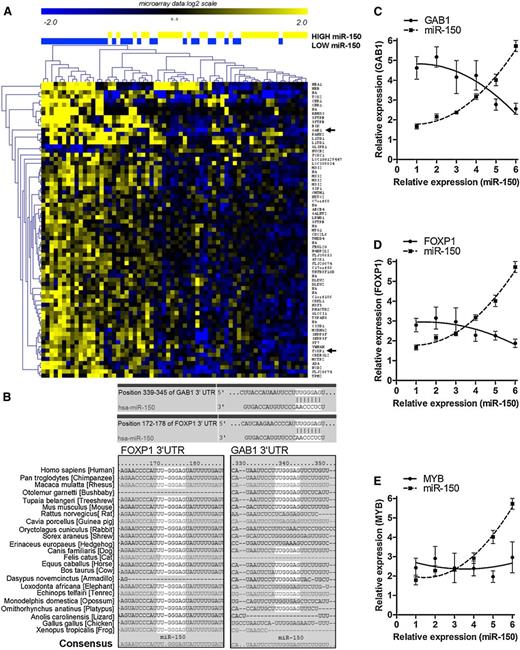
Several genes have been identified as being potentially regulated by miR-150 in various cell types.43-47 However, the genes targeted by a certain miRNA can vary depending upon the cellular context.12,48-50 Because an miRNA can influence the relative stability of its target mRNA,11-14 evaluating for differences in gene expression between cells that express high vs low levels of a given miRNA potentially could identify mRNA(s) that are regulated by that miRNA.11-14 As such, we performed array-based transcriptome analyses of 100 CLL samples to examine for differences in gene expression between CLL cells that expressed high vs low levels of miR-150. For this purpose, we divided the samples into 3 groups based upon their relative expression levels of miR-150. We examined for gene expression differences between samples from the third of the cases having the lowest levels of miR-150 (n = 32) with the third of the cases having the highest expression levels of miR-150 (n = 32), identifying genes with a significance analysis of microarray (SAM) method (MeV) with a fold change of more than 1.5 (false discovery rate <0.1). This analysis identified 58 genes (with 72 probes) that differed in their relative expression levels between these 2 groups of samples (Figure 2A). Most of these genes (55/58) were expressed at relatively lower levels in samples with the high-level expression of miR-150 (Figure 2A; supplemental Table 2). Thirteen of the 58 identified genes (22%) were predicted targets of miR-150 using TargetScan software (supplemental Table 2). We noted that 2 of the 13 genes had evolutionary conserved binding sides for miR-150, namely GAB1 and FOXP1 (Figure 2B). Moreover, FOXP1 and GAB1 were predicted targets of miR-150 using other database tools (DIANAmT, miRnda, miRWalk, PICTAR5, or RNA hybrid). The 7-nucleotide binding site for miR-150 (containing the whole seed sequence) in GAB1 was identical to that found in FOXP1 (Figure 2B).
Identification of miR-150 targets by gene expression analysis. (A) Microarray data for protein-coding gene expression (Affymetrix HG-U133+2 GeneChips) were compared in CLL samples stratified based on low (n = 32) vs high (n = 32) levels of miR-150. The samples from the lowest and highest tercile were compared (SAM in MeV, fold change >1.5, false discovery rate <0.1). This analysis identified differential expression of 58 genes (72 probes) between CLL cells expressing low vs high levels of miR-150. GAB1 and FOXP1 are marked with a black arrow. (B) The evolutionary conserved binding sides for miR-150 in GAB1 and FOXP1 3′UTR (TargetScan). (C-E) The association between miR-150 levels and the expression of mRNAs for GAB1 (C), FOXP1 (D), or c-MYB (E) in a validation cohort of 60 CLL samples. Each point represents the average value for gene expression from 10 CLL samples. The standard deviation about the mean is indicated by error bars.
Identification of miR-150 targets by gene expression analysis. (A) Microarray data for protein-coding gene expression (Affymetrix HG-U133+2 GeneChips) were compared in CLL samples stratified based on low (n = 32) vs high (n = 32) levels of miR-150. The samples from the lowest and highest tercile were compared (SAM in MeV, fold change >1.5, false discovery rate <0.1). This analysis identified differential expression of 58 genes (72 probes) between CLL cells expressing low vs high levels of miR-150. GAB1 and FOXP1 are marked with a black arrow. (B) The evolutionary conserved binding sides for miR-150 in GAB1 and FOXP1 3′UTR (TargetScan). (C-E) The association between miR-150 levels and the expression of mRNAs for GAB1 (C), FOXP1 (D), or c-MYB (E) in a validation cohort of 60 CLL samples. Each point represents the average value for gene expression from 10 CLL samples. The standard deviation about the mean is indicated by error bars.
We observed that the expression level of miR-150 was inversely proportional to that of GAB1 or FOXP1. This also was observed among CLL samples from a validation cohort of 60 patients (Figure 2C-D; supplemental Figure 7) and in normal peripheral CD19+ B cells (supplemental Figure 3). On the other hand, a known target of miR-150, c-MYB,47 was not inversely proportional to miR-150 and expressed at very low levels (median Ct value 32.8), even in samples with low-level expression of miR-150 (Figure 2A,E). The expression levels of c-MYB were close to the detection limit of our real-time polymerase chain reaction assay (median Ct value 32.8, detection limit Ct = ∼35.0). Finally, the levels of c-MYB detected in CLL cells were 34-fold lower than those of GAB1 (median Ct value 27.7) and 415-fold lower than those of FOXP1 (median Ct value 24.1). Altogether, these data suggest that GAB1 and FOXP1, rather than c-MYB, are potential targets of miR-150 in the CLL transcriptome.
Validation of newly identified targets of miR-150 in CLL
To study the relationship between expression of miR-150 and proteins encoded by GAB1 or FOXP1, we examined for GAB1 and FOXP1 protein in CLL cells of each of 16 patients that had relatively low- (n = 8) or high-level expression of miR-150 (n = 8; ∼5- to 10-fold difference in miR-150 levels). We found that cases with relatively low miR-150 had higher levels of GAB1 or FOXP1 (P < .005, fold change >10.0) than cases with high-level expression of miR-150 by immunoblot analysis (Figure 3A). Transfection of the B-cell lines MEC-1 or Raji, or primary CLL cells, with a synthetic miR-150, reduced the levels of both GAB1 and FOXP1 by 28% to 44% and 46% to 80%, respectively (P < .05) (Figure 3B). Moreover, transfection of cells with miR-150 down-modulated either splicing variant of FOXP1, but to varying degrees. In contrast, the levels of GAB1 or FOXP1 did not change in cells transfected with an irrelevant control miRNA (Figure 3B).
Validation of newly defined miR-150 targets. (A) GAB1 or FOXP1 immunoblot analysis in CLL samples of 16 patients selected for low-level (n = 8) vs high-level expression of miR-150 (n = 8). The relative levels of miR-150 expression are indicated (first sample set as 1). The samples were solely chosen based on the differences in miR-150 expression and partially overlapped with samples used in the microarray analysis (7 of 16 samples). The double bands identified by the anti-FOXP1 antibody (JC12 clone) represent 2 splicing variants of FOXP1 as described previously.53 (B) A representative immunoblot for GAB1 or FOXP1 protein in MEC-1 and Raji, primary CLL cells transfected (48 hours) with artificial miR-150 (miR-150, 100 nM), and control miRNA (negative control [Neg. Ctrl], 100 nM). MOCK represents cells treated only with the electroporation pulse. The Blot images were quantified with ImageJ 1.42q (National Institutes of Health), and the GAB1/β-actin and FOXP1/β-actin ratio in MOCK was arbitrarily set at 1. The FOXP1/β-actin rations for its 2 major splicing variants are provided (the ratio of the larger vs the smaller isoform is separated by a slash). (C) The luciferase activity in 293FT cells cotransfected with miR-150 mimic (MIMIC miR-150, 100 nM) or luciferase reporter construct (100 ng) containing the 3′UTR region of GAB1, FOXP1 or GAPDH (GAPDH as control). As a negative control, we used a control miRNA mimic (CTRL Scramble, 100 nM). Luciferase activity was measured 24 hours posttransfection and compared by paired Student t test.
Validation of newly defined miR-150 targets. (A) GAB1 or FOXP1 immunoblot analysis in CLL samples of 16 patients selected for low-level (n = 8) vs high-level expression of miR-150 (n = 8). The relative levels of miR-150 expression are indicated (first sample set as 1). The samples were solely chosen based on the differences in miR-150 expression and partially overlapped with samples used in the microarray analysis (7 of 16 samples). The double bands identified by the anti-FOXP1 antibody (JC12 clone) represent 2 splicing variants of FOXP1 as described previously.53 (B) A representative immunoblot for GAB1 or FOXP1 protein in MEC-1 and Raji, primary CLL cells transfected (48 hours) with artificial miR-150 (miR-150, 100 nM), and control miRNA (negative control [Neg. Ctrl], 100 nM). MOCK represents cells treated only with the electroporation pulse. The Blot images were quantified with ImageJ 1.42q (National Institutes of Health), and the GAB1/β-actin and FOXP1/β-actin ratio in MOCK was arbitrarily set at 1. The FOXP1/β-actin rations for its 2 major splicing variants are provided (the ratio of the larger vs the smaller isoform is separated by a slash). (C) The luciferase activity in 293FT cells cotransfected with miR-150 mimic (MIMIC miR-150, 100 nM) or luciferase reporter construct (100 ng) containing the 3′UTR region of GAB1, FOXP1 or GAPDH (GAPDH as control). As a negative control, we used a control miRNA mimic (CTRL Scramble, 100 nM). Luciferase activity was measured 24 hours posttransfection and compared by paired Student t test.
To test for interaction of miR-150 with the 3′ untranslated region (UTR) of GAB1 or FOXP1, we cotransfected 293FT cells with miR-150, or a control miRNA, and a luciferase reporter–construct containing the 3′ UTR of GAB1 or FOXP1 (Figure 3C). Cotransfection of miR-150 with this luciferase construct decreased the luciferase activity of the transfected cells by about 25% relative to that of cells cotransfected with the luciferase construct and control miRNA (Figure 3C). Such differences are similar to those of luciferase constructs containing the 3′ UTR of other genes that are targeted by a cotransfected miRNA.40 On the other hand, the miR-150 mimic had no effect on the luciferase activity of cells cotransfected with a luciferase-reporter construct containing the 3′ UTR of GAPDH, which contained no predicted binding sites for miR-150. We conclude that miR-150 interacts with the 3′UTR of either GAB1 or FOXP1 mRNA.
Relationship between expression of miR-150, GAB1, or FOXP1 and relative sensitivity to BCR ligation
GAB1 is an adaptor molecule that can recruit phosphoinositide 3-kinase (PI3K) to the plasma membrane after surface immunoglobulin ligation and thereby enhance BCR signaling.51 FOXP1 is a transcription factor that is upregulated upon B-cell activation and found associated with adverse prognosis and the activated B cell (ABC) phenotype of diffuse large B-cell lymphoma (DLBCL).52,53 Because miR-150 can target the genes encoding each of these factors, the expression level of miR-150 could influence the intensity of BCR signaling induced by surface immunoglobulin ligation.
To examine for this, we transfected Raji cells with siRNA specific for GAB1, FOXP1, a synthetic miR-150, or control short RNA. We selected Raji cells because of their relatively low-level expression of miR-150 and high transfection efficacy (>85%, supplemental Figure 1). Transfection with either miR-150 or a siRNA specific for GAB1 significantly reduced expression of GAB1 (Figure 4A), which was not affected by transfection with control short RNA. Similarly, transfection with either miR-150 or siRNA for FOXP1 reduced expression of FOXP1 relative to that noted in cells transfected with control siRNA (Figure 4A).
Expression of miR-150 targets GAB1 or FOXP1 affects BCR signaling. (A) The Raji cell line was transfected with artificial miR-150 (miR-150, 100 nM), control miRNA (miR negative control [Neg. Ctrl], 100 nM), siRNA against GAB1 (siGAB1, 100 nM) or FOXP1 (siFOXP1, 100 nM), and control siRNA (siRNA Neg. Ctrl, 100nM). The band intensities observed in immunoblot analysis were measured using ImageJ 1.42q, and the GAB1/β-actin and FOXP1/β-actin ratio in control was arbitrarily set at 1. The FOXP1/β-actin ratios for the 2 major isoforms of FOXP1 are provided (ratio for larger and smaller variant is separated by a slash). GAB1 and FOXP1 protein levels were analyzed 48 hours posttransfection and β-actin was used as a loading control (representative example of an immunoblot). (B-C) The effect of Raji cell transfection with siRNA against GAB1 (siGAB1, 100 nM) or FOXP1 (siFOXP1, 100 nM) or control siRNA (siNEG CTRL, 100 nM) on calcium influx after anti-μ stimulation (10 μg/mL; indicated by a black arrow). (D) The effect of Raji cell transfection with artificial miR-150 (miR-150 MIMIC, 100 nM) or control miRNA (miR NEG CTRL, 100 nM) on calcium influx after anti-μ stimulation (10 μg/mL; indicated by a black arrow). The calcium influx (FLUO-4 fluorescence intensity) in panels B-D were analyzed continuously for 120 seconds. The acquisition of background fluorescence (30 seconds) was followed by adding anti-μ (final concentration 10 μg/mL; indicated by a black arrow), and data at 6-second intervals were visualized as peak intensity. Error bars represent standard error of the mean of at least 2 independent experiments. Differences were statistically analyzed by 2-way analysis of variance.
Expression of miR-150 targets GAB1 or FOXP1 affects BCR signaling. (A) The Raji cell line was transfected with artificial miR-150 (miR-150, 100 nM), control miRNA (miR negative control [Neg. Ctrl], 100 nM), siRNA against GAB1 (siGAB1, 100 nM) or FOXP1 (siFOXP1, 100 nM), and control siRNA (siRNA Neg. Ctrl, 100nM). The band intensities observed in immunoblot analysis were measured using ImageJ 1.42q, and the GAB1/β-actin and FOXP1/β-actin ratio in control was arbitrarily set at 1. The FOXP1/β-actin ratios for the 2 major isoforms of FOXP1 are provided (ratio for larger and smaller variant is separated by a slash). GAB1 and FOXP1 protein levels were analyzed 48 hours posttransfection and β-actin was used as a loading control (representative example of an immunoblot). (B-C) The effect of Raji cell transfection with siRNA against GAB1 (siGAB1, 100 nM) or FOXP1 (siFOXP1, 100 nM) or control siRNA (siNEG CTRL, 100 nM) on calcium influx after anti-μ stimulation (10 μg/mL; indicated by a black arrow). (D) The effect of Raji cell transfection with artificial miR-150 (miR-150 MIMIC, 100 nM) or control miRNA (miR NEG CTRL, 100 nM) on calcium influx after anti-μ stimulation (10 μg/mL; indicated by a black arrow). The calcium influx (FLUO-4 fluorescence intensity) in panels B-D were analyzed continuously for 120 seconds. The acquisition of background fluorescence (30 seconds) was followed by adding anti-μ (final concentration 10 μg/mL; indicated by a black arrow), and data at 6-second intervals were visualized as peak intensity. Error bars represent standard error of the mean of at least 2 independent experiments. Differences were statistically analyzed by 2-way analysis of variance.
We monitored for changes in intracellular calcium induced with anti-µ treatment by flow cytometry. This demonstrated that the relative level of GAB1 or FOXP1 apparently influenced the magnitude of the calcium flux observed following BCR ligation (Figure 4B-C). The transfection with miR-150 also significantly affected BCR responsiveness, but with higher variability (Figure 4D). The transfection with miR-150 or siGAB1 or siFOXP1 did not have any significant effect on cell viability or the levels of calcium flux induced by treatment with ionomycin (supplemental Figure 1B; supplemental Figure 8). We noted that silencing FOXP1 down-modulated both the basal and anti-µ–induced levels of phosphorylated AKT, whereas silencing GAB1 reduced the levels of phospho-AKT induced by anti-µ (supplemental Figure 9).
To test the relevance of GAB1 and FOXP1 in BCR signaling in CLL, we treated 45 CLL samples with anti-μ and monitored the induced calcium flux by flow cytometry. This allowed us to examine for differences between samples in their response to BCR crosslinking, allowing us to stratify cases as relatively sensitive to BCR ligation (n = 27) or resistant to treatment with anti-µ (n = 18) (see supplemental methods). CLL samples that were more sensitive to BCR ligation had significantly higher levels of GAB1 and FOXP1 mRNAs (Figure 5A-B). On the other hand, cases with relatively high levels of miR-150 were relatively insensitive to BCR ligation (Figure 5C). As expected,4,5,41 most samples that were ZAP-70–positive responded well to treatment with anti-µ. However, there were cases that were ZAP-70–positive that were relatively insensitive to BCR ligation. Such samples had relatively low levels of GAB1 or FOXP1, and relatively high levels of miR-150 (Figure 5D-F). These data reveal that high-level expression of GAB1 and FOXP1 and low-level expression of miR-150 associates with relatively high sensitivity to surface immunoglobulin ligation in CLL. Moreover, high expression of miR-150 might account in part for the relatively low response to surface immunoglobulin ligation observed in some samples that express ZAP-70.
Relationship between expression of miR-150, GAB1, or FOXP1 and the relative sensitivity to BCR ligation in CLL. The association between BCR responsiveness and GAB1 (A), FOXP1 (B), or miR-150 (C) expression. Forty-five CLL samples were stimulated by anti-μ (10 μg/mL) and the calcium flux was assessed by flow cytometry. Below each panel is a “+” or a “−” in the row labeled “BCRsig” on the left to indicate samples that were high responders or low responders, respectively. (D-F) Another row is beneath the panels labeled on the left as “ZAP-70” to indicate the samples that were positive or negative for ZAP-70, as indicated by the “+” or “−”, respectively. n.s., not significant; ZAP-70+, ZAP-70 positive samples.
Relationship between expression of miR-150, GAB1, or FOXP1 and the relative sensitivity to BCR ligation in CLL. The association between BCR responsiveness and GAB1 (A), FOXP1 (B), or miR-150 (C) expression. Forty-five CLL samples were stimulated by anti-μ (10 μg/mL) and the calcium flux was assessed by flow cytometry. Below each panel is a “+” or a “−” in the row labeled “BCRsig” on the left to indicate samples that were high responders or low responders, respectively. (D-F) Another row is beneath the panels labeled on the left as “ZAP-70” to indicate the samples that were positive or negative for ZAP-70, as indicated by the “+” or “−”, respectively. n.s., not significant; ZAP-70+, ZAP-70 positive samples.
Prognostic significance of miR-150, GAB1, or FOXP1
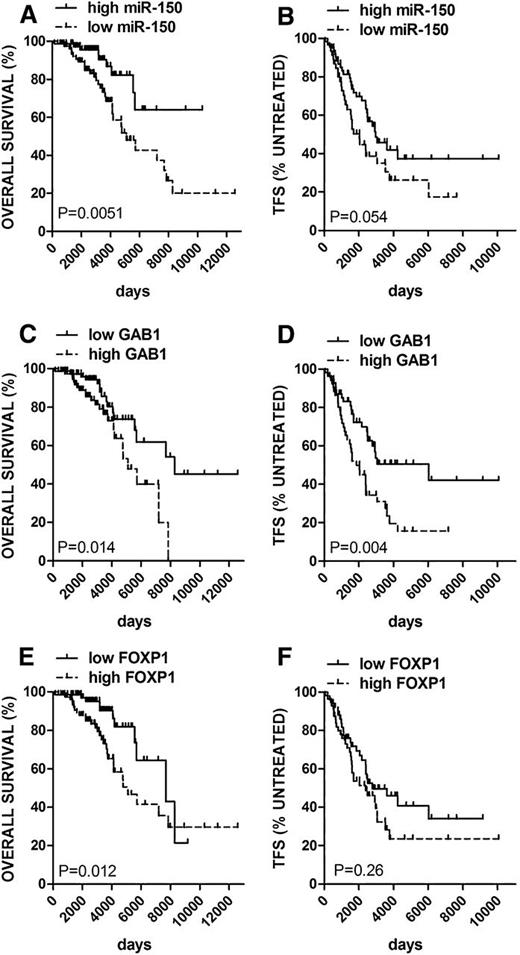
The relationship between expression of GAB1, FOXP1, and miR-150 with the sensitivity to BCR ligation prompted us to investigate the relationship between the relative expression of each of these genes and prognosis in a well-characterized cohort of patients with CLL (n = 168, Table 1). The median follow-up of patients was 8.3 years. During the follow-up period, 63% of patients required therapy (n = 105, median time to therapy 5.6 years) and 23% had died (n = 39, median survival 19.8 years). For the analysis of the relationship between relative gene expression and OS, we only used data from patients who had a stored blood sample available before the last day of follow-up (n = 154) or for TFS before first treatment (n = 107). We divided the cohort by median miR-150 expression (cutoff point identification is depicted in supplemental Figure 10) and performed a univariate Cox regression (Table 2) and also Kaplan-Meier analysis. Compared with cases that had high-level expression of miR-150, the cases with low-level expression of miR-150 had a significantly shorter median OS (13.9 years vs not reached, HR: 2.8 [CI 1.3-5.9]) and shorter median TFS (5.6 vs 8.1 years, HR: 1.7 [CI 1.0-2.8]; Figure 6 A-B; Table 2).
The prognostic significance of miR-150, GAB1, or FOXP1 in a univariate analysis
| . | Univariate Cox regression model of OS . | Univariate Cox regression model of TFS . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n = 154, number of events = 39* . | n = 107, number of events = 58* . | ||||||||||||
| Nondelayed entry model . | Delayed entry model . | Nondelayed entry model . | Delayed entry model . | ||||||||||
| Variable . | HR . | CI . | P . | HR . | CI . | P . | HR . | CI . | P . | HR . | CI . | P . | |
| miR-150 (≤ vs > median) | 2.8 | 1.3-5.9 | .01 | 3.2 | 1.5-6.7 | .003 | 1.7 | 1-2.8 | .057 | 2.3 | 1.4-4 | .002 | |
| GAB1 (> vs ≤ median) | 2.3 | 1.2-4.4 | .02 | 1.7 | 0.9-3.3 | .12 | 2.1 | 1.3-3.6 | .005 | 2.4 | 1.4-4.1 | .002 | |
| FOXP1 (> vs ≤ median) | 2.4 | 1.2-4.8 | .02 | 2.9 | 1.4-5.8 | .004 | 1.4 | 0.8-2.3 | .3 | 1.3 | 0.7-2.1 | .39 | |
| IGHV (unmut. vs mut.) | 12.2 | 4.9-30.5 | <.001 | 7.5 | 3-18.9 | <.001 | 2.9 | 2-4.4 | <.001 | 2.5 | 1.4-4.2 | .001 | |
| ZAP-70 (pos. vs neg.) | 7.3 | 3.5-15.2 | <.001 | 7.7 | 3.3-18 | <.001 | 2.4 | 1.6-3.5 | <.001 | 2.3 | 1.3-3.9 | .002 | |
| CD38 (pos. vs neg.) | 3.0 | 1.6-5.7 | .001 | 2.7 | 1.4-5.2 | .003 | 2.5 | 1.7-3.8 | <.001 | 2.6 | 1.4-4.6 | .002 | |
| Gender (male vs female) | 1.3 | 0.7-2.5 | .5 | 1.2 | 0.6-2.4 | .56 | 1.4 | 1-2.2 | .08 | 1.4 | 0.8-2.5 | .2 | |
| Rai stage | I vs 0 | 2.9 | 0.9-9.2 | .07 | 2.3 | 0.7-7.5 | .16 | 1.4 | 0.8-2.4 | .2 | 0.8 | 0.4-1.7 | .63 |
| II vs 0 | 4.4 | 1.2-15.7 | .02 | 2.5 | 0.7-9.1 | .15 | 3.2 | 1.8-5.8 | <.001 | 1.2 | 0.5-2.6 | .66 | |
| ≥III vs 0 | 12.0 | 3.5-40.9 | <.001 | 7.1 | 2.1-24 | .002 | 5.2 | 2.4-11.1 | <.001 | 3.2 | 0.7-14.4 | .13 | |
| Age (> vs ≤ median) | 3.3 | 1.7-6.6 | <.001 | 2.7 | 1.3-5.7 | .008 | 1.1 | 0.7-1.6 | .75 | 0.9 | 0.5-1.6 | .76 | |
| del17p13 | 2.9 | 1-8.8 | .06 | 3.3 | 1.1-10 | .04 | 1.9 | 1-3.8 | .05 | 2.4 | 1-5.8 | .05 | |
| dell11q23 | 2.6 | 0.9-8 | .09 | 1.6 | 0.5-4.8 | .43 | 2.4 | 1.3-4.4 | .007 | 1.8 | 0.8-4 | .13 | |
| . | Univariate Cox regression model of OS . | Univariate Cox regression model of TFS . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n = 154, number of events = 39* . | n = 107, number of events = 58* . | ||||||||||||
| Nondelayed entry model . | Delayed entry model . | Nondelayed entry model . | Delayed entry model . | ||||||||||
| Variable . | HR . | CI . | P . | HR . | CI . | P . | HR . | CI . | P . | HR . | CI . | P . | |
| miR-150 (≤ vs > median) | 2.8 | 1.3-5.9 | .01 | 3.2 | 1.5-6.7 | .003 | 1.7 | 1-2.8 | .057 | 2.3 | 1.4-4 | .002 | |
| GAB1 (> vs ≤ median) | 2.3 | 1.2-4.4 | .02 | 1.7 | 0.9-3.3 | .12 | 2.1 | 1.3-3.6 | .005 | 2.4 | 1.4-4.1 | .002 | |
| FOXP1 (> vs ≤ median) | 2.4 | 1.2-4.8 | .02 | 2.9 | 1.4-5.8 | .004 | 1.4 | 0.8-2.3 | .3 | 1.3 | 0.7-2.1 | .39 | |
| IGHV (unmut. vs mut.) | 12.2 | 4.9-30.5 | <.001 | 7.5 | 3-18.9 | <.001 | 2.9 | 2-4.4 | <.001 | 2.5 | 1.4-4.2 | .001 | |
| ZAP-70 (pos. vs neg.) | 7.3 | 3.5-15.2 | <.001 | 7.7 | 3.3-18 | <.001 | 2.4 | 1.6-3.5 | <.001 | 2.3 | 1.3-3.9 | .002 | |
| CD38 (pos. vs neg.) | 3.0 | 1.6-5.7 | .001 | 2.7 | 1.4-5.2 | .003 | 2.5 | 1.7-3.8 | <.001 | 2.6 | 1.4-4.6 | .002 | |
| Gender (male vs female) | 1.3 | 0.7-2.5 | .5 | 1.2 | 0.6-2.4 | .56 | 1.4 | 1-2.2 | .08 | 1.4 | 0.8-2.5 | .2 | |
| Rai stage | I vs 0 | 2.9 | 0.9-9.2 | .07 | 2.3 | 0.7-7.5 | .16 | 1.4 | 0.8-2.4 | .2 | 0.8 | 0.4-1.7 | .63 |
| II vs 0 | 4.4 | 1.2-15.7 | .02 | 2.5 | 0.7-9.1 | .15 | 3.2 | 1.8-5.8 | <.001 | 1.2 | 0.5-2.6 | .66 | |
| ≥III vs 0 | 12.0 | 3.5-40.9 | <.001 | 7.1 | 2.1-24 | .002 | 5.2 | 2.4-11.1 | <.001 | 3.2 | 0.7-14.4 | .13 | |
| Age (> vs ≤ median) | 3.3 | 1.7-6.6 | <.001 | 2.7 | 1.3-5.7 | .008 | 1.1 | 0.7-1.6 | .75 | 0.9 | 0.5-1.6 | .76 | |
| del17p13 | 2.9 | 1-8.8 | .06 | 3.3 | 1.1-10 | .04 | 1.9 | 1-3.8 | .05 | 2.4 | 1-5.8 | .05 | |
| dell11q23 | 2.6 | 0.9-8 | .09 | 1.6 | 0.5-4.8 | .43 | 2.4 | 1.3-4.4 | .007 | 1.8 | 0.8-4 | .13 | |
CI, confidence interval (of 95%); HR, hazard ratio. The values in rows miR-150, GAB1, and FOXP1 are boldfaced to highlight results for these genes of interest.
Analysis for miR-150, GAB1, and FOXP1 includes only samples obtained before the last day of follow-up for OS analysis (154/168) or before first therapy for TFS analysis (107/168); the others use all available samples.
Prognostic significance of miR-150, GAB1, or FOXP1. The OS and TFS are depicted using the Kaplan-Meier curves (with log-rank test) in the CLL cohort divided by median miR-150 expression (A-B), GAB1 expression (C-D) and FOXP1 expression (E-F). The cohort of CLL patients was divided by median expression of studied genes, and analysis included only samples obtained before the last day of follow-up for OS analysis (n = 154) or before first therapy for TFS analysis (n = 107).
Prognostic significance of miR-150, GAB1, or FOXP1. The OS and TFS are depicted using the Kaplan-Meier curves (with log-rank test) in the CLL cohort divided by median miR-150 expression (A-B), GAB1 expression (C-D) and FOXP1 expression (E-F). The cohort of CLL patients was divided by median expression of studied genes, and analysis included only samples obtained before the last day of follow-up for OS analysis (n = 154) or before first therapy for TFS analysis (n = 107).
To determine if miR-150 is an independent predictor of OS and TFS, we performed a multivariate analysis that included miR-150 levels and 8 routinely used prognostic markers. Although low-level expression of miR-150 was associated with the expression of ZAP-70 or use of unmutated IGHV (Figure 1C-D), low-level miR-150 had a strong independent prognostic value for OS and TFS (OS HR: 9.6 [CI 1.6-57.8]; TFS HR: 2.4 [CI 1.2-4.7]; Table 3). As a sensitivity analysis, univariate and multivariate use of delayed-entry Cox models to account for the variable timing of the blood draws (see supplemental methods) provided similar results (Tables 2 and 3). In univariate analyses, high-level expression of GAB1 and FOXP1 (>median) each was associated with a significantly shorter OS; high-level GAB1 also associated with shorter TFS (OS GAB1 HR: 2.3 [CI 1.2-4.4]; OS FOXP1 HR: 2.4 [CI 1.2-4.8], TFS GAB1 HR: 2.1 [CI 1.3-3.6]; Table 2; Figure 6C-F). The analysis performed with the use of delayed-entry models confirmed significant predictive association of FOXP1 levels with OS, and GAB1 levels with TFS (Table 2). Altogether, the adverse prognostic impact of low-level expression of miR-150, and the reverse trend for GAB1 and FOXP1 provides support for the relevance of these genes to the biology of neoplastic B cells.
The prognostic significance of miR-150 in a multivariate analysis
| . | Multivariate Cox regression of OS . | Multivariate Cox regression of TFS . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n = 108, number of events = 22* . | n = 76, number of events = 49* . | ||||||||||||
| Nondelayed entry model . | Delayed entry model . | Nondelayed entry model . | Delayed entry model . | ||||||||||
| Variable . | HR . | CI . | P . | HR . | CI . | P . | HR . | CI . | P . | HR . | CI . | P . | |
| miR-150 (≤ vs > median) | 9.6 | 1.6-57.8 | .01 | 6.0 | 1.2-30.8 | .03 | 2.4 | 1.2-4.7 | .014 | 3.6 | 1.8-7.4 | .0004 | |
| IGHV (unmut. vs mut.) | 5.9 | 0.4-85.5 | .19 | 2.4 | 0.1-40.0 | .54 | 1.3 | 0.5-3.5 | .64 | 0.3 | 0.1-1.2 | .099 | |
| ZAP-70 (pos. vs neg.) | 8.9 | 1.5-53.6 | .02 | 28.5 | 2.1-78.3 | .01 | 2.4 | 0.8-7.5 | .11 | 5.5 | 1.6-19.2 | .008 | |
| CD38 (pos. vs neg.) | 1.7 | 0.5-5.5 | .37 | 1.7 | 0.5-5.8 | .41 | 3.2 | 1.4-7.1 | .004 | 3.0 | 1.2-7.5 | .02 | |
| Gender (male vs female) | 2.6 | 0.7-9.5 | .17 | 2.8 | 0.7-10.9 | .14 | 2.1 | 0.9-4.6 | .07 | 2.0 | 0.9-4.7 | .1 | |
| Rai stage | I vs 0 | 5.2 | 0.8-35.8 | .09 | 2.3 | 0.3-18.1 | .42 | 2.9 | 1.3-6.6 | .01 | 1.1 | 0.5-2.6 | .79 |
| II vs 0 | 6.6 | 0.7-63.5 | .10 | 5.0 | 0.4-55.2 | .19 | 7.8 | 2.8-21.6 | .0001 | 2.6 | 0.9-7.3 | .07 | |
| ≥III vs 0 | 3.4 | 0.5-21.0 | .19 | 2.5 | 0.4-15.7 | .34 | 4.4 | 0.8-24.3 | .09 | 2.2 | 0.4-12.6 | .39 | |
| Age (> vs ≤ median) | 2.4 | 0.6-9.2 | .22 | 1.8 | 0.4-7.8 | .46 | 0.5 | 0.3-1.0 | .05 | 0.7 | 0.4-1.4 | .28 | |
| del17p13 | 4.0 | 0.8-19.5 | .09 | 3.0 | 0.5-16.3 | .21 | 1.4 | 0.5-4.3 | .52 | 3.2 | 1.1-9.5 | .03 | |
| del11q23 | 1.2 | 0.3-4.4 | .81 | 1.0 | 0.3-3.4 | .97 | 1.2 | 0.5-2.9 | .75 | 1.1 | 0.4-2.8 | .9 | |
| . | Multivariate Cox regression of OS . | Multivariate Cox regression of TFS . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n = 108, number of events = 22* . | n = 76, number of events = 49* . | ||||||||||||
| Nondelayed entry model . | Delayed entry model . | Nondelayed entry model . | Delayed entry model . | ||||||||||
| Variable . | HR . | CI . | P . | HR . | CI . | P . | HR . | CI . | P . | HR . | CI . | P . | |
| miR-150 (≤ vs > median) | 9.6 | 1.6-57.8 | .01 | 6.0 | 1.2-30.8 | .03 | 2.4 | 1.2-4.7 | .014 | 3.6 | 1.8-7.4 | .0004 | |
| IGHV (unmut. vs mut.) | 5.9 | 0.4-85.5 | .19 | 2.4 | 0.1-40.0 | .54 | 1.3 | 0.5-3.5 | .64 | 0.3 | 0.1-1.2 | .099 | |
| ZAP-70 (pos. vs neg.) | 8.9 | 1.5-53.6 | .02 | 28.5 | 2.1-78.3 | .01 | 2.4 | 0.8-7.5 | .11 | 5.5 | 1.6-19.2 | .008 | |
| CD38 (pos. vs neg.) | 1.7 | 0.5-5.5 | .37 | 1.7 | 0.5-5.8 | .41 | 3.2 | 1.4-7.1 | .004 | 3.0 | 1.2-7.5 | .02 | |
| Gender (male vs female) | 2.6 | 0.7-9.5 | .17 | 2.8 | 0.7-10.9 | .14 | 2.1 | 0.9-4.6 | .07 | 2.0 | 0.9-4.7 | .1 | |
| Rai stage | I vs 0 | 5.2 | 0.8-35.8 | .09 | 2.3 | 0.3-18.1 | .42 | 2.9 | 1.3-6.6 | .01 | 1.1 | 0.5-2.6 | .79 |
| II vs 0 | 6.6 | 0.7-63.5 | .10 | 5.0 | 0.4-55.2 | .19 | 7.8 | 2.8-21.6 | .0001 | 2.6 | 0.9-7.3 | .07 | |
| ≥III vs 0 | 3.4 | 0.5-21.0 | .19 | 2.5 | 0.4-15.7 | .34 | 4.4 | 0.8-24.3 | .09 | 2.2 | 0.4-12.6 | .39 | |
| Age (> vs ≤ median) | 2.4 | 0.6-9.2 | .22 | 1.8 | 0.4-7.8 | .46 | 0.5 | 0.3-1.0 | .05 | 0.7 | 0.4-1.4 | .28 | |
| del17p13 | 4.0 | 0.8-19.5 | .09 | 3.0 | 0.5-16.3 | .21 | 1.4 | 0.5-4.3 | .52 | 3.2 | 1.1-9.5 | .03 | |
| del11q23 | 1.2 | 0.3-4.4 | .81 | 1.0 | 0.3-3.4 | .97 | 1.2 | 0.5-2.9 | .75 | 1.1 | 0.4-2.8 | .9 | |
CI, confidence interval (of 95%); HR, hazard ratio. The values in row miR-150 are boldfaced to highlight results for this gene of interest.
Analysis includes only samples with known FISH status and obtained before the last day of follow up for OS analysis (108/168) or before first therapy for TFS analysis (76/168). The multivariate analysis for samples including those with unknown FISH status obtained before the last day of follow-up for OS analysis (154/168) or before first therapy for TFS analysis (107/168) is provided in supplemental Table 4.
Discussion
We identified miR-150 as being the most abundantly expressed miRNA in CLL. However, we observed significant heterogeneity in the expression levels of this miRNA among CLL cells of different patients. Low-level expression of miR-150 associated with unfavorable clinicobiological and prognostic markers, such as expression of ZAP-70 or use of unmutated IGHV (P < .005). Additionally, our data suggest that the levels of methylation of the upstream region of 1000 nt proximal to miR-150 associate with its expression. We demonstrated that GAB1 and FOXP1 genes represent newly defined direct targets of miR-150 in CLL cells. We also showed that high-level expression of GAB1 and FOXP1 associates with relatively high sensitivity of CLL cells to surface immunoglobulin ligation. High levels of GAB1/FOXP1 and low levels of miR-150 associate with a greater responsiveness to BCR ligation in CLL cells and more adverse clinical prognosis.
It was demonstrated that abundantly expressed, evolutionary-conserved miRNAs are important for immune-cell phenotypes and a can regulate stability of mRNAs (reviewed in O’Connell and Baltimore12 ). Several target genes have previously been described for miR-150 in studies performed on various cell types.43-47 To describe targets regulated by miR-150 in CLL B cells, we performed array-based transcriptome profiling of 100 CLL samples stratified based on miR-150 levels in combination with the computational prediction of its targets. This approach allows for identification of cell type–specific miR targets because of a dominant effect of miRNA on mRNA stability.11-14,48,49 This approach obviated studies using forced expression of the studied miRNA that can lead to shifts in target mRNAs and off-target effects.50,54,55 In our analysis, 95% of genes (55/58) were downregulated in cases with high-level expression of miR-150 (P < .001), suggesting that the effects on mRNA stability of high-level expression of miR-150 were not random. We found that out of the 58 genes, 2 genes (GAB1 and FOXP1) contained evolutionary-conserved binding sides for miR-150. The expression of mRNAs for GAB1 and FOXP1 decreased with increase in miR-150 levels. The higher mRNA levels of GAB1 and FOXP1 in CLL cases with low miR-150 expression were also reflected in their higher protein levels, and transfection of B cells with synthetic miR-150 resulted in their down-modulation. The cotransfection of miR-150 with luciferase construct containing 3′UTR of GAB1 or FOXP1 resulted in a ∼25% reduction in luciferase activity, confirming that these 2 genes represent newly defined evolutionary conserved targets for miR-150. However, we cannot exclude the possibility that some of the other identified differentially expressed genes represent targets of miR-150 that evolved later in evolution and are not conserved in vertebrates. Additionally, it is possible that the spectrum of preferential targets regulated by miR-150, and the extent to which a target mRNA is regulated can vary with the global gene expression changes that are associated with events such as disease progression.38 Nevertheless, neither GAB1/FOXP1 nor miR-150 previously had been identified in the screening of gene networks or miRNAs that change with disease progression.33,38 In our study, the lower levels of miR-150 in cases with higher Rai stage (III-IV) could indicate that miR-150 decreases with disease progression, but more likely reflects that this subgroup of patients contained higher proportion (80%) of cases that used unmutated IGHV and expressed ZAP-70, which are each associated with lower level expression of miR-150.
None of the 58 genes identified by our microarray analysis of CLL cases with high vs low levels of miR-150 overlapped with a known miR-150 target, which illustrates the importance of identifying cell type–specific miRNA targets.50 The only previously validated miR-150 target in B cells is the c-MYB gene, which was identified in a mouse model in which the authors noticed that miR-150 deficiency leads to a phenotype opposite to that caused by deleting c-MYB.47 The B cell–specific deletion of the c-MYB gene leads to a block in B cell development at the pro- to pre-B transition.56 miR-150 regulates c-MYB levels during the transition of pro-B cell to pre-B cells; ectopic expression of miR-150 in B cell progenitors could block B-cell development.47 In our study, the expression of c-MYB was not inversely correlated with miR-150 levels, which is not surprising because the original study had already described that c-MYB levels were not regulated by miR-150 in mature resting B cells.47 The c-MYB is expressed in pro- and pre-B cells; in mature B lymphocytes, its mRNA expression is kept low and controlled at the transcriptional level.56,57 Interestingly, the phenotype of mice with manipulated miR-150 expression supports the regulation of FOXP1 by miR-150 in B cells. The mice ectopically expressing miR-150 in hematopoietic cells share comparable phenotype (pro-B to pre-B transition blockade) not only with c-MYB knockout animals, but also with FOXP1 knockout mice.47,52,56,58 Overall, our data demonstrate that in the context of peripheral blood CLL cell transcriptome, the GAB1 and FOXP1 genes are relevant targets of miR-150. However, it is likely that under circumstances that would activate expression of c-MYB mRNA in CLL, this mRNA could be targeted by miR-150.
The GAB1 and FOXP1 proteins are both known to have important functions in B cells. FOXP1 is a transcription factor that controls B-cell maturation, specifically the expression of genes required for rearranging immunoglobulin subgenes in immature B cells.52 In malignant B cells, it was shown that its aberrant overexpression is present in cells with ABC in DLBCL and associated with worse survival in DLBCL, follicular lymphoma, or mucosa-associated lymphoid tissue lymphoma.53,59-61 FOXP1 protein expression is used in some of the immunohistochemical algorithms that distinguish ABC vs germinal center phenotype in DLBCL.62 In malignant B cells, FOXP1 expression can be induced by activation of nuclear factor-κB or stimulation through the BCR,53,59 and its levels have massive effect on the gene expression network in B cells.63 However, the exact functions of this transcription factor in mature normal or malignant B cells are poorly understood. We found that silencing FOXP1 in B cells reduces their responsiveness to BCR stimulation and in CLL its higher levels are associated with higher responsiveness to BCR stimulation. Furthermore, we noted that silencing FOXP1 down-modulated both the basal and anti-µ–induced levels of phosphorylated AKT. It has been shown recently that other members of the forkhead-box family of proteins regulate basal and induced phospho-AKT levels.64 Additionally, expression of FOXP1 has pleiotropic effects on multiple cell signaling pathways in B cells including BCR signaling and cell activation.63 This could at least partially explain association of its high-level expression with poor prognosis in multiple B-cell malignancies.
The functions of GAB1 in modulating the sensitivity of B cells to anti-µ are better described.51 GAB1 is an adaptor molecule recruiting the PI3K to the B-cell membrane, which allows for the amplification of BCR signaling through activation of AKT.51 The GAB1 is also involved in other pathways where it serves as a docking/scaffolding for PLCγ, Crk, and CrkL proteins (reviewed in Sármay et al65 ). In CLL cells, higher GAB1 levels were associated with strong BCR responsiveness. Silencing of GAB1 in B-cell lines directly affected the amplitude of their response to BCR crosslinking. Importantly, the transfection of B cells with miR-150 recapitulated this phenotype. Overall, this resembles miRNA’s known role in balancing T-cell receptor signaling strength and PI3K signaling in T cells.13,18,19
The significance of miR-150 in CLL B-cell biology is supported by the association of its expression and its targets with the magnitude of BCR signaling observed following surface immunoglobulin ligation. Additionally, lower miR-150 levels associated with shorter OS and TFS (OS HR: 2.8 [CI 1.3-5.9]; TFS HR: 1.7 [CI 1.0-2.8]). The higher levels of FOXP1 and GAB1 in CLL cells associated with shorter OS of these patients (FOXP1 HR: 2.4 [CI 1.2-4.8]; GAB1 HR: 2.3 [CI 1.2-4.4]). This indicates that the regulatory connection between miR-150 and the identified targets is an important one for the behavior of malignant B cells. However, it is possible that miR-150 regulates other genes that also contribute to the biology of malignant B cells.
Conceivably inhibition of BCR signaling might also influence the expression of miRNAs (such as miR-150), which are involved in the regulation of BCR signaling. Also, the expression of such miRNAs may impact on the sensitivity of malignant B cells to inhibitors of BCR signaling. Examining CLL cells of patients obtained before and after initiation of therapy with such inhibitors could help address this hypothesis.
In summary, we have used integrated analysis of miRNA and gene-expression profiling to identify 2 novel targets regulated by miR-150. Regulation of GAB1 and FOXP1 by this miRNA contributes to the sensitivity to BCR ligation in CLL cells and associates with differences in disease prognosis. Our study represents a seminal example of how miRNA can modulate the expression of proteins that contribute to the competency of BCR signaling (Figure 7). It is likely that miR-150 also will be found capable of influencing the expression levels of GAB1 and FOXP1 and BCR signaling, in other B-cell malignancies or normal B cells.
MicroRNA miR-150 modulates the expression of GAB1 and FOXP1 that are involved in BCR-signaling pathway in CLL B cells.
MicroRNA miR-150 modulates the expression of GAB1 and FOXP1 that are involved in BCR-signaling pathway in CLL B cells.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE49896).
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Institutes of Health Method to Extend Research in Time (grant R37 CA049870-23), National Institutes of Health CLL Research Consortium (grant 2P01 CA081534-12A1) , the University of California-San Diego Foundation Blood Cancer Research Fund, South Moravian Programme for Distinguished Researchers II (SoMoPro II) (cofinanced by the European Union and the South-Moravian Region), CZ.1.07/2.3.00/30.0009 (cofinanced by the European Union and Czech Republic), and the EHA Research Fellowship award granted by the European Hematology Association.
Authorship
Contribution: M.M. designed and performed research, analyzed data, and wrote the manuscript; L.C. performed and analyzed the anti-μ stimulation of CLL; L.Z.R. and E.M.G. provided clinical samples and collected the clinical data; H. L. and K.M. performed the statistical analysis of overall survival and treatment-free survival; K.J., E.N.S., and K.A.F. analyzed the methylation of miR-150 upstream region; and T.J.K. designed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thomas J. Kipps, University of California-San Diego, 3855 Health Sciences Dr, La Jolla, CA 92093; e-mail: tkipps@ucsd.edu.


![Figure 3. Validation of newly defined miR-150 targets. (A) GAB1 or FOXP1 immunoblot analysis in CLL samples of 16 patients selected for low-level (n = 8) vs high-level expression of miR-150 (n = 8). The relative levels of miR-150 expression are indicated (first sample set as 1). The samples were solely chosen based on the differences in miR-150 expression and partially overlapped with samples used in the microarray analysis (7 of 16 samples). The double bands identified by the anti-FOXP1 antibody (JC12 clone) represent 2 splicing variants of FOXP1 as described previously.53 (B) A representative immunoblot for GAB1 or FOXP1 protein in MEC-1 and Raji, primary CLL cells transfected (48 hours) with artificial miR-150 (miR-150, 100 nM), and control miRNA (negative control [Neg. Ctrl], 100 nM). MOCK represents cells treated only with the electroporation pulse. The Blot images were quantified with ImageJ 1.42q (National Institutes of Health), and the GAB1/β-actin and FOXP1/β-actin ratio in MOCK was arbitrarily set at 1. The FOXP1/β-actin rations for its 2 major splicing variants are provided (the ratio of the larger vs the smaller isoform is separated by a slash). (C) The luciferase activity in 293FT cells cotransfected with miR-150 mimic (MIMIC miR-150, 100 nM) or luciferase reporter construct (100 ng) containing the 3′UTR region of GAB1, FOXP1 or GAPDH (GAPDH as control). As a negative control, we used a control miRNA mimic (CTRL Scramble, 100 nM). Luciferase activity was measured 24 hours posttransfection and compared by paired Student t test.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/1/10.1182_blood-2013-09-527234/4/m_84f3.jpeg?Expires=1765898035&Signature=tfzs6Kws0F1x8QRLXU06mqqCTZL3-caXj6fSdLZdp0Q-ck5KgBmZpyTlBbfukajEZiFRstgko~3a9r59ibOYE-xhp4rpPsYHGp8aiVJ8f-fdVircqd1e2zRQzN1mBDfUy2yv4RYg7XAnJIgS9Jo0JqL6-mdVQwh6P3EBSI5EwmRfDWK~1dX13YQeunMsSclnYUBrW1CH-oIZfxmyHxGwaatq75hOX0vsD~C4~FtlmmxDrdteC3~ODbv45ypzl9lLbGfZ-MkFK8rPJehgB~Aip15wtQyY3ZUK-Dx-Zb7cpOH1a0LJDzmX7o7kPHtJ-OyKe6KhpdecDHrHQDpUScacXw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Expression of miR-150 targets GAB1 or FOXP1 affects BCR signaling. (A) The Raji cell line was transfected with artificial miR-150 (miR-150, 100 nM), control miRNA (miR negative control [Neg. Ctrl], 100 nM), siRNA against GAB1 (siGAB1, 100 nM) or FOXP1 (siFOXP1, 100 nM), and control siRNA (siRNA Neg. Ctrl, 100nM). The band intensities observed in immunoblot analysis were measured using ImageJ 1.42q, and the GAB1/β-actin and FOXP1/β-actin ratio in control was arbitrarily set at 1. The FOXP1/β-actin ratios for the 2 major isoforms of FOXP1 are provided (ratio for larger and smaller variant is separated by a slash). GAB1 and FOXP1 protein levels were analyzed 48 hours posttransfection and β-actin was used as a loading control (representative example of an immunoblot). (B-C) The effect of Raji cell transfection with siRNA against GAB1 (siGAB1, 100 nM) or FOXP1 (siFOXP1, 100 nM) or control siRNA (siNEG CTRL, 100 nM) on calcium influx after anti-μ stimulation (10 μg/mL; indicated by a black arrow). (D) The effect of Raji cell transfection with artificial miR-150 (miR-150 MIMIC, 100 nM) or control miRNA (miR NEG CTRL, 100 nM) on calcium influx after anti-μ stimulation (10 μg/mL; indicated by a black arrow). The calcium influx (FLUO-4 fluorescence intensity) in panels B-D were analyzed continuously for 120 seconds. The acquisition of background fluorescence (30 seconds) was followed by adding anti-μ (final concentration 10 μg/mL; indicated by a black arrow), and data at 6-second intervals were visualized as peak intensity. Error bars represent standard error of the mean of at least 2 independent experiments. Differences were statistically analyzed by 2-way analysis of variance.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/1/10.1182_blood-2013-09-527234/4/m_84f4.jpeg?Expires=1765898035&Signature=twoOWmF71kjE~Hm7B6fXz3EmgmfBXxKY4VPAOp9c6EVRlFxJoWN-ntKIdh-DHxQZeyKRI4P3yLRUCqXyoP71oQkPrVALNvA6ZNGZ0vB3goU0W4XBXchm~z2WFJgO9C5ko5GjeSPrLPFPSCdcZhj0HiwgAFe37zyQYy9C6lLCDLHCEHHuDSEyYJE~Du4I6lsgw1j-jTWw~QP-aQKUOua2doe4YFLtjo2FUIsQqSdQVfLwAONZCNUy6F-6n~rCSZePGUI-kp52uaMWyRyh0moYNHYHJlHImu99ls4aYt8Mp-2~DzODWj4fiALl0xIF3sLvi137cxvAhmhwGvVzXUjvMg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal