Abstract
Chromosomal abnormalities, immunoglobulin heavy chain variable–region (IGHV) gene mutation status, and ζ-associated protein 70 (ZAP-70) expression levels have independent prognostic relevance in chronic lymphocytic leukemia (CLL); however, their concordance is variable. Because deregulation of microRNAs has been linked to disease initiation and progression in CLL, we studied the value of the microRNAs as a signature for CLL patients with specific chromosomal abnormalities. We identified 32 microRNAs able to discriminate the 11q deletion, 17p deletion, trisomy 12, 13q deletion, and normal karyotype cytogenetic subgroups. The expression values of 9 among the 32 microRNAs (miR-151-3p, miR-34a, miR-29c, miR-29b, miR-155, miR-148a, miR-146a, miR-146b5p, and miR-640) were correlated with gene expression data from the same samples to assess their biologic impact on CLL. In this study we also found that IGHV unmutated, high expression of ZAP-70 protein, and low expression of the miR-223, miR-29c, miR-29b, and miR-181 family were strongly associated with disease progression in CLL cases harboring 17p deletion, whereas in those harboring trisomy 12 only high expression of the miR-181a, among the analyzed parameters, suggested more aggressive disease. Thus, the use of the microRNA-based classifications may yield clinically useful biomarkers of tumor behavior in CLL.
Introduction
Chronic lymphocytic leukemia (CLL) is the most common human leukemia; approximately 10 000 patients are diagnosed annually in the United States. CLL is generally a disease of the elderly. This disease is characterized by an accumulation of small, mature B cells typically displaying the CD5/CD19/CD23 markers.1 The clinical course of CLL is highly variable, and several prognostic markers have been identified to facilitate the clinical management of CLL patients, including immunoglobulin heavy chain variable-region (IGHV) gene mutation status, ζ-associated protein 70 (ZAP-70) expression levels, and cytogenetic abnormalities.2,3 The IGHV gene is a genetic marker whose mutation status is related to the stage of B-cell differentiation. The authors of 2 studies4,5 showed that the presence of unmutated IGHV genes (IGHV homology to the germline ≥ 98%) predicted an inferior clinical course in CLL. ZAP-70 is a tyrosine kinase protein that plays a role in T-cell development and lymphocyte activation. ZAP-70 is not expressed in normal B lymphocytes, but detectable levels are present in leukemic cells. The authors of several studies2,6,7 have demonstrated that high expression of this protein is usually correlated with IGHV unmutated status, suggesting that ZAP-70 is a useful surrogate marker for the IGHV mutation status. Genomic aberrations can be identified in more than 80% of CLL patients, with the most common chromosomal abnormalities including deletion at 13q (> 50%), 11q (18%), trisomy 12 (15%-18%), and 17p (7%-10%). Therefore, 5 prognostic categories have been defined in a statistical model showing poor survival in patients harboring 17p deletion, 11q deletion, or trisomy 12 and better survival for patients with normal karyotype and 13q deletion as the sole abnormality.8
However, because several CLL cases show discordant prognostic factors, the identification of new parameters able to relate disease activity and clinical outcome is important for patient management. Recently, a new class of small noncoding RNAs (17-27 nt in length), microRNAs (miRNAs), have been linked to several types of cancer.9 MiRNAs play an important role in biologic processes by binding to the target messenger RNAs (mRNAs) and either inhibiting their translation or promoting their degradation. A role of miRNAs as prognostic and diagnostic factors already has been established in CLL irrespective of the genetic alterations.10,11 Therefore, in this study we performed a genome-wide miRNA expression analysis in 5 CLL subgroups with different karyotypes to identify possible novel markers for the diagnosis and the treatment of CLL in the future. Moreover, to find miRNAs associated with disease progression, we relate clinical data and miRNA expression values in a cohort of CLL cases with chromosomal aberrations reflecting an unfavorable clinical outcome, and we correlate these to ZAP-70 expression and IGHV mutation status.
Methods
Patients
In the current study, 90 CLL previously untreated patients with cytogenetic abnormalities were enrolled in the CLL Research Consortium upon written informed consent in accordance with the Declaration of Helsinki. The institutional review board of The Ohio State University provided approval for this research. This group included CLL samples from patients with 13q deletion (n = 15), trisomy 12 (n = 33), 11q deletion (n = 9), 17p deletion (n = 22), and normal karyotype (n = 11). Trisomy 12 and 13q deletion were found as sole abnormalities in the CLL cells, whereas most of the CLL B cells with either 11q deletion or 17p deletion showed also 13q deletion. Data for ZAP-70 expression and IGHV mutational status were available for all patients at the time of the enrollment in this study. Cases that showed ZAP-70 expression in less than 20% of the CLL B cells (ZAP-70 negative) occurred as tumors with IGHV homology to germ line genes less than 98% (IGHV mutated). These cases include all CLL samples with 13q deletion and normal karyotype, 12 of 21 samples with 17p deletion, and 17 of 33 samples with trisomy 12. The remaining samples expressed ZAP-70 protein in more than 20% of the cells (ZAP-70 positive) and exhibited IGHV homology to germ line genes greater than 98% (IGHV unmutated). Tables 1, 4, and 6 and supplemental Table 3 (available on the Blood website; see the Supplemental Materials link at the top of the online article) illustrate the molecular and the genetic features of the patients enrolled in this study.
Molecular and genetic features of the B-CLL patients
| Sample no. . | 13q del+ cells, %* . | 17p del+ cells, %* . | Trisomy 12+ cells, %* . | 11q del+ cells, %* . | IGHV homology, % . | ZAP-70+ cells, % . |
|---|---|---|---|---|---|---|
| 12 | 64.5 | – | – | – | < 98 | < 20 |
| 9 | 49.5 | 76 | – | – | ≥ 98 | > 20 |
| 3 | 61 | 12 | – | – | < 98 | < 20 |
| 12 | – | – | 65 | – | ≥ 98 | > 20 |
| 5 | – | – | 55 | – | < 98 | < 20 |
| 9 | 23.5 | – | – | 60 | ≥ 98 | > 20 |
| 11 | – | – | – | – | < 98 | > 20 |
| Sample no. . | 13q del+ cells, %* . | 17p del+ cells, %* . | Trisomy 12+ cells, %* . | 11q del+ cells, %* . | IGHV homology, % . | ZAP-70+ cells, % . |
|---|---|---|---|---|---|---|
| 12 | 64.5 | – | – | – | < 98 | < 20 |
| 9 | 49.5 | 76 | – | – | ≥ 98 | > 20 |
| 3 | 61 | 12 | – | – | < 98 | < 20 |
| 12 | – | – | 65 | – | ≥ 98 | > 20 |
| 5 | – | – | 55 | – | < 98 | < 20 |
| 9 | 23.5 | – | – | 60 | ≥ 98 | > 20 |
| 11 | – | – | – | – | < 98 | > 20 |
B-CLL indicates B-cell chronic lymphocytic leukemia; IGHV, immunoglobulin heavy chain variable-region; and ZAP-70, ζ-associated protein 70.
Values represent the median of the percentage of positive cells from the samples of the same cytogenetic group.
Analysis of ZAP-70, IGHV mutation status, and genomic alteration
Analysis of ZAP-70 and sequence analysis of IGHV were performed as described previously.7 Fluorescent in situ hybridization was performed on interphase nuclei of blood lymphocytes. The FISH panel (Vysis) detected aberrations at 11q22, 12 centromere, 13q14.3, and 17p13.
MiRNA and mRNA expression profiling and bioinformatics analysis
MiRNA microarray profiling was performed as previously described.12 RNA from the sets of samples described in Table 1 (61 patients), Table 4 (10 of which are common to the 61 patients, plus 9 new patients), supplemental Table 3 (3 patients with 13q deletion), and Table 6 (16 new patients) were profiled in 4 separate batches. Average values of the replicate spots of each miRNA were background subtracted, normalized, and further analyzed. Normalization was performed by the use of the quantile method. We selected the miRNAs measured as present in at least as many samples as the smallest class in the dataset (50%). Absent calls were threshold to 3.3 (log2 scale) before statistical analysis, representing the average minimum intensity level detectable in the system. More than 95% of blank probes (ie, negative controls) fall below the threshold value of 3.3. MiRNAs that are differentially expressed in 5 different cytogenetic groups were identified by use of the “class comparison among genes” within Biometric Branch Research (BRB) Array Tools version 3.6.0 developed by Richard Simon and Amy Peng Lam.13 This tool is designed to analyze data by use of the parametric tests t/F tests and random variance t/F tests. The criterion for inclusion of a gene in the gene list is a P value less than a specified threshold value (.05).
The class comparison algorithm in BRB Array Tools was used to determine whether miRNA microarray expression patterns could accurately differentiate between 2 groups. Predictor miRNAs were determined by the prediction analysis of microarrays, which implements nearest shrunken centroids.14 Gene-expression profiling of 61 samples (Table 1) was performed by the use of Affymetrix U133 plus 2.0 GeneChips (Affymetrix). BRB-Array Tools version 3.6.0 also was used to import Affymetrix CEL files, and the robust multichip average method was done. This method uses background correction on the perfect match data and then applies a quantile normalization and summarizes the probe set information by the use of the Tukey median polish algorithm. Genes that are differentially expressed in different tissues were identified with the class comparison analysis as previously described. The criterion for inclusion of a gene in the gene list is a P value less than a specified threshold value (.001). These patient samples also were used for the identification of the genes expression associated with the expression values of the 9 miRNA validated by quantitative real-time polymerase chain reaction (qRT-PCR). The Spearman correlation coefficient and a univariate significance level of α = 0.001 were used for the tests. Analyses were performed with the use of BRB Array Tools version 3.6.0. All microarray data have been deposited on ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/) under accession numbers E-TABM-763 and E-TABM-762.
Statistical analysis
Expression values (obtained by qRT-PCR) from the 61 CLL samples for each of the 9 miRNAs were tested using the Bartlett test to evaluate the homogeneity of the variance among the samples. Kruskal-Wallis or analysis of variance (ANOVA) tests were used to assess whether the 9 miRNAs are differentially expressed among the cytogenetic groups on the basis of the Bartlett test P value. The Kruskal-Wallis test was used for Bartlett test P values less than .05, whereas the ANOVA test was used for a P value greater than .05.
The starting point was the time of the patients enrollment in this study, and the end point was the time to the first-line treatment (TTT). The untreated patients were indicated as censored. For patients with trisomy 12 (Table 6), TTT and miRNAs microarray expression data were used in univariate Cox proportional hazards regression model to identify miRNAs that correlate with the disease progression, defined according to the guidelines from the International Workshop on Chronic Lymphocytic Leukemia.15 The Fisher exact test was used to analyze categoric values. An effect was considered significant at a P value less than .05. We used R 2.6.0 (www.r-project.org) to design box and whisker plots.
RNA extraction and qRT-PCR
RNA from CLL patients was extracted by the use of standard TRIZOL (Invitrogen) methods. Mature miRNA expression was assayed by Taqman MiRNA assay (Applied Biosystem) and normalized on RNU6B (P/N: 4373381) according to the manufacturer's protocol. A total of 5 ng (or 150 ng for miR-640) of total RNA was reverse transcribed. Each sample was analyzed in triplicate. The level of miRNA and mRNA was measured by the use of the threshold cycle (Ct). The amount of target, normalized to an endogenous reference and relative to a calibrator, is given by: 2−ΔCt (Comparative Ct method, Applied Biosystem User Bulletin #2).
Results
MiRNAs differentially expressed among cytogenetic groups of CLL
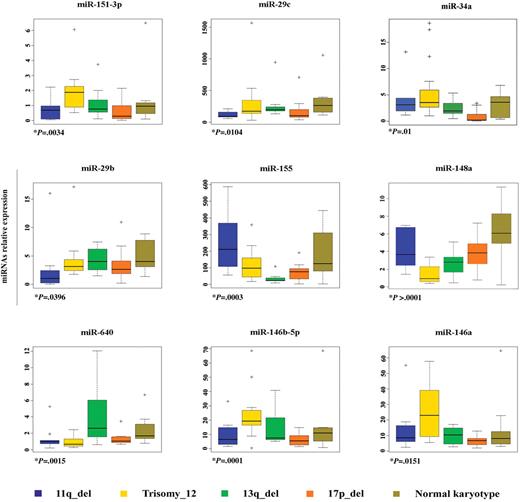
Genome-wide miRNA expression profiling was performed on 61 untreated CLL samples with different karyotypes by the use of a miRNA microarray platform previously described.12 The samples include CLL cells with 13q deletion (n = 12), trisomy 12 (n = 17), 11q deletion (n = 9), 17p deletion (n = 12), and normal karyotype (n = 11). Laboratory findings of the patients are summarized in Table 1. Expression data of 474 human mature miRNAs were first analyzed on the predefined classes, by use of the multiclass comparison within BRB Array Tools, to identify a specific set of genes differentially expressed among the cytogenetic classes. The tool computed 65 miRNAs at a P value of .05 or less, along with a false discovery rate filter of 16% or less (supplemental Table 1).
