In chronic lymphocytic leukemia (CLL), biologic risk factors such as immunoglobulin variable heavy chain gene (VH) mutation status, CD38 expression level, and genomic aberrations have recently been identified, but the relative prognostic impact of the individual parameters is unknown. In the current study, we analyzed VH mutation status by polymerase chain reaction and sequencing (n = 300), genomic aberrations by fluorescence in situ hybridization (+3q, 6q−, +8q, 11q−, +12q, 13q−, t(14q), 17p−) (n = 300), and CD38 expression by triple-color FACS (CD5, CD19, CD38) (n = 157) in a unicentric CLL cohort. The prognostic influence of VH mutation rate and CD38 expression level was tested by maximally selected log-rank statistics. A corrected P value (Pcor) for a cutoff level allowing the best separation of 2 subgroups with different survival probabilities was identified at 97% VH homology (95% confidence interval [CI], 96%-98% homology,Pcor <.001) and at 7% CD38 expression (95% CI, 20%-71% expression, Pcor = .02). In univariate analyses, unmutated VH genes and high CD38 expression levels predicted for shorter survival times. The overall incidence of genomic aberrations was similar in theVH unmutated and VHmutated subgroups. High-risk genomic aberrations such as 17p− and 11q− occurred almost exclusively in the VHunmutated subgroup, whereas favorable aberrations such as 13q− and 13q− as single abnormalities were overrepresented in theVH mutated subgroup. In multivariate analysis, unmutated VH, 17p deletion, 11q deletion, age, WBC, and LDH were identified as independent prognostic factors, indicating a complementary role of VH mutation status and genomic aberrations to predict outcome in CLL.
Introduction
Chronic lymphocytic leukemia (CLL) is characterized by a highly variable clinical course. Some patients die within a few months of diagnosis, whereas others survive prolonged periods without requiring therapy.1,2 To estimate prognosis in CLL, 2 major staging systems, mainly based on tumor load, are used.3 4 These systems define distinct prognostic subgroups, but their ability to predict the outcome of individual patients at the time of diagnosis, in particular in early-stage disease, is limited. Therefore, other factors related to the biology of CLL, such as genetic markers, are currently evaluated for their prognostic impact.
Clonal genomic aberrations can be identified in approximately 80% of CLL patients by fluorescence in situ hybridization (FISH)5; 17p deletion (17p−) and 11q deletion (11q−) are independent prognostic factors identifying subgroups of patients with rapid disease progression and short survival times in multivariate analysis, whereas 13q deletion (13q−) as the sole aberration is associated with favorable outcome.5 In addition, 17p− abnormalities and TP53 mutations have been associated with treatment failure.6-9
Another genetic marker, related to the stage of B-cell differentiation, is provided by the recombination of variable (V), diversity (D), and joining (J) immunoglobulin gene segments and the process of somatic hypermutation, which physiologically occurs in the germinal center (for review, see 10). Although the CLL B cells were originally considered to correspond to antigen-inexperienced pregerminal center lymphocytes, recent data indicated that in approximately half of all cases, somatically mutated variable region heavy chain genes (VH) are present.11 12
In pivotal studies correlating the VH mutation status with survival probability, Hamblin et al13 and Damle et al14 have shown that the presence of unmutatedVH predicted for an inferior clinical course in CLL. In one of these studies, a high CD38 expression level was found to correlate with the presence of unmutated VHgenes and an unfavorable clinical outcome,14 but the relation of CD38 expression to VH mutation status and survival is the subject of controversial discussion.15-18 The objective of this study was to evaluate the relation among genomic aberrations,VH mutation status, and CD38 expression and the relative prognostic impact of these factors in a large series of CLL patients.
Patients and methods
Patients
Between October 1990 and August 1998, 325 consecutive CLL patients from a single institution (Med. Klinik und Poliklinik V, University of Heidelberg) were enrolled in the study and followed with regard to survival. Cytogenetic data have been described previously, together with detailed clinical characteristics.5 Briefly, the diagnosis of CLL required a persistent lymphocytosis of more than 5.0 × 109/L, and immunophenotypic data were available for 314 of the 325 patients: all were CD19+, 298 of 308 tested were CD5+, and 300 of 308 were CD23+. Two hundred forty-eight patients were untreated, 39 patients underwent one chemotherapeutic regimen, and 38 patients underwent 2 or more chemotherapeutic regimens before genetic analysis. Treatment was not in controlled trials, and standard clinical criteria2 were used for the initiation of therapy that was usually alkylator based at first line and mostly fludarabine at second line. Median time from diagnosis to genetic study was 15 months. Based on the availability of archived samples, VH mutation status was evaluated in 300 patients, and the CD38 expression level was evaluated in 157 patients. Among the 300 patients analyzed forVH mutation status, there were 182 men and 118 women. At the time of enrollment, ages ranged from 30 to 87 years (median, 62 years); 156 patients were at Binet stage A, 95 were at Binet stage B, and 48 were at Binet stage C. According to Rai, 59 patients were at stage 0, 42 were at stage 1, 138 were at stage 2, 30 were at stage 3, and 31 were at stage 4. Staging information was incomplete for one patient. Estimated median follow-up and survival times were 69 months and 104 months, respectively. During this study, 107 patients died.
Interphase cytogenetic analysis
Based on conventional cytogenetic studies and data from comparative genomic hybridization, a comprehensive set of DNA probes was developed to diagnose the most frequent genomic aberrations in CLL.5 Our probe set allows screening for the following partial deletions, partial trisomies, and translocations: +(3q26), del(6q21) + (8q24), del(11q22-q23), t(11;14)(q13;q32), +12q13, del(13q14), t(14;18)(q32;q21), t(14q32), and del(17p13), as previously described.5
Identification and sequencing of clonal VDJ rearrangements
Genomic DNA was isolated from cryopreserved cells with the Trizol reagent (Gibco BRL, Eggenstein, Germany). In 273 of 300 patients, VDJ analysis was carried out from the same sample as the FISH analysis, and in the remaining 27 patients follow-up samples were used. Amplification of the VDJ rearrangements was performed by polymerase chain reaction (PCR) using (5′-VH) framework region (FR)I or leader region and (3′-JH) FRIV family-specific consensus oligonucleotide primers.12,19 To reduce the number of PCR reactions, the 5′-VH primers were labeled with fluorescent dyes for subsequent genescan analysis. In 2 multiplex PCR set-ups, the unlabeled (3′-JH) primer mix was combined with labeled mixtures of either VH1/7, 3, and 4 or VH2, 5, and 6 (5′-VH) family-specific primers. To detect whichVH gene family was involved in the clonal VDJ re-arrangement, the 5′-VH primer leading to the amplification of the PCR product was identified by genescan analyses on a genetic analyzer model 377 (Applied Biosystems, Weiterstadt, Germany). PCR was performed in a final volume of 50μL with 10 pmol of each primer, 100 μmol of each deoxyribonucleoside triphosphate (dNTP), and 1.25 U HotStarTaq polymerase (Qiagen, Hilden, Germany) with reaction buffer. Amplification consisted of an initial denaturation step of 15 minutes at 95°C followed by 35 cycles of 94°C for 30 seconds, 65°C for 45 seconds, and 72°C for 1.5 minutes, with a final extension step of 10 minutes. Because of the PCR primer location in the FRI, this region was only partially evaluable with this set-up, and VH sequencing was successfully repeated in 217 of the 300 patients to allow analysis of the entire FRI with the corresponding leader region I primers as described,12except for the VH4 family for which the primer 5′-ATGAAACACCTGTGGTTCTT-3′ was used.
PCR products were spin column-purified (Qiagen) and directly sequenced using dye terminator chemistry (Big Dye Kit; Applied Biosystems) with the appropriate nonlabeled 5′-VH and 3′-JH PCR primers on a genetic analyzer model 377 (Applied Biosystems). If more than one product was detected for one VH family the PCR products were cloned and subsequently sequenced. All cases were verified by sequence analysis of at least one independent PCR product.
The VDJ nucleotide sequences were aligned to current databases (EMBL/GenBank, V-base directory).20 Homology to the nearest germline gene was calculated in relation to the entireVH (n = 217) or to the length of the sequenced fragment available (n = 83) in patients who did not amplify with leader region primers or in whom no DNA was available. CDRIII was defined as the nucleotide sequence between the codon 94, the last codon of FRIII, and codon 102, the first codon of FRIV.
CD38 expression
CD38 expression was analyzed from cryopreserved cells in 157 cases by triple-color fluorescence-activated cell sorting (FACS) analysis. Mononuclear cells had been isolated before freezing by Ficoll-Hypaque density gradient centrifugation. In analogy with Damle et al,14 the cells were stained with anti-CD5, anti-CD19, and anti-CD38 using the following antibody conjugates: anti–CD5-FITC (clone L17F12), anti–CD19-PerCPCy5.5 (clone SJ25C1), and anti–CD38-PE (clone HB-7), (all from Becton Dickinson, Heidelberg, Germany). Isotype-matched negative controls were used in all assays to determine positive from negative cells. In accordance with previous studies,15 -18 the percentage of CD38+ cells was measured in the CD19+/CD5+ fraction. Flow cytometric analyses were performed on a Becton Dickinson FACScan flow cytometer. In 143 cases samples from the same time point as the cytogenetic analysis were studied. To investigate whether CD38 expression remains stable during the course of the disease, samples obtained at several different time points were studied in 12 patients; 2 time points in 6, and 3 time points in another 6 patients.
Statistical analysis
The primary end point was survival from the time of diagnosis. Survival time distributions were plotted using Kaplan-Meier estimates. Median duration of follow-up was calculated according to the method of Korn.21 Testing and estimation of possible cutoff values for the VH gene homology and CD38 expression was done by maximally selected log-rank statistics,22 and 95% CIs of the cut-off values were computed using 1000 bootstrap samples. The proportional hazards regression model of Cox was used to identify differences in survival time distribution from prognostic factors.23 As possible prognostic factors age, sex, stage according to Binet and Rai, hemoglobin (Hb) levels, white blood cell count (WBC), platelet count, serum lactate dehydrogenase (LDH) and alkaline phosphatase (AP) levels, presence or absence of splenomegaly and lymphadenopathy, extent of peripheral lymphadenopathy, greatest lymph-node diameter measured, presence or absence of genomic aberrations (17p−,11q−,+12q,13q−,6q−), CD38 expression 7% or more versus less than 7%, and VH homology 97% or more versus less than 97% were included in the regression model. Regression analysis was repeated with the same parameters, and the classical VH homology cut-off at 98% (ie, separation of 2 groups with VH homology 98% or greater versus less than 98%). To correct for overestimation of the relative risk estimate, shrinkage of the parameter estimates was applied.24 Missing data were estimated using a multiple-imputation technique with 10 random draws. A limited backward-selection procedure was used to exclude redundant or unnecessary variables.25
Groupwise comparisons of distributions of clinical, laboratory, and genetic data were performed with the Kruskal-Wallis test (quantitative) and the Fisher exact test (categorical variables). All tests were 2-sided. An effect was considered statistically significant atP = .05. To provide quantitative information on the relevance of statistically significant results, 95% CIs for hazard ratios were computed. Pairwise correlations were estimated using Pearson product-moment correlation coefficient. Statistical analyses were performed with the StatXact (Cytel Software, Cambridge, MA), S-Plus (Insightful, Seattle, WA), and Design software packages25 and with GraphPad Prism version 3.00 (GraphPad Software, San Diego, CA).
Results
VHmutation status
In all cases clonal VDJ rearrangements were amplifiable using the 5′-VH FRI PCR primers, whereas VDJ amplification with corresponding 5′-VH family specific leader region I primers was successful in 217 patients. Further analysis was therefore based on the sequence of the entireVH, obtained with leader region I primers in 217 patients, and on the sequence of VH lacking part of the FRI, obtained with the 5′-VH FRI primers in the remaining 83 patients. According to the following criteria, the VDJ rearrangements were considered to be nonfunctional: rearrangement of a pseudogene, stop codon, or frameshift in the CDRIII, incomplete rearrangement. Applying these criteria, 265 patients had functional VDJ rearrangement.
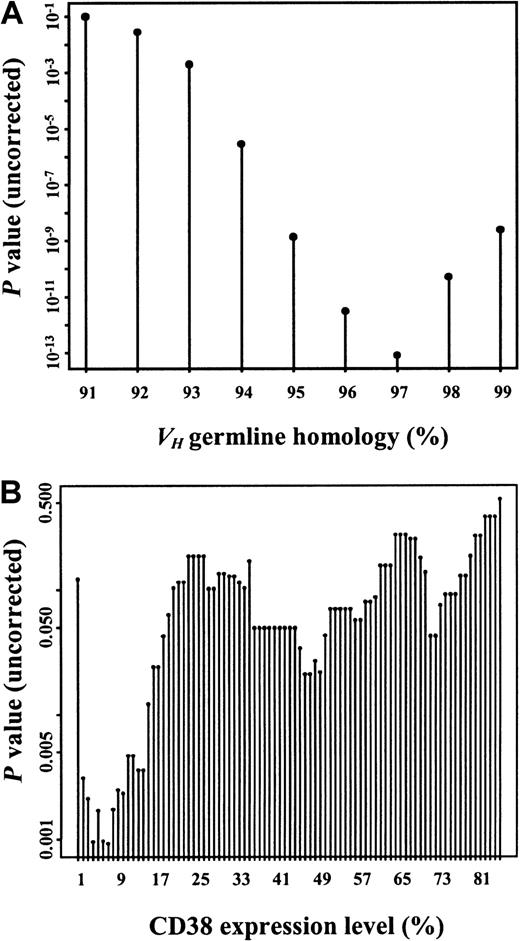
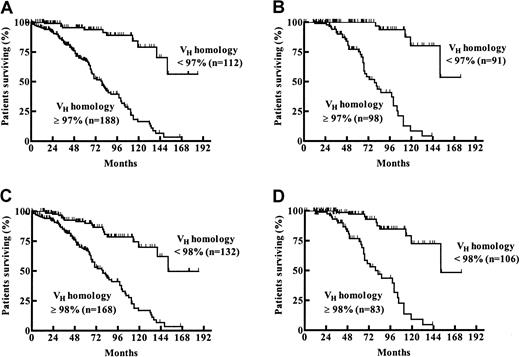
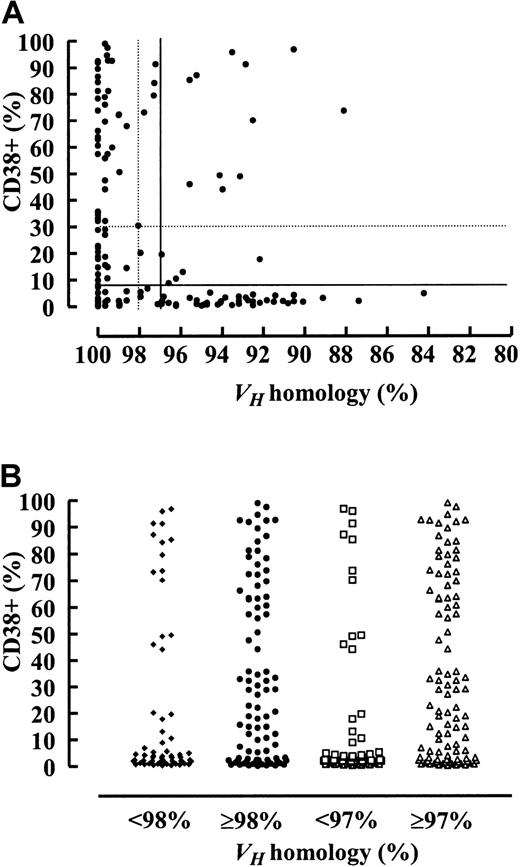
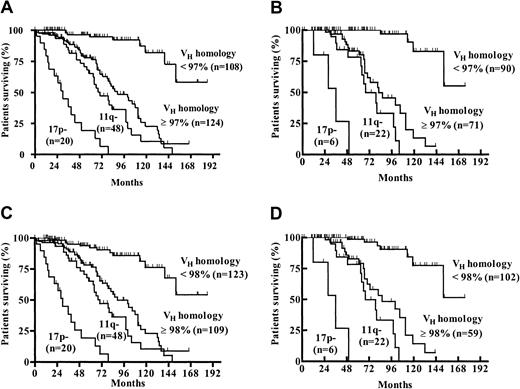
Considering the classical VH homology cutoff value of 98%, 132 (44%) patients had mutated and 168 (56%) patients had unmutated VH genes. The germline homology rate ranged from 84% to 100% (median, 98.97%; interquartile range, 94.58% to 100%). To study the prognostic influence of different VH mutation rates, maximally selected log-rank tests were performed. According to the corrected P(Pcor) of the maximally selected log-rank statistics, the estimated VH homology rate allowing the best separation of 2 subgroups with different survival probabilities was not 98% but was 97% (95% CI, 96%-98%;Pcor <.001) (Figure1A). One hundred eighty-eight (63%) patients had a VH homology of 97% or greater, and 112 (37%) patients had a VH homology less than 97%. Median overall survival (OS) time of the subgroup withVH homology less than 97% could not be estimated. The last observed death was after 152 months of follow-up time with a corresponding estimated survival probability of 56%. Estimated median survival time of the group withVH homology 97% or greater was 79 months (Figure 2A). When only patients diagnosed at Binet stage A were considered (n = 189), 91 (48%) had aVH homology less than 97% and 98 (52%) had aVH homology of 97% or greater. Estimated median OS of the group with VH homology of 97% or greater in the Binet A patients was 79 months. The last death in the group with VH homology less than 97% was observed after 152 months of follow-up time, with a corresponding estimated survival probability of 53% (Figure 2B). The same analyses were performed for the classical VH homology cutoff value of 98%. Estimated median OS of the groups withVH homology of 98% or greater was 79 months for all patients and for Binet A patients, whereas the median OS was 152 months for the groups with VH homology less than 98% for all patients and for Binet A patients (Figure 2C-D).
Maximally selected log-rank statistics were performed for the VH homology rate and the CD38 expression level to test for a possible cut-off value separating 2 groups with different survival distributions.
VH germline homology rate (A) and CD38 expression levels (B) are shown on the x-axes. Corresponding uncorrected P values for the separation of 2 groups with different survival probability are shown on the y-axes. CorrectedP values were Pcor <.001 for theVH homology rate at the corresponding estimate of 97% and Pcor = .02 for CD38 expression at the corresponding estimate of 7%.
Maximally selected log-rank statistics were performed for the VH homology rate and the CD38 expression level to test for a possible cut-off value separating 2 groups with different survival distributions.
VH germline homology rate (A) and CD38 expression levels (B) are shown on the x-axes. Corresponding uncorrected P values for the separation of 2 groups with different survival probability are shown on the y-axes. CorrectedP values were Pcor <.001 for theVH homology rate at the corresponding estimate of 97% and Pcor = .02 for CD38 expression at the corresponding estimate of 7%.
Probability of survival from the date of diagnosis among patients with a VH homology less than 97% or 97% or greater and less than 98% or 98% or greater.
(A) Estimated median survival time for the VHhomology 97% or greater group was 79 months. The last observed death in the VH homology less than 97% group was after 152 months of follow-up time (survival probability, 56%). (B) When only patients diagnosed at Binet stage A were evaluated, the estimated median survival times for the VHhomology 97% or greater and the VH homology less than 97% groups were 79 months versus not reached (last observed death after 152 months of follow-up time; survival probability, 53%). (C) Estimated median survival time for the VHhomology 98% or greater and less than 98% groups were 79 months and 152 months, respectively. (D) When only patients diagnosed at Binet stage A were evaluated, the estimated median survival times for theVH homology 98% or greater andVH homology less than 98% groups were 79 months versus 152 months.
Probability of survival from the date of diagnosis among patients with a VH homology less than 97% or 97% or greater and less than 98% or 98% or greater.
(A) Estimated median survival time for the VHhomology 97% or greater group was 79 months. The last observed death in the VH homology less than 97% group was after 152 months of follow-up time (survival probability, 56%). (B) When only patients diagnosed at Binet stage A were evaluated, the estimated median survival times for the VHhomology 97% or greater and the VH homology less than 97% groups were 79 months versus not reached (last observed death after 152 months of follow-up time; survival probability, 53%). (C) Estimated median survival time for the VHhomology 98% or greater and less than 98% groups were 79 months and 152 months, respectively. (D) When only patients diagnosed at Binet stage A were evaluated, the estimated median survival times for theVH homology 98% or greater andVH homology less than 98% groups were 79 months versus 152 months.
CD38 expression
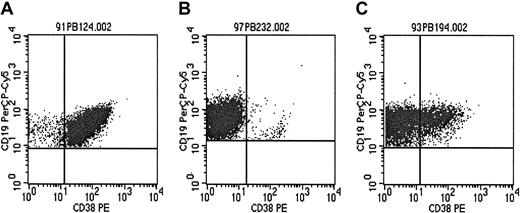
CD38 expression level of the tumor cell population was evaluated in 157 patients. The proportion of CLL cells expressing CD38 above the isotype control level ranged from 0.24% to 98.99%, with a median of 9.99% (interquartile range, 1.99%-57.4%). Examples of representative scatterplots are shown in Figure 3A-C.
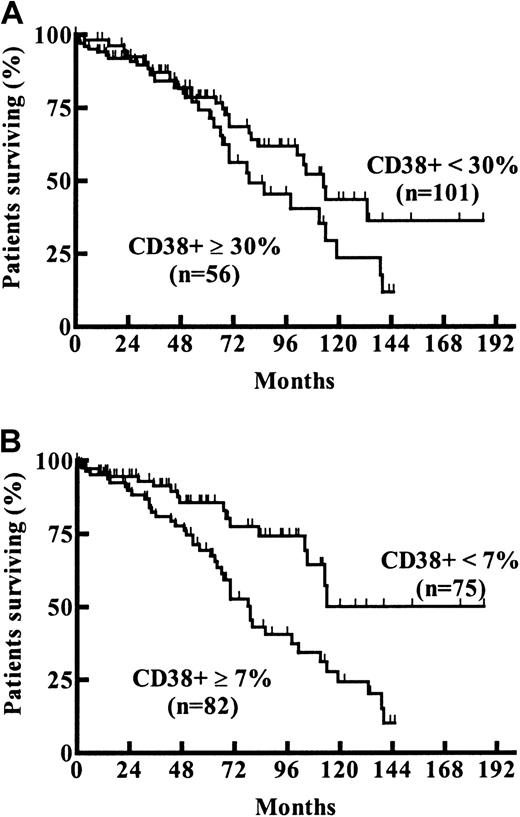
Survival probabilities of patients expressing CD38 in more (56 patients, 36%) or less (101 patients, 64%) than 30% of tumor cells was not significantly different (estimated median OS, 79 vs 113 months;P = .13, log-rank test; Figure4A). Maximally selected log-rank statistics were performed to evaluate a possible cut-off value for CD38 expression with respect to changes in the survival time distribution. Estimated CD38 expression level yielding the best separation of 2 subgroups with different survival probabilities was 7% (95% CI, 20%-71%; Pcor = .02; Figure 1B). Estimated median OS of cases with a CD38 expression less than 7% (75 patients, 48%) was 114 months versus 79 months in the group expressing CD38 in 7% or more (82 patients, 52%) of tumor cells (Figure 4B).
Examples of CD38 expression patterns.
The proportion of CLL cells expressing CD38 varied widely in the patient cohort studied (see also Figure 5). Three representative types of CD38 expression patterns of the CD19+/CD5+cell population of individual patients are shown. (A) Homogeneously positive (97.5% CD38+ cells). (B) Homogeneously negative (1.5% CD38+ cells). (C) Mixed expression pattern with CD38+ and CD38− CLL cells (28.9% CD38+ cells).
Examples of CD38 expression patterns.
The proportion of CLL cells expressing CD38 varied widely in the patient cohort studied (see also Figure 5). Three representative types of CD38 expression patterns of the CD19+/CD5+cell population of individual patients are shown. (A) Homogeneously positive (97.5% CD38+ cells). (B) Homogeneously negative (1.5% CD38+ cells). (C) Mixed expression pattern with CD38+ and CD38− CLL cells (28.9% CD38+ cells).
Probability of survival from the date of diagnosis compared among CLL subgroups with different CD38 expression levels.
Estimated median survival times for the subgroups with (A) CD38+ up to 30% versus greater than 30% were 113 months versus 79 months (P = .13; log-rank test) and for the subgroups with (B) CD38+ up to 7% versus more than 7% were 114 months versus 79 months (Pcor = .02; derived from maximally selected log-rank statistics), respectively.
Probability of survival from the date of diagnosis compared among CLL subgroups with different CD38 expression levels.
Estimated median survival times for the subgroups with (A) CD38+ up to 30% versus greater than 30% were 113 months versus 79 months (P = .13; log-rank test) and for the subgroups with (B) CD38+ up to 7% versus more than 7% were 114 months versus 79 months (Pcor = .02; derived from maximally selected log-rank statistics), respectively.
Table 1 and Figure5A,B summarize the relation and distribution of CD38 expression and the VHmutation status. A high CD38 expression correlated with unmutatedVH, but in at least 34% of patients, depending on the VH homology and CD38 expression cut-off value chosen, CD38 expression failed to predict theVH mutation status.
Correlation between CD38 expression andVH mutation status
| . | CD38 expression No. patients (%) . | |||
|---|---|---|---|---|
| <30% 101 (64) . | ≥30% 56 (36) . | <7% 75 (48) . | ≥7% 82 (52) . | |
| VHhomology | ||||
| <98% | 50 (32) | 15 (10) | 44 (28) | 21 (13) |
| ≥98% | 51 (32) | 41 (26) | 31 (20) | 61 (39) |
| rho value* (95% CI) | 0.22 (0.07-0.37) | 0.34 (0.19-0.48) | ||
| VHhomology | ||||
| <97% | 45 (29) | 11 (7) | 40 (26) | 16 (10) |
| ≥97% | 56 (35) | 45 (29) | 35 (22) | 66 (42) |
| rho value* (95% CI) | 0.25 (0.11-0.39) | 0.35 (0.21-0.50) | ||
| . | CD38 expression No. patients (%) . | |||
|---|---|---|---|---|
| <30% 101 (64) . | ≥30% 56 (36) . | <7% 75 (48) . | ≥7% 82 (52) . | |
| VHhomology | ||||
| <98% | 50 (32) | 15 (10) | 44 (28) | 21 (13) |
| ≥98% | 51 (32) | 41 (26) | 31 (20) | 61 (39) |
| rho value* (95% CI) | 0.22 (0.07-0.37) | 0.34 (0.19-0.48) | ||
| VHhomology | ||||
| <97% | 45 (29) | 11 (7) | 40 (26) | 16 (10) |
| ≥97% | 56 (35) | 45 (29) | 35 (22) | 66 (42) |
| rho value* (95% CI) | 0.25 (0.11-0.39) | 0.35 (0.21-0.50) | ||
Pearson product-moment correlation coefficient.
Relation between CD38 expression andVH mutation status in the CLL cohort.
The proportion of CD38+ CLL cells is shown on the y-axes.VH mutation status is shown on the x-axes as percentage value of homology to the nearest germline gene (A) and according to the most important subgroups (B). Low CD38 expression levels correlated with high VH mutation rates, and high CD38 expression levels correlated with lowVH mutation rates. However, in at least 34% of patients, depending on the cut-off value used (see “Results” and Table 1 for details), the CD38 expression level failed to predict theVH mutation status.
Relation between CD38 expression andVH mutation status in the CLL cohort.
The proportion of CD38+ CLL cells is shown on the y-axes.VH mutation status is shown on the x-axes as percentage value of homology to the nearest germline gene (A) and according to the most important subgroups (B). Low CD38 expression levels correlated with high VH mutation rates, and high CD38 expression levels correlated with lowVH mutation rates. However, in at least 34% of patients, depending on the cut-off value used (see “Results” and Table 1 for details), the CD38 expression level failed to predict theVH mutation status.
CD38 expression level was analyzed in follow-up samples of 12 patients over a median time interval of 29 months (range, 8 to 60 months) to investigate whether the CD38 expression level remains constant over time. All but one patient received chemotherapy, consisting of alkylating agents or purine analogs, or both, within this period. The median change in the proportion of CD38-expressing cells was 8% (range, 0%-51%). Among the 12 patients, 3 exhibited initial CD38 expression levels less than 7% and 7% or more in follow-up samples. Four patients exhibited CD38 expression levels less than 30% or 30% or more at different time points. Of these, CD38 expression changed from less than 30% to 30% or more in 2 patients and from 30% or more to less than 30% in another 2 patients. Thus, at least 3 of 12 patients, depending on the cutoff chosen, would have been classified in different prognostic subgroups at different time points according to the CD38 expression level.
Relation of VHmutation status and genomic aberrations
By interphase cytogenetics, 246 (82%) of 300 patients exhibited genomic aberrations. Table 2 lists these aberrations in order of decreasing frequency and shows the comparison of specific genomic aberrations according to theVH mutation status. The overall incidence of genomic abnormalities in the VH mutated and unmutated subgroups was similar, as was the incidence of trisomy 12q (Table 2). Prognostically favorable aberrations, such as 13q deletion or 13q deletion as a single aberration, were significantly more frequent in the VH mutated group. High-risk genomic aberrations such as 17p deletion and 11q deletion were detected almost exclusively in VH unmutated patients. However, only approximately one third of the VHunmutated patients did show high-risk genomic aberrations (11q or 17p deletion) (Table 2).
Incidence of genomic abnormalities in 300 CLL according to the VH mutation status
| Aberration . | All cases no. patients (%) . | VH homology <97% 112 (37) . | VH homology ≥97% 188 (63) . | P* . | VH homology <98% 132 (44) . | VH homology ≥98% 168 (56) . | P . |
|---|---|---|---|---|---|---|---|
| 13q deletion | 167 (56) | 75 (67) | 92 (49) | .003 | 86 (65) | 81 (48) | .004 |
| 13q deletion single | 109 (36) | 60 (54) | 49 (26) | <.001 | 66 (50) | 43 (26) | <.001 |
| 12q trisomy | 52 (17) | 20 (18) | 32 (17) | .88 | 20 (15) | 32 (19) | .44 |
| 11q deletion | 50 (17) | 2 (2) | 48 (26) | <.001 | 5 (4) | 45 (27) | <.001 |
| 17p deletion | 20 (7) | 2 (2) | 18 (10) | .008 | 4 (3) | 16 (10) | .03 |
| 17p or 11q deletion | 68 (22) | 4 (4) | 64 (34) | <.001 | 9 (7) | 59 (35) | <.001 |
| Clonal abnormalities | 246 (82) | 87 (78) | 159 (85) | .16 | 105 (80) | 141 (84) | .37 |
| Aberration . | All cases no. patients (%) . | VH homology <97% 112 (37) . | VH homology ≥97% 188 (63) . | P* . | VH homology <98% 132 (44) . | VH homology ≥98% 168 (56) . | P . |
|---|---|---|---|---|---|---|---|
| 13q deletion | 167 (56) | 75 (67) | 92 (49) | .003 | 86 (65) | 81 (48) | .004 |
| 13q deletion single | 109 (36) | 60 (54) | 49 (26) | <.001 | 66 (50) | 43 (26) | <.001 |
| 12q trisomy | 52 (17) | 20 (18) | 32 (17) | .88 | 20 (15) | 32 (19) | .44 |
| 11q deletion | 50 (17) | 2 (2) | 48 (26) | <.001 | 5 (4) | 45 (27) | <.001 |
| 17p deletion | 20 (7) | 2 (2) | 18 (10) | .008 | 4 (3) | 16 (10) | .03 |
| 17p or 11q deletion | 68 (22) | 4 (4) | 64 (34) | <.001 | 9 (7) | 59 (35) | <.001 |
| Clonal abnormalities | 246 (82) | 87 (78) | 159 (85) | .16 | 105 (80) | 141 (84) | .37 |
Fisher exact test.
VHmutation status, genomic aberrations, clinical data, and survival
Based on a VH homology cutoff value of 97%, the proportional-hazards regression model identified 5 significant prognostic factors: VH homology of 97% or greater (P < .001), 17p deletion (P < .001), age (P < .001), WBC (P < .001), and LDH (P < .001). The 11q deletion was identified as an additional independent factor in multivariate analysis when a VH homology of 98% was used as cutoff value. Hazard ratios, together with their 95% confidence limits, are shown in Table 3. Disease stage was not selected as a significant independent factor whenVH mutation status and genomic aberrations were included in the multivariate analysis. However, the relevance of stage according to Binet as a prognostic factor was evident if tested alone (P < .001).
Cox regression analysis of survival time from diagnosis in 300 patients with CLL
| Variable . | Hazard ratio for death (95% CI) atVH homology cut-off value . | |
|---|---|---|
| 97% . | 98% . | |
| VH unmutated | 5.27 (2.67-10.40) | 2.91 (1.71-4.95) |
| 17p deletion | 5.53 (2.98-10.26) | 8.19 (4.29-15.65) |
| 11q deletion | — | 1.78 (1.07-2.96) |
| Age | 1.49 (1.18-1.87) | 1.68 (1.33-2.13) |
| LDH | 1.26 (1.10-1.44) | 1.2 (1.05-1.38) |
| WBC | 1.11 (1.06-1.17) | 1.11 (1.05-1.16) |
| Variable . | Hazard ratio for death (95% CI) atVH homology cut-off value . | |
|---|---|---|
| 97% . | 98% . | |
| VH unmutated | 5.27 (2.67-10.40) | 2.91 (1.71-4.95) |
| 17p deletion | 5.53 (2.98-10.26) | 8.19 (4.29-15.65) |
| 11q deletion | — | 1.78 (1.07-2.96) |
| Age | 1.49 (1.18-1.87) | 1.68 (1.33-2.13) |
| LDH | 1.26 (1.10-1.44) | 1.2 (1.05-1.38) |
| WBC | 1.11 (1.06-1.17) | 1.11 (1.05-1.16) |
Hazard ratios and CIs are computed for 10-year increments in age; 50 IU/L increments in LDH; and for 20,000 per cubic millimeter increments in the WBC. Estimated shrinkage factor for the effect of unmutated VH was 0.956 (VH homology ≥97%) and 0.935 (VH homology ≥98%), yielding a corrected estimated hazard ratio of 4.90 (VH homology ≥97%) and 2.71 (VH homology ≥98%).
Table 4 shows the clinical and laboratory data for the patients in the subgroups 17p deletion,VH homology 97% or greater, andVH homology less than 97% at the time of enrollment. Distribution of age and sex was comparable in the 3 subgroups. Patients with 17p deletion and VHhomology of 97% or greater had more advanced disease, whereas patients with VH homology less than 97% had the highest proportion at Binet stage A. The subgroups with 17p deletion andVH homology of 97% or greater were more likely to have extensive splenomegaly and lymphadenopathy. Moreover, fever, night sweats, or weight loss (B symptoms) were more frequent in the groups with 17p deletion or VH homology of 97% or greater. These subgroups also had lower Hb levels, lower platelet counts, higher WBC, and higher LDH and AP levels. As indicated by the treatment-free intervals, patients with 17p deletion,VH homology 97% or greater and less than 97% showed significant differences in their rates of disease progression (median treatment-free intervals, 3.5 vs 33 vs 105 months).
Comparison of the clinical and laboratory data among the three genetic subgroups
| Parameter . | 17p− . | VH ≥97% . | VH<97% . | P . |
|---|---|---|---|---|
| No. patients | 20 | 170 | 110 | |
| Median age, y | 64 | 63 | 61 | .124-151 |
| Male sex, % | 75 | 60 | 59 | .41‡ |
| Stage, % | ||||
| Binet A | 25 | 43 | 71 | <.001‡ |
| Binet B | 40 | 37 | 23 | |
| Binet C | 35 | 20 | 6 | |
| Rai 0 | 0 | 14 | 33 | <.001‡ |
| Rai 1 | 5 | 16 | 13 | |
| Rai 2 | 45 | 45 | 48 | |
| Rai 3 | 20 | 13 | 4 | |
| Rai 4 | 30 | 12 | 4 | |
| White blood count, ×109/L | 52.3 | 39.3 | 26.8 | .024-151 |
| Hemoglobin, g/L | 125 | 131 | 136 | .0014-151 |
| Platelet count, ×109/L | 148 | 171 | 188 | .0044-151 |
| Lactate dehydrogenase, IU/L | 250 | 180 | 156 | <.0014-151 |
| Alkaline phosphatase, IU/L | 159 | 127 | 117 | .0034-151 |
| Albumin, g/L | 43 | 45 | 46 | .104-151 |
| Splenomegaly, % | 89 | 67 | 54 | .003‡4-150 |
| Mediastinal lymphadenopathy, % | 25 | 12 | 0 | <.001‡ |
| Abdominal lymphadenopathy, % | 65 | 61 | 27 | <.001‡ |
| Peripheral lymphadenopathy, cm24-150 | 11 | 4 | 0 | <.0014-151 |
| Largest lymph node diameter, cm | 3 | 2.85 | 1 | <.0014-151 |
| B symptoms, % | 37 | 20 | 8 | .001‡ |
| Time from diagnosis to first treatment, mo | 3.5 | 33 | 105 | <.0014-153 |
| Parameter . | 17p− . | VH ≥97% . | VH<97% . | P . |
|---|---|---|---|---|
| No. patients | 20 | 170 | 110 | |
| Median age, y | 64 | 63 | 61 | .124-151 |
| Male sex, % | 75 | 60 | 59 | .41‡ |
| Stage, % | ||||
| Binet A | 25 | 43 | 71 | <.001‡ |
| Binet B | 40 | 37 | 23 | |
| Binet C | 35 | 20 | 6 | |
| Rai 0 | 0 | 14 | 33 | <.001‡ |
| Rai 1 | 5 | 16 | 13 | |
| Rai 2 | 45 | 45 | 48 | |
| Rai 3 | 20 | 13 | 4 | |
| Rai 4 | 30 | 12 | 4 | |
| White blood count, ×109/L | 52.3 | 39.3 | 26.8 | .024-151 |
| Hemoglobin, g/L | 125 | 131 | 136 | .0014-151 |
| Platelet count, ×109/L | 148 | 171 | 188 | .0044-151 |
| Lactate dehydrogenase, IU/L | 250 | 180 | 156 | <.0014-151 |
| Alkaline phosphatase, IU/L | 159 | 127 | 117 | .0034-151 |
| Albumin, g/L | 43 | 45 | 46 | .104-151 |
| Splenomegaly, % | 89 | 67 | 54 | .003‡4-150 |
| Mediastinal lymphadenopathy, % | 25 | 12 | 0 | <.001‡ |
| Abdominal lymphadenopathy, % | 65 | 61 | 27 | <.001‡ |
| Peripheral lymphadenopathy, cm24-150 | 11 | 4 | 0 | <.0014-151 |
| Largest lymph node diameter, cm | 3 | 2.85 | 1 | <.0014-151 |
| B symptoms, % | 37 | 20 | 8 | .001‡ |
| Time from diagnosis to first treatment, mo | 3.5 | 33 | 105 | <.0014-153 |
All values correspond to the time of enrollment.
Median sum of the products of the diameters of the largest cervical, axillary, and inguinal lymph nodes.
Median values are given for quantitative variables:
Kruskal-Wallis test; ‡Fisher exact test;
log-rank test.
Based on the results of the regression model, 4 genetic subgroups were defined in which each patient was allocated to one category only: patients with a 17p deletion regardless of theirVH mutation status, patients with an 11q deletion and no 17p deletion regardless of theirVH mutation status, patients with unmutatedVH but not a 17p or 11q deletion, and patients with mutated VH but not a 17p or 11q deletion. As illustrated in Figure 6, the estimated median survival times for these categories were 30 months for 17p deletion, 70 months for 11q deletion, and 89 months for unmutatedVH (homology cut-off at 97% or 98%), whereas in the group with mutated VH, the last observed death was at 152 months, corresponding to an estimated survival probability of 58% at less than 97% and of 54% at less than 98% homology cut-off, respectively. The impact of the genetic risk categories held also true when only patients diagnosed at Binet stage A were evaluated (Figure 6).
Survival probabilities.
Survival probabilities from the date of diagnosis among patients in the following genetic categories: 17p− (17p deletion regardless ofVH mutation status), 11q− (11q deletion and no 17p deletion, regardless of VH mutation status), unmutated VH (VH homology 97% or greater or 98% or greater and no 17p or 11q deletion), and mutated VH (VH homology less than 97% or less than 98% and no 17p or 11q deletion). Median survival times for the respective genetic subgroups were as follows: A (all patients and 97% VH homology cut-off): 17p− 30 months, 11q− 70 months, VH 97% or greater 89 months, and VH less than 97% not reached (58% survival at 152 months). B (Binet stage A patients and 97% VH homology cut-off): 17p− 36 months, 11q− 68 months, VH 97% or greater 82 months, and VH less than 97% not reached (55% survival at 152 months). C (all patients and 98% VHhomology cut-off): 17p− 30 months, 11q− 70 months,VH 98% or greater 89 months, andVH less than 98% not reached (54% survival at 152 months). D (Binet stage A patients and 98%VH homology cut-off): 17p− 36 months, 11q− 68 months, VH 98% or greater 86 months, andVH less than 98% not reached (52% survival at 152 months).
Survival probabilities.
Survival probabilities from the date of diagnosis among patients in the following genetic categories: 17p− (17p deletion regardless ofVH mutation status), 11q− (11q deletion and no 17p deletion, regardless of VH mutation status), unmutated VH (VH homology 97% or greater or 98% or greater and no 17p or 11q deletion), and mutated VH (VH homology less than 97% or less than 98% and no 17p or 11q deletion). Median survival times for the respective genetic subgroups were as follows: A (all patients and 97% VH homology cut-off): 17p− 30 months, 11q− 70 months, VH 97% or greater 89 months, and VH less than 97% not reached (58% survival at 152 months). B (Binet stage A patients and 97% VH homology cut-off): 17p− 36 months, 11q− 68 months, VH 97% or greater 82 months, and VH less than 97% not reached (55% survival at 152 months). C (all patients and 98% VHhomology cut-off): 17p− 30 months, 11q− 70 months,VH 98% or greater 89 months, andVH less than 98% not reached (54% survival at 152 months). D (Binet stage A patients and 98%VH homology cut-off): 17p− 36 months, 11q− 68 months, VH 98% or greater 86 months, andVH less than 98% not reached (52% survival at 152 months).
Discussion
VH mutation status, CD38 expression, genomic aberrations, and clinical data were analyzed in a series of 300 CLL patients. In this study we confirm that CLL patients with unmutatedVH have significantly shorter survival times than patients with mutated VH.13 14Interestingly, in our series the estimated VHhomology rate yielding the best separation of 2 subgroups with different survival probabilities was not the classical cut-off value of 98% but was 97% VH homology to the nearest germline gene. This finding is still compatible with published data because the 95% CI of the analysis ranged from 96% to 98% homology. We cannot rule out that this effect is attributable to methodological aspects because in patients in whom there was no amplification with leader region primers (n = 83), the amplification was performed with FRI primers, and, therefore, the first part of the FRI was not amenable for sequence homology evaluation. However, in a separate analysis of the 217 patients amplifiable with leader region I 5′-VH primers, a cut-off value of 97%VH homology was obtained by maximally selected log-rank statistics (data not shown). On the other hand, one should note that the classical 98% homology cut-off definition for mutatedVH was based on germline polymorphism considerations and was not derived from the analysis of survival probability in CLL patients according to VHmutation status. The biologic significance of this finding is not clear and will be further studied.
The proportion of CLL cells expressing CD38, a parameter proposed to serve as a surrogate marker for VH mutation status,14 was investigated in our series in 157 CLL patients. High CD38 expression levels correlated with unmutatedVH genes and low CD38 expression levels with mutated VH genes, but, as described by others, in approximately one third of patients the CD38 expression level did not predict the VH mutation status.15-17 In agreement with Thunberg et al,16 there was no significant difference in the estimated median survival times when using the 30% cut-off value for CD38 positivity. However, by maximally selected log-rank statistics the best separation of 2 prognostic subgroups was achieved for a cut-off value of 7% CD38 positivity. Of note is the wide 95% CI of the analysis, ranging from 20% to 71%. As in the current study, a lower CD38 cut-off value, namely 20%, was also used recently by Ibrahim et al,18 to demonstrate an inferior outcome of CD38+ CLL patients. In their series the prognostic impact of CD38 expression was significant in multivariate analysis (P = .0258), but 17p deletion, 11q deletion, andVH mutation status, which are the strongest adverse prognostic factors in our study, were not included as parameters in their model.
As shown by Ibrahim et al,18 who found a significant CD38 expression change in 1 of 6 patients, at least 25% of patients in our study would have been classified in different CD38 expression subgroups at different time points. Chemotherapy was initiated in 11 of the 12 patients showing variability in CD38 expression. Although at the 7% cut-off 3 patients and at the 30% cut-off 2 patients exhibited increasing CD38 expression, in 2 patients CD38 expression decreased from greater than 30% to less than 30%. The variation observed among different studies on the CD38 expression in CLL might have been caused by methodological aspects. Therefore, an interlaboratory survey, coordinated by N. Chiorazzi (North Shore-Long Island Research Institute, NY), is being performed to evaluate this issue.
Apart from the VH mutation status, another genetic parameter of prognostic significance in CLL is provided by genomic aberrations.5 In a previous study applying chromosome banding, a higher rate of trisomy 12 was observed in theVH unmutated CLL subgroup.13 In this study the lower incidence of trisomy 12 coincided with a lower overall ability to detect genomic aberrations in theVH mutated compared with the unmutated subgroup, a fact likely reflecting the different in vitro proliferative potential.13 In the current study interphase FISH, a more sensitive technique not dependent on in vitro proliferation, was used. The overall incidence of genomic aberrations and the incidence of trisomy 12 were similar in the VH mutation subgroups. In contrast, high-risk genomic aberrations such as 17p deletion and 11q deletion occurred significantly more frequently in the unmutated VH subgroup, whereas favorable prognostic markers such as 13q deletion and 13q deletion as a single aberration were overrepresented in the mutatedVH subgroup. The unbalanced distribution of genomic aberrations might point toward a distinct biologic background of the CLL subgroups defined by VH mutation status and may in part explain their different clinical behaviors.
To evaluate the relative prognostic impact of VHmutation status, CD38 expression level, genomic aberrations, and clinical, as well as laboratory parameters, multivariate analysis was performed. VH homology of 97% or more (P < .001), 17p deletion (P < .001), age (P < .001), WBC (P < .001), and serum LDH (P < .001) were identified as significant prognostic factors at the 97% VH homology cut-off value. Deletion 11q entered the model as an additional independent prognostic factor on the use of the classical 98% VHhomology cut-off. Clinical staging provided a significant separation of prognostic subgroups with respect to their survival time distributions but did not contribute additional information regarding survival probability in the knowledge of VH mutation status and genomic aberrations. This may be illustrated by Figure 2. Patients with VH mutated CLL were frequently diagnosed at Binet stage A (91 of 112 at the less than 97% and 106 of 132 at the less than 98% VH homology cutoffs) and showed a good outcome. However, among patients diagnosed at Binet stage A, unmutated VH status had the power to identify a large subgroup (98 of 189 at the 97% or greater and 83 of 189 at the 98% or greater VH homology cutoffs) with an inferior outcome.
In this study 17p deletion, 11q deletion, and unmutatedVH status were the strongest adverse prognostic factors, indicating that the clinical heterogeneity of CLL1-4 might have a biologic basis. Genomic aberrations and VH mutation status appeared to have a complementary role in estimating prognosis. A risk model derived from multivariate regression analysis allowed the definition of 4 subgroups of CLL patients based on 17p deletion, 11q deletion, andVH mutation status. The subgroups appear to be characterized by markedly different survival probabilities and may allow a prediction of the clinical course of CLL patients at an early stage of disease. Therefore, the value of genetic risk parameters in relation to other factors, such as Rai and Binet staging, lymphocyte doubling time, bone marrow infiltration pattern, β2-microglobulin, thymidine kinase, and so on is being evaluated in prospective multicenter trials of the German CLL Study Group (GCLLSG). Based on these trials future treatment strategies using biologic risk stratification at the time of diagnosis for individual patients may be envisaged.
We thank Annett Habermann, Katrin Scherer, and Andreas Bühler for excellent experimental assistance and Dr Lutz Edler for statistical advice. We thank Drs Freda Stevenson, David Oscier, Terry Hamblin, Nick Chiorazzi, and Guillaume Dighiero for open discussions during the project.
Supported by grants from Wilhelm Sander-Stiftung (2001.004.1), Deutsche Krebshilfe (10-1289-St I), University of Ulm (P.679), and BMBF (01KW9934, 01KW9938, and NGFN/KB-6).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hartmut Döhner, Department of Internal Medicine III, University of Ulm, Robert-Koch-Strasse 8, 89081 Ulm, Germany; e-mail: hartmut.doehner@medizin.uni-ulm.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal