Established adverse prognostic factors in chronic lymphocytic leukemia (CLL) include CD38 expression, relative lack ofIgVH mutation, and defects of theTP53 gene. However, disruption of the p53 pathway can occur through mechanisms other than TP53 mutation, and we have recently developed a simple screening test that detects p53 dysfunction due to mutation of the genes encoding either p53 or ATM, a kinase that regulates p53. The present study was conducted to examine the predictive value of this test and to establish the relationship between p53 dysfunction, CD38 expression, and IgVHmutation. CLL cells from 71 patients were examined forIgVH mutation, CD38 expression, and p53 dysfunction (detected as an impaired p53/p21 response to ionizing radiation). Survival data obtained from 69 patients were analyzed according to each of these parameters. Relative lack ofIgVH mutation (less than 5%; n = 45), CD38 positivity (antigen expressed on more than 20% of malignant cells; n = 19), and p53 dysfunction (n = 19) were independently confirmed as adverse prognostic factors. Intriguingly, all p53-dysfunctional patients and all but one of the CD38+ patients had greater than 5% IgVH mutation. Moreover, patients with p53 dysfunction and/or CD38 positivity (n = 31) accounted for the short survival of the less mutated group. These findings indicate that the poor outcome associated with having less than 5%IgVH mutation may be due to the overrepresentation of high-risk patients with p53 dysfunction and/or CD38 positivity within this group, and that CD38− patients with functionally intact p53 may have a prolonged survival regardless of the extent of IgVH mutation.
Introduction
B-cell chronic lymphocytic leukemia (CLL) is the most common leukemia in the Western world and is characterized by the accumulation of clonal mature B lymphocytes in the blood, bone marrow, and secondary lymphoid tissues.1,2 One of the most intriguing features of the disease is its clinical heterogeneity. Thus, whereas some CLL patients survive for decades without requiring any treatment, others die of drug-resistant disease within a year or two of presentation.1 2
Sequencing of the B-cell receptor heavy chain gene (IgVH) of CLL lymphocytes has revealed extensive mutation in some cases.3 BecauseIgVH mutation underlies the process of affinity maturation and occurs in the germinal center of secondary lymphoid follicles, such hypermutated cases have been considered to be clonal expansions of postgerminal center (memory) B cells.3 In contrast, the malignant lymphocytes from other patients haveIgVH genes that are less mutated; these cases have been considered to represent clonal expansions of pregerminal center (naive) B cells.3
Considerable interest was generated by the recent demonstration by 2 independent groups that the extent of IgVHmutation can predict the clinical outcome in CLL.4 5 Thus, patients with relatively unmutated IgVH genes have a significantly shorter survival than patients with more extensiveIgVH mutation.
CD38 is a transmembrane glycoprotein that is widely expressed on a range of hemic and nonhemic cell types.6 In normal mature B cells, its expression is confined to the germinal center of secondary lymphoid follicles.7 There has been a considerable amount of interest (and controversy) in the notion that CD38 expression in CLL is associated with short survival and relative lack of IgVH mutation.4,5 7-11
p53 is a transcription factor that is activated by DNA breaks and which can induce apoptosis or cell-cycle arrest.12,13 By orchestrating the repair or elimination of cells with damaged DNA, p53 maintains the integrity of the genome and thereby prevents clonal progression.14 The protein also contributes to the cytotoxicity of many anticancer agents,15 including purine analogues commonly used in the treatment of CLL.16-18Unsurprisingly, TP53 mutations are associated with an adverse clinical outcome in a range of cancers, including CLL.19-21
Identification of p53 defects in the routine clinical setting has been improved recently by the development in this laboratory of a simple functional test that examines the p53 response of malignant cells to ionizing radiation.22 The test has demonstrated 2 types of p53 dysfunction in CLL, which together affect up to one quarter of cases overall. One defect is associated with the TP53mutation and the other with inactivation of the gene encoding ATM, a kinase that regulates p53. Intriguingly, low ATM protein levels—an imperfect surrogate for ATMmutations23—are, like TP53 mutations, associated with an adverse clinical outcome.24
The aims of the present study were to establish the predictive value of the p53 function test and to examine the relationship between p53 dysfunction, CD38 positivity, and relative lack ofIgVH mutation.
Patients, materials, and methods
CLL patients
All of the patients in the present study had typical CLL (mature lymphocytes expressing CD19, CD5, CD23, and weak, clonally restricted surface immunoglobulin), and most were selected on the basis of having a lymphocyte count of more than 50 × 109/L. The median age at diagnosis was 65 years, the male to female ratio was 1.65, and the distribution of clinical stage was as follows: 42% stage A, 17% stage B, and 41% stage C. Peripheral blood samples were obtained with informed consent at a median interval of 48 months after diagnosis, with no significant differences between the subgroups analyzed. Mononuclear cells were prepared by centrifugation over Lymphoprep (Gibco, Paisley, United Kingdom) and cryopreserved in liquid nitrogen.
IgVHmutation analysis
Total RNA was extracted from CLL cells using Trizol reagent (Life Technologies, Paisley, Scotland), and 1-μg aliquots were reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Promega, Southampton, United Kingdom) and an oligo(dT)15 primer. The resulting cDNAs were used to isolate clonally expressed VHDHJHsequences (covering the entire length of the gene to CDR3) in 2 sets of VH gene family–specific polymerase chain reactions (PCRs). In the first set of reactions, the sense primers were consensus sequences derived from framework region 1 (FWR1)3 of each of the 7 families. In the second set of 6 reactions, the sense primers were from the leader regions (families 1 and 7 having a common primer).25 In all reactions, the same combination of 3 constant region–specific (α, γ, μ) antisense primers was employed.3 The 50-μL reactions (20 pmol of each primer, 1.5 mM MgCl2, 100 μM of each dNTP, and 2.5 U Taq polymerase in supplied buffer from Promega) were cycled 30 times using a touch-down protocol,26 with the annealing temperature reduced from 63°C to 57°C over the first 12 cycles, and then analyzed by electrophoresis. PCR products for a given family, which were duplicated in both sets of reactions, were cloned. Between 2 and 4 clones derived from each PCR were sequenced commercially with a vector-specific primer. The sequences were accepted as representing the CLL cell clone if there was identity between and within the FWR1 and leader sequence–derived PCR clones. The VH gene alleles were identified and the degree of sequence divergence from the nearest germline counterpart was determined using V-BASE.27
Detection of CD38 expression
This was performed by fluorescence-activated cell sorting analysis of mononuclear cell preparations that had been double stained with phycoerythrin-conjugated anti-CD38 and fluorescein isothiocyanate–conjugated anti-CD19 (both from Becton Dickinson, San Jose, CA), as previously described.10
Detection of functional defects of the p53 pathway
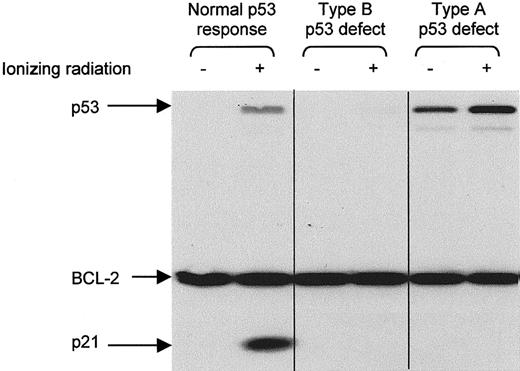
CLL lymphocytes were exposed to ionizing radiation (5 Gy) and cultured in RPMI supplemented with 10 mg/mL bovine serum albumin. After 10 to 18 hours, the cells were lysed in radioimmunoprecipitation assay (RIPA) buffer and the lysates were examined for p53, p21, and BCL-2 proteins by Western blotting, as previously described.22 Type A p53 dysfunction (due toTP53 mutation) is characterized by increased baseline levels of p53 and impaired radiation-induced p21 up-regulation. Type B p53 dysfunction (due to ATM mutation) is characterized by impaired up-regulation of both p53 and p21. BCL-2 was used as a protein control because it is highly expressed in CLL cells,28 is unaffected by radiation,22 and is extracted in RIPA buffer from live cells only (unpublished observations, 2001), thereby controlling for the possibility that p53/p21 up-regulation was impaired because the cells were dead.
Analysis of patient outcome
Clinical information was retrieved retrospectively from case notes. Patient survival was analyzed using Kaplan-Meier survival curves and the log-rank test. Disease-specific survival was obtained by censoring those deaths that could not be explained by CLL or its treatment. Differences between patient groups in the time interval from diagnosis to blood collection were analyzed using the Mann-Whitney U test. The χ2 test or Fisher exact test was used to compare features between different patient groups. Statistical significance was accepted atP < .05.
Results
Profile of p53 dysfunction, CD38 expression, andIgVHmutation
CLL cells from 71 patients were examined for p53 dysfunction, CD38 expression, and IgVH mutation. Nineteen patients (26%) were found to have p53 dysfunction, detected as an impaired p53/p21 response to ionizing radiation. Eight (11%) had the type A defect associated with TP53 mutation, whereas 11 (15%) had the type B defect associated with ATM mutation (Figure1). The proportion of B cells expressing CD38 varied from 0 to 82%, and the extent ofIgVH mutation varied from 0 to 12.4%.
Detection of p53 dysfunction by Western blotting.
CLL cells were analyzed for p53 and p21 expression 10 to 18 hours after exposure to ionizing radiation (5 Gy). BCL-2 was used as a control for protein loading. Type A p53 dysfunction (due to TP53mutation) is characterized by increased baseline levels of p53 and impaired radiation-induced up-regulation of p21. Type B p53 dysfunction (due to ATM mutation) is characterized by impaired up-regulation of both p53 and p21.
Detection of p53 dysfunction by Western blotting.
CLL cells were analyzed for p53 and p21 expression 10 to 18 hours after exposure to ionizing radiation (5 Gy). BCL-2 was used as a control for protein loading. Type A p53 dysfunction (due to TP53mutation) is characterized by increased baseline levels of p53 and impaired radiation-induced up-regulation of p21. Type B p53 dysfunction (due to ATM mutation) is characterized by impaired up-regulation of both p53 and p21.
To confirm that p53 dysfunction, IgVH mutation, and CD38 expression had prognostic value within the present study, we separated patients according to each of these 3 criteria and constructed Kaplan-Meier curves. Survival data were available for 69 of the 71 patients.
Adverse prognostic impact of p53 dysfunction
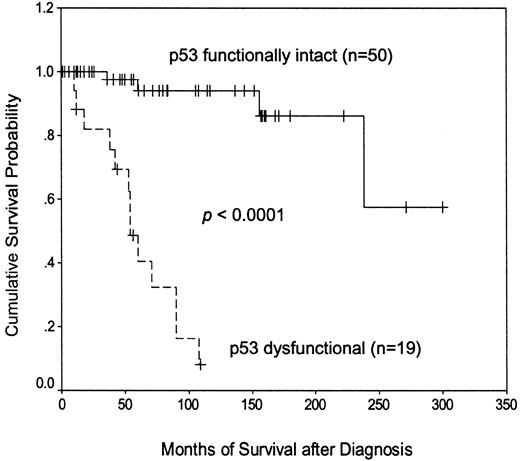
Separating patients according to their p53 functional status (Figure 2) revealed that those with demonstrable dysfunction of the p53 pathway had a much shorter disease-specific survival than those with functionally normal p53 (median, 54 months versus not reached). Subdividing the p53 dysfunctional cases (not shown) revealed that patients with the type A p53 defect, associated with TP53 mutation, had a significantly shorter survival than patients with the type B defect, associated with ATM mutation (median, 38 versus 90 months;P = .0003). These findings indicate that the p53 function test is a powerful predictor of outcome in CLL.
Kaplan-Meier plot of disease-specific survival according to p53 functional status.
Kaplan-Meier plot of disease-specific survival according to p53 functional status.
Adverse prognostic impact of relative lack ofIgVHmutation
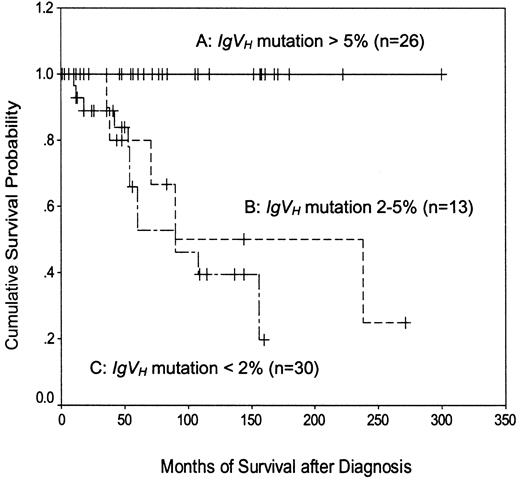
Normal CD5+ B cells have less than 5%IgVH mutation, whereas CLL cells may have less than or greater than 5%.3 However, most previous studies of IgVH mutation in CLL have employed a cutoff of 2% to define “mutated” and “unmutated” subgroups. In the present study, therefore, patients were divided into 3 groups: those with less than 2%, those with 2% to 5%, and those with greater than 5% IgVH mutation (Figure3). As expected,4 5 patients with less than 2% mutation had a shorter disease-specific survival than those with greater than 5% mutation (median, 90 months versus not reached; P < .0001). The survival of patients with 2% to 5% mutation (median, 90 months) was similar to that of patients with less than 2% mutation (P = .43) and significantly shorter than that of patients with greater than 5% mutation (P = .0015). These data confirm the adverse prognostic importance of having relatively unmutated IgVHgenes and indicate that in the present study, a cutoff value of 5% was better than 2% for defining CLL subgroups on the basis of this criterion.
Kaplan-Meier plot of disease-specific survival according to the extent of IgVH mutation.
A versus B, P = .0015; A versus C,P < .0001; B versus C, P = .43.
Kaplan-Meier plot of disease-specific survival according to the extent of IgVH mutation.
A versus B, P = .0015; A versus C,P < .0001; B versus C, P = .43.
Adverse prognostic impact of CD38 positivity
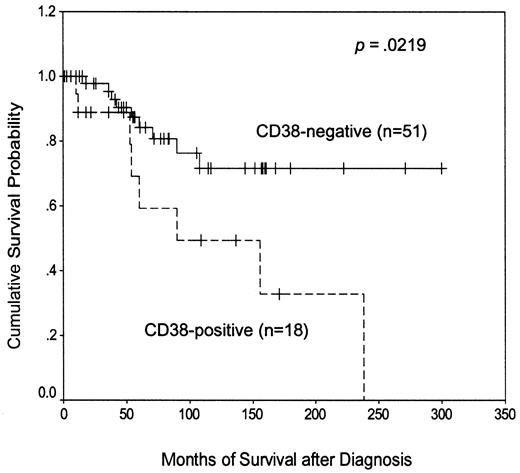
Most previous reports of CD38 expression in CLL have defined CD38+ cases as those in which the antigen is expressed on at least 30% of the malignant cells, although in one recent large report,11 a lower threshold value of 20% was used. However, these considerations were not relevant to the present study because no patient had between 20% and 30% CD38 positivity. Arbitrarily taking the lower cutoff value (Figure4), we found that CD38+patients had a significantly shorter disease-specific survival than the CD38− group (median, 90 months versus not reached). These data therefore extend our previous observations10 and confirm the adverse prognostic importance of CD38 positivity in the present study.
Kaplan-Meier plot of disease-specific survival according to CD38 expression.
Having confirmed that p53 dysfunction, relative lack ofIgVH mutation, and CD38 positivity each had an adverse impact on patient survival in the present study, we next examined the relationships between these prognostic factors (Figure5).
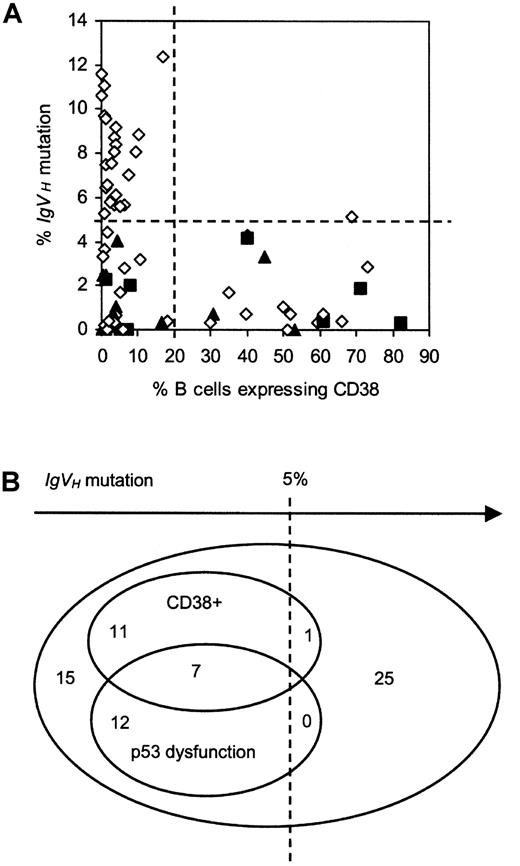
Relationship between p53 dysfunction, CD38 expression, and IgVH mutation.
(A) 2-D plot. Patients with functionally intact p53 are represented by open diamonds, those with type A p53 dysfunction by closed squares, and those with type B p53 dysfunction by closed triangles. Lines have been drawn to indicate the cutoff values used to define CLL subgroups on the basis of IgVH mutation and CD38 expression. (B) Venn diagram showing the relationship.
Relationship between p53 dysfunction, CD38 expression, and IgVH mutation.
(A) 2-D plot. Patients with functionally intact p53 are represented by open diamonds, those with type A p53 dysfunction by closed squares, and those with type B p53 dysfunction by closed triangles. Lines have been drawn to indicate the cutoff values used to define CLL subgroups on the basis of IgVH mutation and CD38 expression. (B) Venn diagram showing the relationship.
Association between CD38 expression and relative lack ofIgVHmutation
In keeping with previous findings from this and other laboratories,4 10 there was a skewed relationship between CD38 positivity and IgVH mutation (Figure 5). Thus, 18 of the 19 CD38+ patients (95%) had less than 5%IgVH mutation. Moreover, the single CD38+ patient exceeding this threshold value did so by only a narrow margin, having 5.12% mutation. In summary, CD38+cases constituted 40% (18 of 45) of the group with less than 5%IgVH mutation and only 4% (1 of 26) of the group with greater than 5% mutation.
Association between p53 dysfunction and relative lack ofIgVHmutation
Intriguingly, there was also a skewed relationship between p53 dysfunction and IgVH mutation (Figure 5). Thus, every p53-dysfunctional patient had less than 5%IgVH mutation. These cases therefore constituted 42% (19 of 45) of the group with less than 5% mutation and none of the group with greater than 5% mutation. There was no difference in the extent of IgVH mutation between cases with the type A versus the type B defect.
Lack of association between CD38 positivity and p53 dysfunction
There was no apparent association between p53 dysfunction and CD38 expression (Figure 5). Thus, 7 of the 19 p53-dysfunctional patients (37%) were CD38+, compared with 12 of the 52 patients with functionally normal p53 (23%). Conversely, among the 19 CD38+ patients, p53 dysfunction was present in 37% (7 of 19), as compared with 23% (12 of 52) of the CD38−cases.
Together, patients with p53 dysfunction and/or CD38 positivity (n = 31) accounted for 67% (30 of 45) of the group with less than 5% IgVH mutation but only 4% (1 of 26) of the group with greater than 5% IgVH mutation. In light of these observations, it was of interest to examine the extent to which patients with p53 dysfunction and/or CD38 positivity accounted for the short survival of the group with less mutatedIgVH genes.
Contribution of p53 dysfunction and CD38 positivity to the short survival of patients with less than 5%IgVHmutation
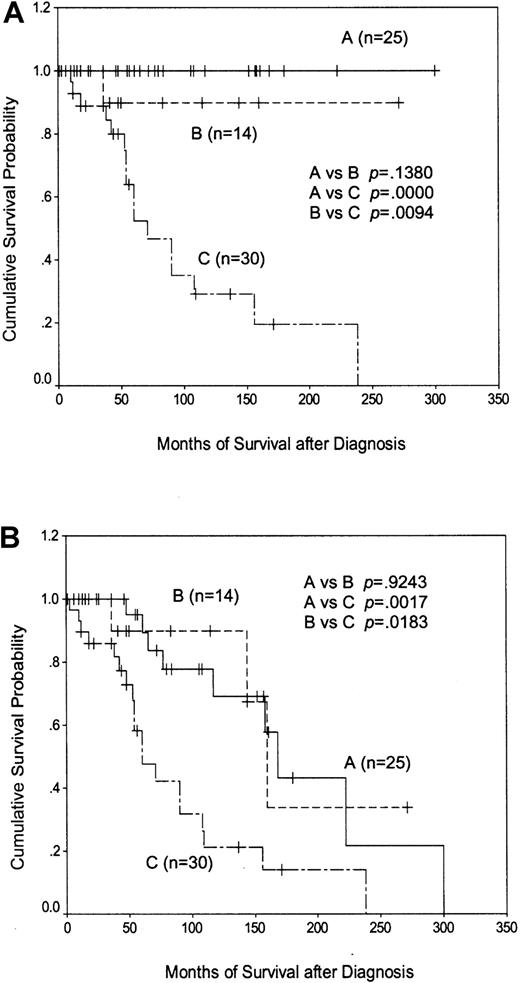
Patients with CD38 positivity and/or p53 dysfunction had a considerably worse outcome than patients with neither of these features (median survival, 71 months versus not reached;P < .0001). Importantly, separating the CD38−, p53-intact patients according toIgVH status showed that those with less than 5%IgVH mutation survived as well as those with greater than 5% mutation, with only one disease-related death occurring within the former group (Figure6A). A similar relationship between the survival curves was obtained when disease-unrelated deaths were included in the analysis (Figure 6B). These data suggest that the poor outcome associated with having less than 5%IgVH mutation is due to the overrepresentation of patients with p53 dysfunction and/or CD38 positivity within this group, and that CD38− patients with functionally intact p53 have a prolonged disease-specific survival that does not depend on the extent of IgVH mutation.
Kaplan-Meier plots of patient survival according to CD38/p53 status and the extent of IgVHmutation.
(A) shows disease-specific survival, whereas (B) shows overall survival. Line A represents CD38−/p53-intact patients with greater than 5% IgVH mutation; line B represents CD38−/p53-intact patients with less than 5%IgVH mutation; and line C represents patients with p53 dysfunction and/or CD38 positivity.
Kaplan-Meier plots of patient survival according to CD38/p53 status and the extent of IgVHmutation.
(A) shows disease-specific survival, whereas (B) shows overall survival. Line A represents CD38−/p53-intact patients with greater than 5% IgVH mutation; line B represents CD38−/p53-intact patients with less than 5%IgVH mutation; and line C represents patients with p53 dysfunction and/or CD38 positivity.
Discussion
The aims of the present study of CLL were to validate our screening test for p53 dysfunction and to examine the relationship between p53 dysfunction, IgVH mutation, and CD38 expression. An abnormal p53 function test was found to be strongly predictive of short survival, with the type A defect (due toTP53 mutation) being associated with a particularly poor outcome. When cases of CLL were separated according to theirIgVH status, patients with 2% to 5%IgVH mutation were found to have a survival similar to that of patients with less than 2% mutation and considerably shorter than that of patients with greater than 5% mutation. Regarding CD38, patients expressing the antigen on more than 20% of their malignant cells had a survival disadvantage as compared with patients with a lower level of expression.
Having confirmed within our patient group the predictive value of p53 dysfunction, IgVH mutation, and CD38 expression, we next examined the relationship between these prognostic factors. As expected, nearly all of the CD38+ patients had relatively unmutated IgVH genes. Intriguingly, the same was found to be true of patients with p53 dysfunction. Although some patients were simultaneously CD38+ and p53 dysfunctional, these 2 prognostic factors were not associated with one another. Because high-risk patients with p53 dysfunction and/or CD38 positivity represented about two thirds of the group with less than 5%IgVH mutation, it seemed possible that they might account for its short survival, and this was indeed found to be so.
That CLL patients with p53 dysfunction have a short survival is entirely in keeping with the established importance of the p53 pathway in maintaining genomic integrity14 and mediating the action of certain cytotoxic agents, including purine analogues.15-18 It is also consistent with the adverse prognostic significance in CLL of deletions of 17p13 and 11q22-23, which involve the TP53 and ATM loci, respectively.29 Indeed, impairment of the p53 pathway may represent a single functional end point through which these (and possibly other) seemingly unrelated prognostic factors exert their deleterious effects.
In contrast, it is unclear why CD38 positivity is associated with a short patient survival. It is possible that expression of CD38 confers to CLL cells a more malignant cellular phenotype. This seems plausible given that the antigen has an important role as a modulator of intracellular signaling30,31 and that cross-linking of CD38 up-regulates BCL-2 and inhibits apoptosis in normal mature B cells.32 33
Our observation that CLL patients with greater than 5%IgVH mutation were nearly all CD38−is consistent with the absence of CD38 on normal B cells that have undergone the germinal-center reaction.7 However, it is less obvious why some tumor samples with less than 5%IgVH mutation were CD38+ and others CD38−. Some of the latter cases—especially those with anIgVH mutation rate of zero—may be clonal expansions of naive mature B cells, which do not express CD38.7 The CD38+ patients with less than 5%IgVH mutation, particularly those with zero mutation, are more difficult to explain in terms of maturation arrest because CD38 expression is a feature of germinal-center B cells, tumors of which have extensively mutated IgVHgenes.7 However, all of the above speculations need to take into account the fact that CD38 is expressed on only a proportion of CLL cells in a given patient and that the antigen may be induced by cytokines such as interferon-α.34
Regarding the association between relatively unmutatedIgVH genes and p53 dysfunction, there are, we suggest, a number of possible explanations. First, dysfunction of the p53 pathway might impair the capacity of the CLL precursor cell to undergo somatic hypermutation. Second, CLL lymphocytes with relatively unmutated IgVH genes might be selectively prone to the acquisition of genetic changes that result in p53 dysfunction. Third, p53 dysfunction might selectively facilitate the clonal expansion of CLL precursor cells with relatively unmutatedIgVH genes. Fourth, the phenotype of tumor cells with extensively mutated IgVH genes might be altered in the presence of p53 dysfunction, such that the disease is not recognizable as CLL. At present, it is unclear which, if any, of these possibilities is correct.
Our survival data led us to employ a cutoff of 5% for defining CLL subgroups on the basis of IgVH mutation. This choice of threshold value was further supported by the observed relationships between IgVH mutation, CD38 expression, and p53 dysfunction. In contrast to our empirical approach, most previous studies have aimed to separate patients on the basis of whether the IgVH gene is in true germline configuration. This theoretical approach is attractive because it has implications for the identity of the normal counterpart cell and whether antigen encounter has occurred. To separate “mutated” from “unmutated” cases, a cutoff of 2% (rather than zero)IgVH mutation has historically been used to avoid mistaking unknown polymorphisms for true somatic mutations acquired during the affinity maturation response to antigen.35 This approach is clearly a compromise because many cases of CLL with up to 2% divergence from the accepted germline sequence are likely to have proper IgVH mutations. There are also problems in using IgVH mutation to define whether antigen encounter has occurred. For example, T-independent antigen stimulation can produce memory B cells with unmutated IgVHgenes.36 Furthermore, only thoseIgVH mutations resulting in amino acid changes within the antigen binding site are functionally important in terms of the affinity maturation response37; mutations of the intervening framework regions are of unclear significance and cannot be taken to indicate that affinity maturation—and therefore antigen encounter—has taken place.
These considerations clearly raise important questions concerning the biologic meaning of IgVH mutation in CLL and howIgVH subgroups should be defined. Indeed, it could be argued that CLL might be better regarded as a single entity with a continuously variable phenotype. This model predicts that, prior to clonal expansion, the precursor cell undergoes a varying amount ofIgVH mutation, the extent of which is governed by the presence and nature of the stimulus (T-dependent versus T-independent antigen and/or intrinsic oncogenic signaling mimicking antigen) and the integrity of the cellular machinery required for hypermutation to occur. We speculate that these determining factors, and not the extent of somatic mutation per se, define the clinical behavior of the disease.
It is important to note that the clinical samples upon which the present observations are based were not all obtained at disease presentation. Because it is unknown to what extent p53 dysfunction and CD38 expression may be acquired with time, caution should be exercised when applying our data to newly diagnosed patients. However, the diagnosis of CLL as a clinical event is of dubious relevance to the natural history of the disease. For example, it is universally accepted that in many patients, the disease is likely to have been present for many years prior to its initial detection and that diagnosis is frequently made by accident. Nevertheless, in the present study, the interval between diagnosis and blood collection was not significantly different between any of the subgroups analyzed. Furthermore, the observed frequencies of CD38 positivity (27%) and p53 dysfunction of each type (11% and 15% for the type A and B defects, respectively) were no greater than expected from any previous reports. Therefore, although we cannot formally exclude the possibility that p53 dysfunction or CD38 expression may have been acquired after diagnosis in the occasional patient, the above considerations suggest that such an occurrence is rare and unlikely to have had a significant impact on the findings.
In summary, the present study provides a plausible explanation for the poor clinical outcome of CLL patients with relatively unmutatedIgVH genes and demonstrates the marked heterogeneity of such patients. From a clinical point of view, it will be important to conduct prospective studies to establish the value of p53 dysfunction and CD38 positivity in predicting the survival of newly diagnosed patients and the response to different therapies. It will also be important to gauge the extent to which these 2 features may be acquired over time, particularly in patients with less than 5%IgVH mutation. Nevertheless, the existing data have unambiguous implications for patients with established p53 dysfunction and/or CD38 positivity. Indeed, the very poor outlook of patients with p53 dysfunction—particularly with the type A defect—would make them ideal candidates for clinical trials of intensive therapy (eg, hemopoietic stem cell transplantation). It would also be rational to target p53-dysfunctional patients for nongenotoxic therapies likely to act independently of p53 (eg, corticosteroids or immunotherapy with antibodies to CD20 or CD52). From a scientific point of view, it will be important to explain the skewed relationship between IgVH mutation, CD38 expression, and p53 dysfunction because this is likely to provide considerable insight into the pathogenesis of this common yet enigmatic disease.
Prepublished online as Blood First Edition Paper, April 17, 2002; DOI 10.1182/blood-2001-11-0066.
Supported by grants from The Leukaemia Research Fund (United Kingdom) and the Royal Liverpool & Broadgreen University Hospitals Trust R&D Fund.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Andrew R. Pettitt, Department of Haematology, Royal Liverpool University Hospital, Liverpool, L7 8XP United Kingdom; e-mail: andrew.pettitt@rlbuh-tr.nwest.nhs.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal