Abstract
There is a demand to understand B-cell lymphoma pathogenesis better, to identify new markers, and to define multiple lymphoproliferative disorders more accurately. MicroRNAs (miRNAs) are regulators of protein translation, comprising a group of more than 1500 short noncoding single-strand RNA molecules of approximately 22 nucleotides in length. They are easily detectable in fresh or paraffin-embedded diagnostic tissue and serum. Expression of individual miRNAs and miRNA signatures allows specific cell-differentiation stages to be identified, and is a powerful diagnostic and prognostic method. Here we review what is known about the pathogenic relevance of miRNAs, and use of miRNAs for the diagnosis and prognosis of B-cell lymphomas. Most of the published data concern chronic lymphocytic lymphoma and diffuse large B-cell lymphoma, and implicate miRNAs in the pathogenesis of these diseases. They identify miRNAs that could be used for diagnosis, prognosis, or prediction of response to specific therapies.
Introduction
B-cell lymphomas make up a heterogeneous group of lymphoproliferative disorders that originate from B cells and whose pathogenesis is still largely unknown. Classification of B-cell lymphomas has essentially been based on the recognition of characteristic gene translocations that deregulate the expression of oncogenes or tumor suppressor genes.1 B-cell lymphoma classification has made it possible to recognize and successfully treat a range of disorders, but a significant proportion of B-cell lymphoma patients still fail to respond to therapy.
Although the accuracy with which the various B-cell disorders are recognized has improved through the use of new immunohistochemical and molecular markers, there is still a demand to understand lymphoma pathogenesis better, to identify new markers, and to define the range of lymphoproliferative disorders more accurately. Lymphoma classification is largely based on the assumption that lymphoma cells derive from specific cell populations, thereby making it possible to correlate the phenotype of particular lymphoma types with that of the normal counterpart B-cell subpopulations. Nevertheless, there is a growing understanding that lymphoma-specific markers also recapitulate the history of lymphoma pathogenesis, expressing additional markers that provide information about specific molecular events.
The potential number of specific molecular markers for lymphoma diagnosis and prognostication has improved in the last few years thanks to the recognition of microRNAs (miRNAs) as central players in B-cell differentiation and oncogenesis. miRNAs are a recently identified class of noncoding single-strand RNA molecules of 21-23 nt that function as posttranscriptional regulators of gene expression, targeting their corresponding messenger RNAs for degradation or translational repression.
MicroRNAs
miRNAs were first described in 1993 in Caenorhabditis elegans,2,3 although it took several years to confirm the supposition that miRNAs are essential components of cell molecular machinery.4-6 There has been a rapid increase in the number of publications concerning miRNAs in the last 5 years, with the number of published articles doubling approximately every 2 years. The discovery that miRNAs are evolutionary conserved4 highlights their importance and, in fact, they are now known to have a key role in protein expression control,7 and consequently, in the decisions about cell differentiation and growth.8 miRNA deregulation is now recognized as being involved in the development of all major complex diseases.9 Data from more than 1500 human miRNAs are currently available in the Sanger database (http://www.mirbase.org/). Nevertheless, our understanding of the role of these miRNAs is incomplete, and much is left to discover. miRNA transcription and maturation have been reviewed recently.10,11 In brief, miRNAs can be found in almost any part of the DNA, including intronic and exonic regions of other genes, in repeated and imprinted sequences. They are transcribed as long transcripts of variable length or as clusters of miRNAs (pri-miRNAs) by RNA polymerase II, individually cleaved in the nucleus by Drosha, capped and polyadenylated (called at this time pre-miRNAs, with a length of ∼ 70 nt), and exported to the cytoplasm by exportin 5, where Dicer is in charge of cutting the double-strand (ds) miRNAs. The ds-miRNAs become separated, and, although there are some exceptions, in most cases only 1 strand is functional. The RISC enzyme binds the ss-miRNA and helps it to find its complementary sequence, which is generally found at 3′UTR on messenger RNA. This results in the modulation of expression of the corresponding protein through RNA degradation (a mechanism that is probably similar to si-RNA) or protein translation blocking/inhibition.12,13
In 2002, Calin et al drew our attention to the relevance of miRNAs in lymphoid malignancies.5 Since then, several studies have confirmed their importance in determining B-cell differentiation, the essential role they play in the lymphoma molecular orchestra, and their potential as diagnostic, prognostic, and predictor markers. This review focuses on the most frequent lymphoma types and mechanistically confirmed miRNAs in an attempt to summarize what is known with respect to B-cell lymphoma.
MicroRNAs in B-cell differentiation
B-cell differentiation is a highly regulated process in which uncommitted hematopoietic precursors are differentiated into antibody-secreting plasma cells by a multistep process. A network of transcriptional regulators determines every step along this pathway, in response to signals from the cell microenvironment. The role of miRNAs in B-cell differentiation is being progressively revealed, mainly through the use of genetically modified mice. Although there are a considerable number of studies contributing to a vast list of miRNAs implicated in B-cell differentiation, everything suggests that our knowledge remains incomplete. Table 1 shows the principal miRNAs that are known to be involved in the different stages of B-cell maturation.
Stage-specific miRNAs involved in B-cell differentiation
| . | Naive B cells . | Germinal center B cells . | Memory B cells . | Plasma cells . |
|---|---|---|---|---|
| ⇑ | miR-18131 | miR-17-5p16,32 | miR-22316,32 | |
| mi-34a24 | miR-181b25,32 | |||
| miR-22316,31,32 | miR-125b 31,33 | |||
| miR-155 26 | ||||
| ⇓ | miR-22331-33 | miR-181b31-33 | miR-181b16,32 | |
| miR-15022,31,33 | miR-17-5p16,32 | miR-17-5p32 | ||
| miR-3016,32 |
Naive B cells migrate to lymphoid organs, where they are transformed in the context of T cell–dependent germinal centers (GCs). Within these centers, B cells undergo somatic hypermutation (SHM) of immunoglobulin-variable genes and Ig class-switch recombination (CSR), and then specific antigen-reactive GC B cells differentiate into the major effector B cells of the adaptive immune system: memory and plasma cells. The process is probably more complex because GC-independent memory B cells have also been found, and exposure to the GC environment may be consecutively repeated for some B-cell subpopulations.14 The role of specific transcription factors in mature B-cell differentiation has been the subject of many studies, but specific miRNAs and other regulators of these transcription factors during mature B-cell differentiation are largely unknown.
It seems that no miRNA species is specific to a single cell type15 and that miRNAs regulate hematopoiesis by modulating multiple signaling pathways that are cell-type and context-specific.15 However, some well-characterized miRNAs have been found to exert a key role in various steps of B-cell development. For example, Zang and coworkers, simultaneously analyzing miRNA and mRNA expression profile data, identified a potential regulatory role for miRNAs in discrete stages of mature B-cell differentiation, and specifically identified a direct role for the miRNA-mediated regulation of oncogenes and key transcription factors in B-cell differentiation (C-MYB, C-MYC, BCL6, LMO2, and PRDM1 [Blimp1]),16 a finding that has been matched to some extent in the study of B-cell malignancies.17
The first evidence of the importance of miRNAs in hematopoiesis differentiation was reported in Science in 2004. The study found that MiR-181 was preferentially expressed in the B-lymphoid cells of mouse bone marrow, and its ectopic expression in hematopoietic stem/progenitor cells led to an increased fraction of B-lineage cells.18
Other miRNAs have been demonstrated to play a determinant role in the lineage differentiation decision: for instance, high expression levels of the cluster of miR-23a (miR-23a, miR-27a, and miR-24) and miR-125b reduce the differentiation to B-cell lymphocyte lineage in favor of myeloid differentiation19,20
Moreover, some miRNAs have been found to play a role in early B-cell development, such as miR-150, whose up-regulation in hematopoietic progenitors has been shown to reduce the normal quantity of mature B cells, suggesting a blockage in the maturation process at the pro-B cell stage.21 A more comprehensive study further clarified the role of miR-150 during B-cell development, showing that miR-150 regulates the expression of C-MYB, a transcription factor that is highly expressed in lymphocyte progenitors, down-regulated on maturation, and up-regulated again after activation of the mature cells during B-cell maturation.22 The miR-17-92 cluster regulates survival of early B-cell progenitors by repressing the expression of proapoptotic genes (Bim) at the pro- to pre-B cell transition, as shown by Ventura.23
Finally, miR-34a, which is expressed at low levels in early B cells, regulates B-cell differentiation from pro-B cell to pre-B cell. Experiments in which the expression of miR-34a has been forcedly introduced confirmed the existence of a blockage at the pro-B cell stage as a consequence of Foxp1 protein down-regulation, a transcriptional regulator required for B-cell differentiation, which is a direct target of miR-34a. In the same work, knockdown of miR-34a results in increased amounts of Foxp1 and mature B cells.24
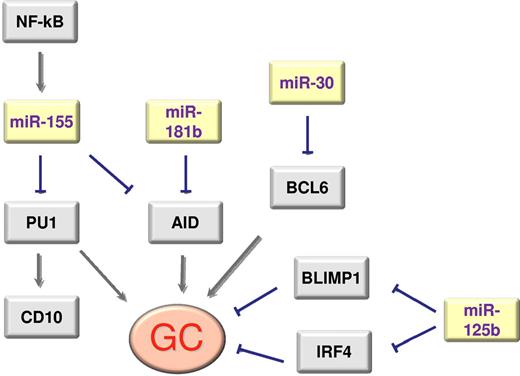
GC reaction is also finely regulated by several miRNAs, in particular by miR-181b and miR-155, which are chief controller miRNAs of GC reactions. These 2 miRNAs share the activation-induced cytidine deaminase (AID)25,26 as a target. AID expression is required during the B-cell somatic hypermutation process of immunoglobulin genes and its loss leads to the impairment of class switching27 : consequently, expression of miR-181b and miR-155 impairs SHM and CSR.
The importance of miR-155 in GC regulation is further documented by the fact that miR-155 negatively regulates the expression of CD10,28 a marker of GC cells. Moreover, miR-155 has also been found to regulate specific B-cell subpopulation maturation in mice, where loss of miR-155 leads to a reduction in IgG1 switching by targeting the PU.1 transcription factor.29 Another miRNA with a role in the final step of B-cell maturation in mice is miR-125b, whose overexpression impairs B-cell transition to plasma cells.30
Specific B-cell subpopulations
GC B cells are distinguished by the expression of a set of transcription factors including BCL6, LMO2, and the absence of plasma cell markers such as PRDM1 and XBP1. A full picture of the epigenetic regulation of the expression of these genes has yet to emerge, but some of the pieces of the puzzle are becoming clear. Thus, miR-155 plays a role in class switching29 and AID26,31 expression together with miR-181b,25 while BCL6 and PRDM1/BLIMP1 are regulated by the miR-30 family, miR-9 and let-7a.32 Malumbres and coworkers have also demonstrated that GC-enriched miR-125b down-regulates the expression of IRF4 and PRDM1/BLIMP1, and that memory B cell–enriched miR-223 down-regulates the expression of LMO2.33
Activated B cells seem to display some specific miRNA markers. Thus, increased miR-155 expression seems to play a role, depending on NF-kB activity, in silencing PU1 and CD10.28 Marginal zone B cells are distinguished by the expression of miR-223,16,34 an miRNA that also distinguishes memory B cells. The main relationships between miRNAs and genes regulating GC differentiation are summarized in Figure 1.
Scheme of the main interactions between genes and miRNAs that regulate GC differentiation.
Scheme of the main interactions between genes and miRNAs that regulate GC differentiation.
B-cell lymphomas and miRNAs
Burkitt lymphoma
The first confirmed discovery in Burkitt lymphoma (BL) was the loss of miR-155,35,36 an miRNA subsequently found also to be important in B-cell development.29 The finding is of particular relevance because miR-155 is expressed by diffuse large B-cell lymphoma (DLBCL), an extremely useful distinction in the clinical milieu.17 In addition, miR-155 has been shown to suppress AID-mediated MYC-IGH translocation,31 a finding that links the loss of miR-155 to the presence of MYC-IGH translocation, the hallmark of BL. Finally, because miR-155 negatively regulates the expression of CD10,28 its low expression in BL may be related to the universal expression of CD10 in BL.
Burkitt lymphoma is thus characterized by the deregulated expression of C-MYC, as a consequence of translocations involving C-MYC (8q24) and immunoglobulin genes. C-MYC regulates a large set of miRNAs that coordinate to regulate the expression of C-MYC.37 Thus, BL is characterized by the unbalanced activity of C-MYC versus miRNAs,37 thereby forming an intricate set of autoregulatory loops that include other transcription factors and multiple miRNAs.38 Some of the miRNAs involved in this autoregulatory loop are let-7a, e and f, miR-34b, miR-98, miR-331, and miR-363.37,39,40
Furthermore, miR-17-92 is a direct transcriptional target of C-MYC, and experiments in a mouse model of B-cell lymphomas have shown cooperation between these 2 oncogenes.41 As might be expected, miR-17-92 is expressed by BL cases.17 Several epidemiologic subtypes of BL (endemic, sporadic, HIV-associated) share a homogenous microRNA profile distinct from that of DLBCL,42 which confirms the potential relevance of this signature in the diagnosis of BL.
At the present time, it is unclear whether C-MYC–negative cases diagnosed as BL should be included within the borderline BL/DLBCL cases or whether they represent bona fide BL cases. The description of a characteristic miRNA signature for these cases lacking C-MYC translocation suggests that they may be a discrete family of cases deserving specific recognition.40
Diffuse large B-cell lymphoma
DLBCL is the most common type of non-Hodgkin lymphoma (NHL), accounting for approximately 30% of them. Despite their relatively common morphology, DLBCL cases have striking clinical and molecular variability, which is also reflected in their miRNA signature. DLBCL gene expression studies have identified 3 major types, all of which have characteristic signatures, genetic alterations, and prognoses: GC, non-GC, and primary mediastinal large B-cell lymphoma (PMLBCL).8
In contrast to BL, DLBCL cells have a 10- to 30-fold higher miR-155 copy number than do normal circulating B cells.43 Significantly higher levels of miR-155 are present in DLBCLs with a non-GC cell phenotype than with the GC phenotype.43 Other GC and non-GC miRNA markers have been added to the list.33,44,45
miR-18a, miR-181a, and miR-222 have been shown to predict overall survival (OS) and progression-free survival (PFS) in rituximab cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP)–treated DLBCL,46 and this finding has been confirmed for miR-222 in at least 2 independent studies,33,45 wherein the expression of an miRNA signature was found to be correlated with OS and PFS, independent of the conventional International Prognostic Index (IPI) and GC/non-GC markers.
More layers of complexity are being added to our understanding of lymphoma pathogenesis. It was recently reported that EBV miRNAs are also active players in the lymphoma choreography, where they exploit the miRNA machinery to modulate and/or subvert virus-host cell interactions, and regulate the expression of BCL6 in tumor samples, thereby enabling NF-kB activation,47 or suppressing the expression of CXCL-11/I-TAC, as an immunomodulatory mechanism in EBV-related lymphomas.48
Follicular lymphoma
Follicular lymphoma (FL) is characterized by a unique miRNA signature containing a subset of miRNAs whose expression is correlated with biologic features of the tumor and its response to chemotherapy.49 The FL miRNA signature includes miR-20a/b and miR-194, which target CDKN1A and SOCS2, potentially contributing to tumor cell proliferation and survival.49 Interestingly, some of the miRNAs (let-7, miR-30) that regulate the expression of BCL6 and BLIMP1/PRDM1 are induced after follicular dendritic cells (FDCs) contact.32
Although most FLs feature a t(14;18)(q32;q21) translocation, a subset of cases lack this. miRNA profiling of these t(14;18)–negative cases has revealed a distinct miRNA profile that is associated with an increased proliferative capacity and a “late” GC B-cell phenotype.50
Chronic lymphocytic leukemia
miRNA studies in chronic lymphocytic leukemia (CLL) have notably contributed to our understanding of the disease and to a better knowledge of the role of miRNAs in cancer pathogenesis.5 In 2002, Calin et al reported the loss of the miR-15a/16-1 cluster from the 13q14 locus as a seminal finding in CLL pathogenesis.5 miRNAs in this cluster regulate the expression of BCL2 and apoptosis, which partially explains the stronger expression of BCL2 and diminished apoptosis that distinguishes CLL cells.51,52 Subsequently, Klein and coworkers demonstrated that the miR-15a/16-1 cluster also negatively regulates the expression of multiple genes involved in the G0/G1-S phase transition in human and mouse B cells.53 miRNAs also influence CLL diversity because miR-29 and miR-181 regulate TCL1,54 miR-221/222 regulate p27,55 miR-106b regulates p73,56 and miR-155 is activated by MYB,57 while miR-34a regulates E2F1 and MYB.58 Transgenic mice that overexpress miR-29 (which targets TCL1) in B cells develop a CLL with an indolent phenotype, indicating that miR-29 might function as a tumor suppressor in CLL.59 Interestingly, Visone and coworkers recently demonstrated that miR-181b expression decreased as the disease progresses, suggesting that it would be worth evaluating as an important tool for monitoring the course of CLL.60 Croce and coworkers have shown that Eμ-mmu-miR155 transgenic mice develop a preleukemic pre-B cell proliferation that is evident in spleen and bone marrow, followed by a B-cell malignancy with features of lymphoblastic leukemia/lymphoma, suggesting that miR155 induces polyclonal expansion, followed by secondary genetic changes leading to full transformation.61 A more detailed description of the CLL findings is beyond the limits of this review, but 2 very recent articles have addressed the use of plasma miRNA expression levels and feedback circuitry in identifying novel parameters for prognosis and prediction.62,63
Mantle cell lymphoma
Mantle cell lymphoma (MCL) is a relatively homogeneous tumor whose miRNA signature has been identified by several groups.64-66 Several miRNAs target cell-cycle genes (Cyclin D1 is regulated by miR-16-167 and oncomiR-168 ; CDK6 is regulated by miR-2964 ), NF-kB (miR-17/9266 and miR-26a65 ), or chromatin modifiers (miR-92b and miR-96 regulate PRMT569 ).
Marginal zone lymphoma, MALT type
miRNA studies in mucosa-associated lymphoid tissue (MALT) lymphoma are particularly difficult because the representation of the neoplastic cells in the tumor samples is always diluted by the reactive lymphocytes or the remnants of the infiltrated epithelium. A study comparing tumors diagnosed in the conjunctiva with adjacent normal conjunctival tissue revealed up-regulation of miR-150 and miR-155, and down-regulation of miR-184, miR-200a, b, and c, and miR-205.70 Further in vitro experiments with miR-200a, b, and c demonstrated their capacity to down-regulate Cyclin E2, alone or in combination.
Another study of gastric MALT lymphoma revealed that a high level of expression of miR-223 is a marker of MALT stratification and that this is correlated with increased E2A+ expression, higher clinical stages, and a less frequent response to Helicobacter pylori eradication therapy.71 Large B-cell lymphomas that originate in the stomach, and which are presumably derived from the MALT, exhibit a C-MYC miRNA signature, composed of 27 miRNAs transcriptionally repressed by C-MYC. In particular, miR-34a loss deregulates the expression of FOXP1, a marker of activated B cells that is usually present in gastric DLBCL cases.72
Nodal marginal zone lymphoma
Studies of nodal marginal zone lymphoma (NMZL) seem to confirm that these tumors have distinctive features that distinguish them from FL cases. Compared with FL, NMZL cases show increased expression of miR-221, miR-223, and let-7f, a signature that is very similar to that exhibited by memory B cells and cells isolated from the normal marginal zone. Up-regulation of miR-223 and miR-221, which target the GC-related genes LMO2 and CD10, could be partially responsible for the expression of a marginal zone signature.34
Splenic marginal zone lymphoma
Splenic marginal zone lymphoma (SMZL) is the lymphoma of the splenic white pulp, whose morphology resembles the normal organization of the splenic lymphoid follicle. The 7q32 deletion is found in approximately 40% of such cases. This chromosomal region contains a cluster of miRNAs (including miR-29a and 29b-1) and is commonly lost in these tumors.73,74 A miRNA signature has been identified in SMZL, including 5 miRNAs that are overexpressed in SMZL, miR-21, miR-155, and miR-146a, while 7 miRNAs, including miR-139, miR-345, miR-125a, and miR-126, had significantly reduced expression. Furthermore, miR-26b, an miRNA known for its suppressor properties, was significantly down-regulated in SMZLs arising in Hepatitis C virus (HCV)–positive patients.75 Some of these miRNAs were confirmed in an independent study that found increased expression of miR-21 to be associated with an adverse outcome.74,75
Is the expression of specific miRNAs or miRNA signatures useful for lymphoma diagnosis?
The most frequent lymphoma types have been studied, and the miRNAs that distinguish each of these tumors have been fairly exhaustively identified. This has enabled lymphoma pathogenesis to be understood and has neatly demonstrated the complexity of the lymphoma molecular machinery (Table 2). The addition of miRNA evaluation in lymphoma diagnostics might be of specific interest in the evaluation of differential diagnosis of uncertain cases. For instance, several groups have used formalin fixed paraffin-embedded (FFPE) samples from independent series of patients to show that miR-155 is lost in BL cases and expressed in DLBCL samples.17,36,42,76 Nevertheless, we await practical applications of these data, particularly in those areas where accurate markers for lymphoma diagnosis are required, such as the recognition of intermediate BL/DLBCL cases.
Validated miRNA targets in B-cell lymphoma types
BL indicates Burkitt lymphoma; CLL, chronic lymphocytic leukemia; AID, activation-induced cytidine deaminase; DLBCL, diffuse large B-cell lymphoma; FL, follicular lymphoma; MCL, mantle cell lymphoma; MZL, marginal zone lymphoma; MALT, mucosa associated lymphoid tissue; and NMZL, nodal marginal zone lymphoma.
Beyond the recognition of markers for histopathologic diagnosis of tumor samples, some emerging findings suggest the evaluation of serum/plasma miRNAs expression levels as a desirable noninvasive method for monitoring minimal residual disease, for patient survival stratification,77 and for predicting response to therapy.78 There are clear advantages to having easy access to the sample required by this strategy; in fact, patient compliance would considerably benefit from this approach. Detection of circulating miRNAs in serum/plasma samples has been investigated in different tumor types, including NHL, where loss of miR-92a in comparison with nontumor controls has been identified and correlated with relapse.79 miRNA plasma expression was also investigated in DLBCL,80,81 where miR-155 was one of the markers that gave the best results. It was expressed at higher levels in patients than in controls. In addition, there is a correlation between low plasma expression of miR-20a and shorter survival in CLL.63 However, a limitation arising from high variability or the low yield of RNA from serum/plasma has been documented,82 and thus, optimization of the RNA extraction/purification protocol and data normalization process are to be welcomed in the near future.
Promising data are also emerging that connect the expression of specific miRNAs with prognosis or response to specific therapies. Most of these data concern CLL and DLBCL, the 2 tumor types for which the most information has been accumulated. Table 3 summarizes the most common findings; among these, 4 different groups have shown that the expression of miR-222 distinguishes DLBCL cases that respond favorably to R-CHOP treatment.33,45,46,83 Nevertheless, most of these studies show that the capacity for predicting therapy response is greater when multiple markers are used (multi-miRNA models), rather than single markers. Furthermore, miRNAs can be incorporated into combined prognostic models that integrate robust clinical (IPI) and consolidated biologic markers (gene translocations and cell of origin classification).
miRNAs associated with prognosis and response to therapy
| Lymphoma/miRNAs . | Clinical variability . | References . |
|---|---|---|
| CLL | ||
| miR-34a | Chemotherapy-refractory disease, p53 inactivation, impaired DNA damage response, and apoptosis resistance | 84,,–87 |
| miR-21, miR-222 | Early survival stratification, fluradabine resistance | 88,123 |
| miR-181b | Disease progression | 60 |
| miR-15a/16 | Drug sensitivity in mouse models | 122 |
| DLBCL | ||
| miR-222 | Favorable response to R-CHOP | 33,45,46,83 |
| FL | ||
| A signature of 23 miRNAs | Cell proliferation and tumor response | 49 |
| MCL | ||
| Oncomir-1 | Radioresistance and chemoresistance | 124,125 |
| Lymphoma/miRNAs . | Clinical variability . | References . |
|---|---|---|
| CLL | ||
| miR-34a | Chemotherapy-refractory disease, p53 inactivation, impaired DNA damage response, and apoptosis resistance | 84,,–87 |
| miR-21, miR-222 | Early survival stratification, fluradabine resistance | 88,123 |
| miR-181b | Disease progression | 60 |
| miR-15a/16 | Drug sensitivity in mouse models | 122 |
| DLBCL | ||
| miR-222 | Favorable response to R-CHOP | 33,45,46,83 |
| FL | ||
| A signature of 23 miRNAs | Cell proliferation and tumor response | 49 |
| MCL | ||
| Oncomir-1 | Radioresistance and chemoresistance | 124,125 |
CLL indicates chronic lymphocytic leukemia; DLBCL, diffuse large B-cell lymphoma; FL, follicular lymphoma; MCL, mantle cell lymphoma; and R-CHOP, rituximab cyclophosphamide, doxorubicin, vincristine, and prednisone.
The findings of several studies performed in CLL also coincide with respect to the association of low levels of expression of miR-34a with chemotherapy-refractory disease, p53 inactivation, impaired DNA damage response, and apoptosis resistance.84 The expression of miR-34a is also associated with treatment-free survival,85 proliferation,86 p53 abnormalities,87 17p deletions,88 and the expression of E2F1 and B-MYB.58 Again, the prognostic and predictive value seems to be greater when multiple markers, rather than individual miRNAs, are used.
The role of viral miRNAs
miRNAs are also coded by other organisms, including viruses.89,90 Two of these are especially interesting when studying B-cell lymphomas: Epstein-Barr virus (EBV) and Kaposi sarcoma virus (KSHV), both of which are members of the human herpesvirus family. EBV infection is frequently documented in BL1,91 (mainly in the endemic subtype) and less frequently in DLBCL cases.92 KSHV infection has been associated with primary effusion lymphoma and other less frequent lymphoma types.93,94 These miRNAs not only modulate the expression of viral proteins,95,96 but also modulate protein expression of the host organism (an example being ebv-miR-BART5, which targets the p53 up-regulated modulator of apoptosis, PUMA97 ). This confers on them an additional role in lymphoma pathogenesis. For instance, ebv-mir-BHRF1, an miRNA codified by EBV, promotes B-cell transformation98 and proliferation and prevents apoptosis,99 while ebv-miR-BART9 and ebv-miR- BART17-5p are able to modulate BCL6 expression.47 Moreover, miR-K12-11, an miRNA codified by KSHV, shows high homology with human miR-155, especially in the seed region. Thus, in 2007, 2 works proposed miR-K12-11 to be an orthologous of miR-155, and validated the hypothesis proposed a few years before90 that these 2 miRNAs could share at least some of their targets,100,101 such as: IKBKE, which is involved in regulating the NF-kB cascade; XAF1 (also known as BIRC4BP) and LDOC1, which are both involved in apoptosis process; FOS, a gene that is involved in cell proliferation and differentiation; and BACH-1, a gene involved in transcription regulation. More recent studies also directly attributed an important role to miR-K12-11 in B-cell proliferative disorders and splenic B-cell expansion in a mouse model.102
Interestingly, mdv1-mir-M4, an orthologue of miR-155, has also been found in chicken, where a role in lymphomagenesis has been established.103,104 EBV protein can induce the expression of human miRNAs. For instance, the EBV-encoded latent membrane protein 1 (LMP1) can induce expression of miR-146a105 and miR-155.106 Finally, EBV infection also modulates host miRNA expression.107
Mechanisms of miRNA deregulation
Underlying the findings of altered miRNA expression level, there is the not fully understood issue of the causes leading to miRNA deregulation. The studies undertaken to clarify this enabled the identification of a series of mechanisms that contribute to the changes in miRNA expression levels.
The existence of some recurrently acquired genetic alterations may be a cause of miRNA loss. It is the case of 13q14 and 7q21 deletion in CLL and SMZL, respectively, where the clusters of miR-15a/16 and miR-29a and 29b-1 are located.5,73 Findings correlated with translocations and miRNA deregulation have been found in acute lymphoblastic leukemia in miR-125b-1 that shows translocation t(11;14)(q24;q32).108 Among those frequently translocated in lymphoma regions, miR-1204 was identified within the region of 8q24109 (frequently translocated in BL), but its role in B-cell lymphoma pathogenesis is currently not known. Efforts have also been made to investigate the association of the miRNA expression profile with DNA losses and gains found in comparative genomic hybridization (CGH) studies, especially in CLL,110 DLBCL,111 and MCL.66,112 Nevertheless, copy number alterations alone cannot explain many of the changes in miRNA expression level.
Epigenetics is another mechanism that has a great effect on the regulation of miRNA expression. DNA hypermethylation is responsible for the decrease of miRNA in NHL. It is the case of loss of expression of miR-124a113 and miR-203114 in a variety of hematologic malignancies, of miR-9 in some BL cases,115 and of miR-139 and miR-582 (among others) in CLL.116
In addition, conventional transcription factors can also regulate miRNA expression. The most interesting case that may be relevant in B-cell lymphomas is the regulation provided by C-MYC on the miRNA cluster miR-17-92.117 In the same study, the expression of cluster miR-17-92 induced by C-MYC was correlated with E2F1 transcription factor expression inhibition, thereby establishing a meticulous mechanism for regulating cell proliferation.117 C-MYC is a transcription factor that is frequently overexpressed in lymphomas, especially in BL, and the overexpression of miRNAs induced by C-MYC has been documented in this lymphoma type.118
Taken together, all these mechanisms account for an important part of miRNA deregulation, but other minor mechanisms (for instance corruption of the miRNA maturation process119 ) are probably also responsible of miRNA expression level changes.
miRNA-targeted therapy
Recognition of the role of miRNA in cancer pathogenesis has fueled interest in the development of clinical trials and preclinical studies targeting specific miRNAs.10,120 Particularly appealing for the recognition of miRNAs as targets is the fact that miRNAs appear to deregulate many genes simultaneously. These genes frequently appear to be functionally related, being involved in the activation of specific oncogenic pathways.65,120 Therefore, targeting oncogenic or key tumor suppressor miRNAs could restore the expression of multiple functionally related genes that are involved in the same oncogenic pathway.
Two main approaches to miRNA therapy have been applied (reviewed by Garzon et al120 ): miRNA down-regulation (basically by antagomiRs) and miRNA replacement. In principle, miRNA replacement could be of more general use because the most frequent miRNA changes identified in cancer samples are miRNA losses. Kota and coworkers have recently demonstrated that systemic administration of a lost miRNA in a mouse model of hepatocellular carcinoma using adeno-associated virus results in inhibition of cancer cell proliferation, induction of tumor-specific apoptosis, and protection from disease progression without toxicity.121 miRNA replacement therapy has been hampered by the lack of robust nonviral delivery methods for in vivo administration, but some promising results in colon cancer, using polyethylenimine-mediated delivery of unmodified miRNAs, have recently been reported by Ibrahim and coworkers.
Salerno and coworkers have reported some interesting results from lymphoma, demonstrating that the addition of exogenous miR-15a and miR-16 in a CLL model could lead to the accumulation of resting B cells in non-New Zealand Black B cells and the New Zealand Black-derived malignant B-1 cell line, by restoration of cell-cycle control, partially depending on the reduced expression of Cyclin D1.122
Nevertheless, the feasibility of miRNA-based treatment remains an area where reviews are more common than original research articles, and where the exploration of new approaches to therapy delivery is still a challenge.
Acknowledgments
This work was supported by grants from the Ministerio de Sanidad y Consumo (RTICC), the Asociación Española contra el Cancer (AECC), and the Ministerio de Ciencia e Innovación (SAF 2008-03 871 and FI08/00 038; L.D.L.), Spain. M.S.-B. is supported by a Contract Miguel Servet from Fondo Investigaciones Sanitarias.
Authorship
Contribution: L.D.L., N.M., and M.A.P. reviewed the bibliography, and planned and wrote the manuscript; and S.M.-M., M.P.-V., and M.S.-B. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nerea Martinez, PhD, Cancer Genomics Laboratory, IFIMAV-Fundación Marqués de Valdecilla, Avda Cardenal Herrera Oria s/n, 39011-Santander, Spain; e-mail: ifimav.nmartinez@fmdv.org.