We further used the class comparison within BRB Array Tool with a “one versus all of the others” approach to search for miRNAs with expression patterns highly correlated with each karyotype. The analysis performed at P values less than .05 identified 19, 29, 40, 46, and 19 miRNAs differentially expressed in CLL cases harboring 11q deletion, 17p deletion, trisomy 12, 13q deletion, and normal karyotype, respectively, compared with all other samples (supplemental Table 2). Among the genes obtained by performing the previously mentioned analyses, the expression values of 9 miRNAs (miR-155, miR-29b, miR-151-3p, miR-29c, miR-34a, miR-640, miR-148a, miR-146a, and 146b-5p) were associated with 1 specific karyotype after that validation by qRT-PCR was undertaken in all sets of the samples used (Table 2; Figure 1); thus, these genes were selected for further studies. Several miRNAs whose expression values were undetectable or not validated by qRT-PCR assay were excluded from our selection. Moreover, the 2 cytogenetic subgroups, 17p deletion and trisomy 12, included cases with mutated and unmutated IGHV genes (see Table 1). Interestingly, we observed low expression of the miR-151-3p, miR-29c, and miR-34a in CLL cells harboring 17p deletion and with IGHV unmutated genes but not in the same cytogenetic group with IGHV mutated genes when they were compared with the other CLL subclasses. We encountered a similar situation for miR-640 in CLL with trisomy 12. Conversely, we observed no difference in the expression values of miR-146a, miR-146b-5p, and miR-148a in the samples bearing trisomy 12 with IGHV mutated and unmutated as well (supplemental Figure 1).
miRNAs differentially expressed among CLL samples with different karyotypes
| CLL subtype/miRNAs . | Multiclass comparison* . | Class prediction† . | Average difference in threshold cycle‡ . | Chromosome map . | |||
|---|---|---|---|---|---|---|---|
| P . | FDR . | Fold change . | P . | Fold change . | P . | ||
| 17p deletion | |||||||
| miR151-3p | <.001 | .0012 | 0.23 | <.001 | 0.43 | .009 | 8q24.3 |
| miR-29c | .032 | .1191 | 0.7 | .007 | 0.67 | .05 | 1q32.2 |
| miR-34a | .017 | .0862 | – | – | 0.38 | .009 | 1p36.23 |
| 11q deletion | |||||||
| miR-29b | .029 | .1168 | 0.74 | .045 | 0.35 | .001 | 7q32.3 |
| miR-155 | .008 | .0487 | – | – | 2.28 | .05 | 21q21.3 |
| Trisomy 12 | |||||||
| miR-640 | <.001 | .0003 | 0.21 | <.001 | 0.39 | .002 | 19p13.11 |
| miR-148a | .010 | .0587 | 0.23 | <.001 | 0.3 | .001 | 7p15.2 |
| miR-146b-5p | .010 | .0587 | 1.94 | <.001 | 2.22 | .002 | 10q24.32 |
| miR-146a | .0021 | .0257 | 2.57 | <.001 | 1.99 | .006 | 5q33.3 |
| Normal karyotype | |||||||
| miR-148a | .010 | .0587 | – | – | 2.1 | .007 | 7p15.2 |
| 13q deletion | |||||||
| miR-155 | .034 | .1263 | – | – | 0.2 | .001 | 21q21.3 |
| miR-640 | <.001 | .0003 | 3.86 | .002 | 2.8 | .05 | 19p13.11 |
| CLL subtype/miRNAs . | Multiclass comparison* . | Class prediction† . | Average difference in threshold cycle‡ . | Chromosome map . | |||
|---|---|---|---|---|---|---|---|
| P . | FDR . | Fold change . | P . | Fold change . | P . | ||
| 17p deletion | |||||||
| miR151-3p | <.001 | .0012 | 0.23 | <.001 | 0.43 | .009 | 8q24.3 |
| miR-29c | .032 | .1191 | 0.7 | .007 | 0.67 | .05 | 1q32.2 |
| miR-34a | .017 | .0862 | – | – | 0.38 | .009 | 1p36.23 |
| 11q deletion | |||||||
| miR-29b | .029 | .1168 | 0.74 | .045 | 0.35 | .001 | 7q32.3 |
| miR-155 | .008 | .0487 | – | – | 2.28 | .05 | 21q21.3 |
| Trisomy 12 | |||||||
| miR-640 | <.001 | .0003 | 0.21 | <.001 | 0.39 | .002 | 19p13.11 |
| miR-148a | .010 | .0587 | 0.23 | <.001 | 0.3 | .001 | 7p15.2 |
| miR-146b-5p | .010 | .0587 | 1.94 | <.001 | 2.22 | .002 | 10q24.32 |
| miR-146a | .0021 | .0257 | 2.57 | <.001 | 1.99 | .006 | 5q33.3 |
| Normal karyotype | |||||||
| miR-148a | .010 | .0587 | – | – | 2.1 | .007 | 7p15.2 |
| 13q deletion | |||||||
| miR-155 | .034 | .1263 | – | – | 0.2 | .001 | 21q21.3 |
| miR-640 | <.001 | .0003 | 3.86 | .002 | 2.8 | .05 | 19p13.11 |
All analyses were performed on 61 CLL samples with different karyotypes.
CLL indicates chronic lymphoid leukemia; FDR, false discovery rate; and miRNA; microRNA.
P and FDR are the results of the multiclass comparison among groups analysis within Biometric Research Branch Array Tool, Version 3.6.0, performed with the use of microRNAs expression patterns (obtained by microarray) from the 5 cytogenetic subgroups.
Fold change is the fold-ratio of geometric means of miRNA expressions between 1 karyotype and all of the others considered as 2 classes. Fold change and P are the results of the Class Prediction analysis within BRB-Array tool.
Fold change is the fold-ratio of the 2−ΔCt average of miRNA expression (obtained by quantitative real-time polymerase chain reaction) between 1 karyotype and all of the others considered as 2 classes. P was calculated using the Student t test; tails = 2.
miRNAs differentially expressed among cytogenetic groups of CLL. MiRNAs' relative expression in CLL samples with different karyotypes was determined by Taqman qRT PCR assay. Each data sample was normalized to the endogenous reference RNU6B by use of the 2−Δct method. The relative expression values were used to design box and whisker plots. Crosses in the boxes indicate outlier points. +P value is the result of either Kruskal-Wallis or ANOVA. The ANOVA test was used only for miR-29b. The homogeneity of the samples variance was defined by use of the Bartlett test.
miRNAs differentially expressed among cytogenetic groups of CLL. MiRNAs' relative expression in CLL samples with different karyotypes was determined by Taqman qRT PCR assay. Each data sample was normalized to the endogenous reference RNU6B by use of the 2−Δct method. The relative expression values were used to design box and whisker plots. Crosses in the boxes indicate outlier points. +P value is the result of either Kruskal-Wallis or ANOVA. The ANOVA test was used only for miR-29b. The homogeneity of the samples variance was defined by use of the Bartlett test.
Prediction of CLL cytogenetic subgroups
We further used a different algorithm, prediction analysis of microarray, to identify a small set of miRNAs able to predict the 5 cytogenetic subgroups. The tool computed 32 miRNAs as best predicting 11q deletion, 17p deletion, trisomy 12, 13q deletion, and normal karyotype by using this platform (Table 3). The cross-validate probabilities performed on the training set of the 61 CLL samples are shown in supplemental Figure 2A. We applied the same algorithm by using the miRNAs signature composed of 32 miRNAs on an independent available set of 29 samples, including CLL cells with 13q deletion (n = 3), 17p deletion/IGHV unmutated (n = 1), 17p deletion/IGHV mutated (n = 9), and trisomy 12 (n = 16; supplemental Table 3). We found a correct classification for all CLL with 13q deletion and for several CLL cases with trisomy 12 (supplemental Figure 2B); however, we reached a better result for predicting the latter subgroup excluding 3 (miR-29b, miR34a, miR-142-5p) of the 32 miRNAs (supplemental Figure 2C). CLLs harboring the 17p deletion and with IGVH mutated genes were mostly classified as normal-karyotype CLL; this finding is consistent with the observation that 17p deletion cases with IGHV mutated have a better clinical course if compared with the IGHV unmutated subgroup.
miRNAs predictors of the cytogenetic subgroups in CLL
| miRNA . | Prediction analysis of microarray score* . | Chromosome map . | ||||
|---|---|---|---|---|---|---|
| 11q deletion . | 13q deletion . | 17p deletion . | Normal karyotype . | Trisomy 12 . | ||
| miR-370 | 0.19 | 0.55 | −1.22 | 0.28 | 0.19 | 14q32.31 |
| miR-16-2* | −1.04 | 0.11 | 0.24 | 0.16 | 0.20 | 3q26.1 |
| miR-572 | 0.76 | 0.53 | −0.99 | −0.36 | 0.15 | 4p15.33 |
| miR-130a* | −0.93 | 0.84 | 0.60 | −0.03 | −0.51 | 11q12.1 |
| miR-590-5p | 0.26 | 0.83 | −0.80 | −0.35 | 0.07 | 7q11.23 |
| miR-151-3p | −0.40 | 0.31 | −0.82 | 0.35 | 0.35 | 8q24.3 |
| miR-640 | −0.51 | 0.68 | 0.41 | 0.32 | −0.70 | 19p13.11 |
| miR-548a-3p | 0.62 | −0.03 | −0.20 | −0.26 | 0.00 | 6p22.3 |
| miR-556-3p | 0.44 | −0.60 | 0.00 | 0.17 | 0.08 | 8q22.3 |
| miR-148a | 0.23 | 0.18 | 0.36 | 0.10 | −0.57 | 1q23.3 |
| miR-29b-1* | 0.06 | 0.01 | −0.57 | −0.09 | 0.42 | 7p15.2 |
| miR-585 | 0.17 | −0.22 | 0.22 | −0.57 | 0.28 | 7q32.3 |
| miR-499-5p | −0.03 | −0.55 | 0.28 | 0.16 | 0.11 | 5q35.1 |
| miR-99b | 0.29 | 0.55 | 0.00 | −0.28 | −0.36 | 20q11.22 |
| miR-146a | −0.06 | −0.23 | −0.36 | −0.14 | 0.54 | 5q33.3 |
| miR-200c | −0.08 | 0.51 | −0.27 | −0.40 | 0.14 | 12p13.31 |
| miR-498 | 0.22 | −0.50 | −0.06 | 0.13 | 0.20 | 19q13.41 |
| miR-377 | 0.14 | −0.43 | 0.10 | 0.13 | 0.08 | 14q32.31 |
| miR-146b-5p | −0.12 | −0.03 | −0.32 | −0.18 | 0.43 | 10q24.32 |
| miR-593 | −0.29 | 0.43 | −0.08 | −0.13 | −0.01 | 7q32.1 |
| miR-205 | −0.33 | 0.39 | 0.42 | −0.03 | −0.38 | 1q32.2 |
| miR-136 | 0.40 | −0.24 | −0.04 | −0.01 | 0.00 | 14q32.31 |
| miR-193a-5p | 0.19 | −0.28 | −0.24 | −0.17 | 0.37 | 17q11.2 |
| miR-543 | 0.14 | −0.37 | −0.03 | 0.26 | 0.05 | 14q32.31 |
| miR-29c | −0.16 | 0.17 | −0.33 | 0.20 | 0.07 | 1q32.2 |
| miR-29a* | 0.31 | −0.25 | −0.04 | 0.00 | 0.04 | 7q32.3 |
| miR-532-3p | −0.04 | −0.06 | 0.21 | 0.31 | −0.28 | Xp11.23 |
| miR-29b | −0.30 | 0.18 | −0.25 | 0.17 | 0.09 | 7q32.3 |
| miR-34a | 0.27 | −0.22 | −0.23 | −0.01 | 0.17 | 1p36.23 |
| miR-202* | 0.23 | −0.26 | 0.01 | −0.26 | 0.22 | 10q26.3 |
| miR-142-5p | −0.22 | 0.24 | 0.13 | 0.02 | −0.16 | 17q22 |
| miR-155 | 0.21 | −0.10 | 0.18 | 0.08 | −0.22 | 21q21.3 |
| miRNA . | Prediction analysis of microarray score* . | Chromosome map . | ||||
|---|---|---|---|---|---|---|
| 11q deletion . | 13q deletion . | 17p deletion . | Normal karyotype . | Trisomy 12 . | ||
| miR-370 | 0.19 | 0.55 | −1.22 | 0.28 | 0.19 | 14q32.31 |
| miR-16-2* | −1.04 | 0.11 | 0.24 | 0.16 | 0.20 | 3q26.1 |
| miR-572 | 0.76 | 0.53 | −0.99 | −0.36 | 0.15 | 4p15.33 |
| miR-130a* | −0.93 | 0.84 | 0.60 | −0.03 | −0.51 | 11q12.1 |
| miR-590-5p | 0.26 | 0.83 | −0.80 | −0.35 | 0.07 | 7q11.23 |
| miR-151-3p | −0.40 | 0.31 | −0.82 | 0.35 | 0.35 | 8q24.3 |
| miR-640 | −0.51 | 0.68 | 0.41 | 0.32 | −0.70 | 19p13.11 |
| miR-548a-3p | 0.62 | −0.03 | −0.20 | −0.26 | 0.00 | 6p22.3 |
| miR-556-3p | 0.44 | −0.60 | 0.00 | 0.17 | 0.08 | 8q22.3 |
| miR-148a | 0.23 | 0.18 | 0.36 | 0.10 | −0.57 | 1q23.3 |
| miR-29b-1* | 0.06 | 0.01 | −0.57 | −0.09 | 0.42 | 7p15.2 |
| miR-585 | 0.17 | −0.22 | 0.22 | −0.57 | 0.28 | 7q32.3 |
| miR-499-5p | −0.03 | −0.55 | 0.28 | 0.16 | 0.11 | 5q35.1 |
| miR-99b | 0.29 | 0.55 | 0.00 | −0.28 | −0.36 | 20q11.22 |
| miR-146a | −0.06 | −0.23 | −0.36 | −0.14 | 0.54 | 5q33.3 |
| miR-200c | −0.08 | 0.51 | −0.27 | −0.40 | 0.14 | 12p13.31 |
| miR-498 | 0.22 | −0.50 | −0.06 | 0.13 | 0.20 | 19q13.41 |
| miR-377 | 0.14 | −0.43 | 0.10 | 0.13 | 0.08 | 14q32.31 |
| miR-146b-5p | −0.12 | −0.03 | −0.32 | −0.18 | 0.43 | 10q24.32 |
| miR-593 | −0.29 | 0.43 | −0.08 | −0.13 | −0.01 | 7q32.1 |
| miR-205 | −0.33 | 0.39 | 0.42 | −0.03 | −0.38 | 1q32.2 |
| miR-136 | 0.40 | −0.24 | −0.04 | −0.01 | 0.00 | 14q32.31 |
| miR-193a-5p | 0.19 | −0.28 | −0.24 | −0.17 | 0.37 | 17q11.2 |
| miR-543 | 0.14 | −0.37 | −0.03 | 0.26 | 0.05 | 14q32.31 |
| miR-29c | −0.16 | 0.17 | −0.33 | 0.20 | 0.07 | 1q32.2 |
| miR-29a* | 0.31 | −0.25 | −0.04 | 0.00 | 0.04 | 7q32.3 |
| miR-532-3p | −0.04 | −0.06 | 0.21 | 0.31 | −0.28 | Xp11.23 |
| miR-29b | −0.30 | 0.18 | −0.25 | 0.17 | 0.09 | 7q32.3 |
| miR-34a | 0.27 | −0.22 | −0.23 | −0.01 | 0.17 | 1p36.23 |
| miR-202* | 0.23 | −0.26 | 0.01 | −0.26 | 0.22 | 10q26.3 |
| miR-142-5p | −0.22 | 0.24 | 0.13 | 0.02 | −0.16 | 17q22 |
| miR-155 | 0.21 | −0.10 | 0.18 | 0.08 | −0.22 | 21q21.3 |
CLL indicates chronic lymphocytic leukemia; and miRNA, microRNA.
Log ratio of expression relative to the shrunken centroids.
Biologic impact of the 9 miRNAs associated with the karyotype on CLL
Protein expression can be modulated by the regulatory activity of miRNAs that can operate by binding directly to their target and either inhibiting their translation or degrading the mRNA target gene. Through these mechanisms, miRNAs affect directly and indirectly the expression of several genes. On these bases, we assessed the influence of the 9 miRNAs on CLL. Specifically, we investigated whether observed changes in miRNAs were correlated with changes in the expression of genes represented in the Affymetrix mRNA microarray performed on the 61 samples. We found that the expression levels of 464, 262, 323, 281, 306, 181, 631, 461, 651, and 324 genes significantly (P < .001) correlated with expression values of the miR-155, miR-29b, miR-151-3p, miR-29c, miR-34a, miR-640, miR-223, miR-148a, miR-146a, and miR-146b-5p, respectively (supplemental Table 4). We observed that some miRNAs, whose expression values were associated with trisomy 12, correlated with integrins and metalloproteinase with a recognized role in transendothelial migration (TEM) in CLL.16-18 MiR-146a positively correlated with IGTB2 and IGTA4 as well as miR-146b-5p with IGTB2, whereas decreased expression of miR-148a was associated with increased expression of IGTA5 and MMP2 (negative correlation). To support this result we also referred to the Gene Ontology project (www.geneontology.org) to evaluate the function of the molecules correlating with these genes. We found that a greater number of molecules involved in biologic mechanisms such as cell migration (n = 7), cell adhesion (n = 23), and cell motility12 were associated with miR-146a than with the other miRNAs, thereby suggesting a putative role of the miR-146a in these processes in CLL. Moreover, gene expression data also demonstrated high levels of ITGA4 and ITGB2 in trisomy 12 compared with other cytogenetic groups, which is consistent with our previous finding (supplemental Table 5).
Finally we referred to Targetscan,19 miRanda,20 and Pictar21 databases to assess which of the negatively correlated genes were predicted to be direct targets of the corresponding miRNAs associated with the karyotypes. A total of 95 predicted target genes were found (supplemental Table 5; highlighted rows). Among them, TM6SF1 was validated by Skalsky et al22 as target of miR-155 in HEK293 cell line. Decreased levels of the DNMT1 mRNA were linked to the overexpression of the miR-29b23 in acute myeloid cells, whereas DNAJB11 was validated as target of the miR-29b (not tested for miR-29c) in HEK293 cell line.24 Therefore, these genes are strongly suggested as direct targets of the miRNAs member of CLL signature.
Association between miRNAs expression, IGHV mutation status, ZAP-70 expression level, and time to initial therapy in CLL bearing the 17p deletion
CLLs harboring 17p deletion occur as IGHV unmutated/ZAP-70–positive tumors in 10% and as IGHV mutated/ZAP-70–negative tumors in 3% of all CLL cases.25 To examine whether ZAP-70 expression level and IGHV mutation status have a prognostic value in this cytogenetic subgroup, we related these factors to clinical outcomes in a set of CLL patients bearing the 17p deletion (n = 19). Among them, 8 samples had CLL B cells expressing ZAP-70 and with unmutated IGHV gene status, whereas the other 11 had CLL B cells lacking ZAP-70 and with mutated IGHV gene status. The patients enrolled in this study were assembled in 2 groups that showed different clinical behavior. According to published data, the 17p deletion predicts a disease progression within 12 to 24 months,26-28 which is identified with the beginning of the treatment.15 Therefore, patients receiving a treatment over time periods ranging from 0 to 24 months since the time of the enrollment in this study were considered as 17p aggressive, whereas all of the others including patients treated or not in a time longer than 24 months (median time of 35 months) were considered as 17p indolent (Table 4). The treatment histories between the 2 subgroups differed significantly in that relatively more patients expressing ZAP-70 protein and with unmutated IGHV genes than patients lacked ZAP-70 protein and with mutated IGHV genes required therapy for the CLL patients harboring 17p deletion (Fisher exact test, P < .005). Finally, we used the miRNA microarray platform to identify miRNAs associated with more aggressive disease in CLLs bearing the 17p deletion. The results of the class comparison analysis between the subgroups defined in the previous analysis demonstrated that the miR-223, miR-29b, miR-29c, and the miR-181 family are down-regulated in 17p-aggressive cases (Table 5). The expression of 4 representative mature miRNAs was confirmed by Taqman MiRNAs assay (supplemental Figure 3).
Genetic, molecular, and clinical data of patients with 17p deletion
| Subgroup/patient no. . | 17p del (% positive cells) . | 13q del (% positive cells) . | IGHV % homology . | ZAP-70+ cells, % . | TTT, mos . | Therapy . |
|---|---|---|---|---|---|---|
| 17p aggressive | ||||||
| 16 | 84.0 | 52.0 | 99.3 | 50.2 | 0 | Yes |
| 17 | 86.0 | 49.5 | 99.6 | 76.8 | 0 | Yes |
| 100 | 7.5 | 59.0 | 88.4 | 3.8 | 24 | Yes |
| 20 | 96.0 | 0.0 | 99.6 | 36.4 | 0 | Yes |
| 21 | 76.0 | 70.0 | 99.3 | 36.5 | 0 | Yes |
| 22 | 93.0 | 94.5 | 99.6 | 84.8 | 0 | Yes |
| 23 | 61.0 | 15.0 | 99.3 | 42.6 | 0.5 | Yes |
| 24 | 69.5 | 70.0 | 99.6 | 53.7 | 6 | Yes |
| 114 | 9.0 | 40.0 | 90.6 | 6.0 | 0 | Yes |
| 17p indolent | ||||||
| 26 | 12.0 | 47.0 | 95.4 | 0.6 | 61 | No |
| 28 | 28.0 | 81.0 | 89.9 | 13.2 | 55 | No |
| 102b | 8.0 | 15.0 | 94.0 | 6.3 | 74 | No |
| 103 | 11.0 | 0.0 | 93.0 | 6.4 | 55 | Yes |
| 107 | 14.0 | 90.0 | 94.7 | 2.2 | 30 | No |
| 108 | 86.5 | 0.0 | 89.0 | 4.2 | 25 | No |
| 110 | 8.0 | 62.0 | 90.9 | 11.7 | 24 | No |
| 111 | 29.0 | 0.0 | 92.0 | 0.7 | 24 | No |
| 112 | 14.0 | 15/38 | 97.6 | 4.3 | 27 | No |
| 19 | 61.0 | 2.0 | 99.6 | 51.5 | 39 | No |
| Subgroup/patient no. . | 17p del (% positive cells) . | 13q del (% positive cells) . | IGHV % homology . | ZAP-70+ cells, % . | TTT, mos . | Therapy . |
|---|---|---|---|---|---|---|
| 17p aggressive | ||||||
| 16 | 84.0 | 52.0 | 99.3 | 50.2 | 0 | Yes |
| 17 | 86.0 | 49.5 | 99.6 | 76.8 | 0 | Yes |
| 100 | 7.5 | 59.0 | 88.4 | 3.8 | 24 | Yes |
| 20 | 96.0 | 0.0 | 99.6 | 36.4 | 0 | Yes |
| 21 | 76.0 | 70.0 | 99.3 | 36.5 | 0 | Yes |
| 22 | 93.0 | 94.5 | 99.6 | 84.8 | 0 | Yes |
| 23 | 61.0 | 15.0 | 99.3 | 42.6 | 0.5 | Yes |
| 24 | 69.5 | 70.0 | 99.6 | 53.7 | 6 | Yes |
| 114 | 9.0 | 40.0 | 90.6 | 6.0 | 0 | Yes |
| 17p indolent | ||||||
| 26 | 12.0 | 47.0 | 95.4 | 0.6 | 61 | No |
| 28 | 28.0 | 81.0 | 89.9 | 13.2 | 55 | No |
| 102b | 8.0 | 15.0 | 94.0 | 6.3 | 74 | No |
| 103 | 11.0 | 0.0 | 93.0 | 6.4 | 55 | Yes |
| 107 | 14.0 | 90.0 | 94.7 | 2.2 | 30 | No |
| 108 | 86.5 | 0.0 | 89.0 | 4.2 | 25 | No |
| 110 | 8.0 | 62.0 | 90.9 | 11.7 | 24 | No |
| 111 | 29.0 | 0.0 | 92.0 | 0.7 | 24 | No |
| 112 | 14.0 | 15/38 | 97.6 | 4.3 | 27 | No |
| 19 | 61.0 | 2.0 | 99.6 | 51.5 | 39 | No |
TTT indicates time to treatment; and ZAP-70, ζ-associated protein 70.
miRNAs that are differentially expressed in CLL 17p-aggressive compared with CLL 17p-indolent groups
| miRNA . | Fold change* . | P† . |
|---|---|---|
| Down-regulated | ||
| miR-29b | 0.53 | <.001 |
| miR-181b | 0.59 | .004 |
| miR-181c | 0.1 | .012 |
| miR-181d | 0.11 | .016 |
| miR-342-3p | 0.66 | .017 |
| miR-223 | 0.44 | .024 |
| miR-181a | 0.15 | .028 |
| miR-29c | 0.56 | .03 |
| miR-29b-1* | 0.74 | .04 |
| miR-367 | 0.16 | .049 |
| Up-regulated | ||
| miR-130b* | 1.44 | .011 |
| miR-129-3p | 11.63 | .013 |
| miR-632 | 1.15 | .021 |
| miR-768-5p | 1.36 | .021 |
| miR-638 | 1.58 | .028 |
| miR-453 | 7.72 | .047 |
| miRNA . | Fold change* . | P† . |
|---|---|---|
| Down-regulated | ||
| miR-29b | 0.53 | <.001 |
| miR-181b | 0.59 | .004 |
| miR-181c | 0.1 | .012 |
| miR-181d | 0.11 | .016 |
| miR-342-3p | 0.66 | .017 |
| miR-223 | 0.44 | .024 |
| miR-181a | 0.15 | .028 |
| miR-29c | 0.56 | .03 |
| miR-29b-1* | 0.74 | .04 |
| miR-367 | 0.16 | .049 |
| Up-regulated | ||
| miR-130b* | 1.44 | .011 |
| miR-129-3p | 11.63 | .013 |
| miR-632 | 1.15 | .021 |
| miR-768-5p | 1.36 | .021 |
| miR-638 | 1.58 | .028 |
| miR-453 | 7.72 | .047 |
CLL indicates chronic lymphoid leukemia; miRNA, microRNAs.
Fold change is the fold-ratio of geometric means of miRNA expressions from 17p-aggressive and 17p-indolent subgroups.
P and fold change are the results of the class comparison analysis within Biometric Research Branch Array Tool.
High expression levels of miR-181a are associated with disease progression in CLL patients showing trisomy 12
Trisomy 12 abnormality occurs with almost the same incidence in CLL patients with B cells bearing the mutated/or unmutated IGHV genes and expressing or not ZAP-70 protein. Survival analysis on our samples demonstrated that these 2 factors are not predictive of the clinical outcome in this cytogenetic subgroup (data not showed), confirming previous data.29 Therefore, we examined the role of the miRNAs in CLL cells showing trisomy 12. Sixteen patients were analyzed with median follow-up time of 28 months for untreated patients who were alive or 19 months for patients who received the first-line treatment (Table 6). Thus, to identify miRNAs associated with the disease progression in this cytogenetic group, we used a Cox regression model. We related TTT and expression values of the 9 miRNAs (miR-146a, miR-221, miR-222, miR-181a, miR-23a, miR-23b, miR-24a, miR-155, and miR-29c) previously demonstrated from our group to be predictive of the clinical behavior in CLLs irrespective of the karyotype.11 The tool computed only one miRNA, miR-181a, as significantly (P < .05) associated with TTT (hazard ratio = 2), thereby suggesting that high expression of miR-181a could be associated with short time to treatment in this group of patients.
Clinical and molecular characteristic of patients with trisomy 12
| Characteristic . | Value . |
|---|---|
| Level of ZAP-70 expression, no. patients | |
| ≤ 20% | 11 |
| > 20% | 5 |
| IGVH mutation status, no. patients | |
| Unmutated (≥ 98% homology) | 5 |
| Mutated (< 98% homology) | 11 |
| Trisomy 12–positive cells (%), median | 60 |
| Therapy not begun | |
| No. patients | 7 |
| Median time since sample data, mos | 28 |
| Therapy begun | |
| No. patients | 9 |
| Median TTT, mos | 19 |
| Characteristic . | Value . |
|---|---|
| Level of ZAP-70 expression, no. patients | |
| ≤ 20% | 11 |
| > 20% | 5 |
| IGVH mutation status, no. patients | |
| Unmutated (≥ 98% homology) | 5 |
| Mutated (< 98% homology) | 11 |
| Trisomy 12–positive cells (%), median | 60 |
| Therapy not begun | |
| No. patients | 7 |
| Median time since sample data, mos | 28 |
| Therapy begun | |
| No. patients | 9 |
| Median TTT, mos | 19 |
IGVH indicates immunoglobulin heavy chain variable-region; TTT, time to treatment; and ZAP-70, ζ-associated protein 70.
Discussion
The authors of several studies11,30,31 have shown that miRNAs play a critical role in the pathogenesis and the progression of CLL. We performed a genome-wide miRNA profiling on samples from CLL patients with B cells harboring various chromosomal abnormalities. In this study we demonstrate that, by using this platform, expression data of 32 miRNAs can distinguish subtypes of CLL with the most common genomic aberrations. This signature could be used to predict CLLs harboring the 17p deletion because it discriminates between cases with unmutated and mutated IGHV genes. In both training and validation sample sets, the signature classified CLL harboring 17p deletion with IGHV mutated as normal karyotype CLL, which is considered as an indolent form of CLL. This finding is consistent with the clinical course of these patients, suggesting an important integrative diagnostic value of this miRNA signature to assess the tumor burden of CLL cases harboring 17p deletion. The expression values of 9 among the 32 miRNAs, namely, miR-155, miR-29b, miR-151-3p, miR-29c, miR-34a, miR-640, miR-148a, miR-146a, and miR-146b-5p, were validated by qRT-PCR and associated with a specific karyotype. Several others, listed in the bioinformatics analysis, were undetectable or not validated by qRT-PCR as the result of cross-hybridization inherent to miRNA microarray method. Hence, we selected the 9 miRNAs for further studies. The expression pattern of these miRNAs supports in part, the results of a previous study,11 in that the miRNAs associated with the adverse cytogenetic subgroups also were associated with the aggressive forms of CLL based on the ZAP-70 positivity. The discrepancy of some miRNAs could be attributable to different platforms and cytogenetic cohort of samples used. The novel data that we present in this article show deregulation of miRNAs that do not map to the altered chromosomal regions corresponding to the karyotype with whom they are associated. Regarding miR-16, we can argue that the reason we do not find this miRNAs as the most down-modulated in CLL with 13q deletion is that almost all samples with 17p and 11q deletion also have 13q deletion.
Most of the miRNAs in our signature have an important role in B-cell differentiation and leukemogenesis. MiR-155 has been reported to be up-regulated in leukemia and lymphomas.32,33 Murine models also demonstrated that enhanced expression of this gene in B-cell precursors leads initially to a pre-B lymphoproliferative disease that progresses to a B-cell malignancy.34 Low expression of miR-29b and miR-29c in CLL with 11q deletion compared with other aggressive CLLs was demonstrated by Pekarsky et al.30 In this study we found low expression of both miRNAs and, in particular, of miR-29c, also in CLLs with 17p deletion. These data are consistent with a recent study35 in which the authors showed miR-29c expression levels to be significantly lower in poor prognostic CLL subgroups. The importance of these miRNAs in CLL could be ascribed to their target T-cell leukemia/lymphoma 1, which is overexpressed in the aggressive form of B-cell CLL. Deregulation of T-cell leukemia/lymphoma 1 may be causal event in CLL pathogenesis because enhanced expression of this gene in murine B cells leads to B-cell CLL.36 Low expression of miR-34a has been associated with both 17p deletion and chemotherapy resistance in CLL,37,38 supporting our data. Also noteworthy is miR-146a, which we demonstrated to be up-regulated in CLL with trisomy 12 compared with other CLLs. It has been suggested that increased expression of miR-146 is linked to tumorigenesis by inappropriate regulation of the inflammatory response.39
We further demonstrated that the 9 miRNAs have a biologic impact on CLL as shown by the correlation between miRNAs values and expression data of multiple genes. Particularly, we observed that the miR-146a correlates with several molecules involved in cell migration, cell motility, and cell adhesion and that increased expression of miR-146a is significantly associated with increased values of genes encoding the integrins ITGA4 and ITGB2, which we also demonstrated to be up-modulated in CLL with trisomy 12. These 2 genes represent the subunit of the heterodimers α4β1 and αLβ2, involved in TEM and motility of malignant cells during tissue invasion in CLL.16 In addition, CLLs that migrate beneath marrow stromal display greater levels of surface ITGA4 than nonmigrated cells.17 Therefore it is tempting to speculate that miR-146a is involved in TEM and cell migration. However, defining the biologic role of the miRNAs members of the signature was not intended point of this study; we only provide preliminary data that need to be validated by further studies.
We also aimed at providing new markers of disease behavior in cases that exhibit discordant prognostic factors. We analyzed the association between IGHV mutation status, ZAP-70 expression level, miRNAs, and additional independent genetic risk factors such as trisomy 12 and 17p deletion. In CLL patients with B cells harboring trisomy 12, IGHV mutation status, and ZAP-70 expression levels are not representative of the clinical outcome, as previously reported.29 Hence, we relate expression values of miRNAs and clinical data by using a Cox proportional hazards model. We found that high expression of miR-181a was associated with disease progression in our set of CLL patients with trisomy 12.
With respect to the CLL subgroup harboring 17p deletion, the authors of several studies28,40,41 have demonstrated that this genomic alteration occurs most frequently in CLL cases with unmutated IGHV genes and that express ZAP-70 protein and that the 17p deletion manifests in a rapid progression of the disease. We took into account the clinical behavior of CLL patients with B cells harboring the 17p deletion, mutated IGHV genes, and lacked ZAP-70 protein. Analysis performed on 2 subgroups, 17p indolent and 17p aggressive, predefined on the base of the clinical outcome, suggests that 17p deletion very likely indicates a high-genetic risk if it occurs in B cells expressing ZAP-70 protein and with unmutated IGHV, whereas CLL patients with mutated IGHV and lacked ZAP-70 protein, regardless of the presence of the 17p deletion, have a better clinical course. This finding may be attributable to the lower proportion of cells with the genomic abnormality in the latter case, as reported by Catovsky et al42
Moreover, analysis of miRNA expression values between the predefined subgroups showed that the miR-223, the miR-29b, the miR-29c, and the miR-181 family are down-regulated in 17p-aggressive cases compared with 17p-indolent cases. This finding suggests new markers to assess the disease course of CLL patients bearing the 17p deletion. Our findings on the impact of miR-29c and miR-223 are supported by a previous study by Stamatopoulos et al,35 who investigated the prognostic impact of miR-29c and miR-223 in a cohort of 110 patients with a median follow-up of 72 months. Noteworthy, miR-181a expression levels had opposite trend in the progression of the CLL depending on the karyotype. Indeed, it seems that low expression levels of miR-181a are associated with more aggressive form of CLL harboring 17p deletion and less aggressive form of CLL with trisomy 12. We are sensitive of basing our conclusion on such a limited cohort of patients; however, we will validate this test cohort in a large validation cohort in the near future.
In conclusion, this study indicates the importance of considering more than one parameter to assess the tumor burden in CLL. We are not advocating the replacement of standard clinical and molecular markers; rather, we suggest the addition of the miRNA expression evaluation for more accurate management of CLL patient care.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This work was performed with the help of National Cancer Institute grant RO1 CA111953 (to C.M.C).
National Institutes of Health
Authorship
Contribution: R.V. and C.M.C. designed the research; L.Z.R. and T.J.K. contributed patient samples; L.Z.R. contributed data; R.V., J.P., V.B., S.C., and H.A performed the research; C.T., S.V., M.F., M.N., and B.A. performed bioinformatics and statistical analysis; R.V. and A.V. analyzed data; and R.V., A.V., and C.M.C. and wrote the paper. All authors critically reviewed and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
L.Z.R., T.J.K., and C.M.C. are members of the CLL Research Consortium. A complete list of CLL Research Consortium participants is available in the supplemental Appendix.
Correspondence: Carlo M. Croce, MD, Department of Molecular Virology, Immunology and Medical Genetics and Comprehensive Cancer Center, The Ohio State University, 460 West 12th Ave, Columbus, OH 43210; e-mail: carlo.croce@osumc.edu.