Abstract
Mature B-cell differentiation provides an important mechanism for the acquisition of adaptive immunity. Malignancies derived from mature B cells constitute the majority of leukemias and lymphomas. These malignancies often maintain the characteristics of the normal B cells that they are derived from, a feature that is frequently used in their diagnosis. The role of microRNAs in mature B cells is largely unknown. Through concomitant microRNA and mRNA profiling, we demonstrate a potential regulatory role for microRNAs at every stage of the mature B-cell differentiation process. In addition, we have experimentally identified a direct role for the microRNA regulation of key transcription factors in B-cell differentiation: LMO2 and PRDM1 (Blimp1). We also profiled the microRNA of B-cell tumors derived from diffuse large B-cell lymphoma, Burkitt lymphoma, and chronic lymphocytic leukemia. We found that, in contrast to many other malignancies, common B-cell malignancies do not down-regulate microRNA expression. Although these tumors could be distinguished from each other with use of microRNA expression, each tumor type maintained the expression of the lineage-specific microRNAs. Expression of these lineage-specific microRNAs could correctly predict the lineage of B-cell malignancies in more than 95% of the cases. Thus, our data demonstrate that microRNAs may be important in maintaining the mature B-cell phenotype in normal and malignant B cells.
Introduction
Naive B cells migrate through the circulation to lymphoid organs, where they undergo the T cell–dependent germinal center (GC) reaction. Adaptive immunity is acquired as specific antigen-reactive GC B cells differentiate into the major effector B cells of the adaptive immune system: memory cells and plasma cells (Figure 1A). Although the role of specific transcription factors in mature B-cell differentiation has been examined,1-5 mechanisms regulating such transcription factors during mature B-cell differentiation are largely unknown.
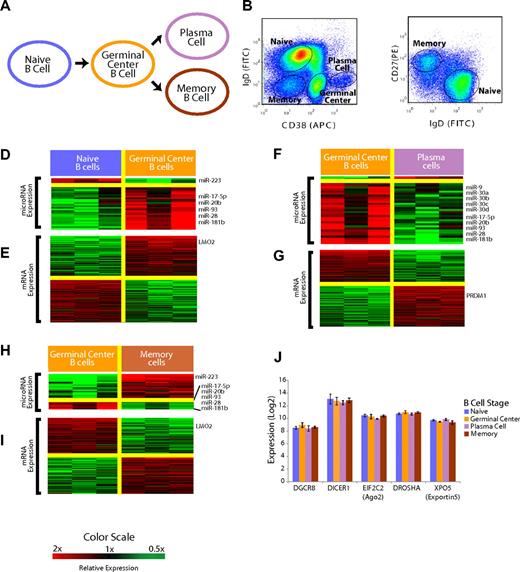
Mature B-cell subsets demonstrate distinct miRNA profiles. (A) Overall schema of mature B-cell differentiation. (B) Selection of the B-cell subsets with the use of flow cytometry. Cells were previously gated on CD19+ cells. Naive and memory B cells were distinguished from GC and plasma cells based on surface CD38 and IgD expression. (C) Distinction of naive and memory B cells based on IgD and CD27 expression with the use of flow cytometry. (D) Relative expression of miRNA in the naive to GC B-cell transition. miRNAs that were, on average, at least 2-fold differentially expressed at a false discovery rate of less than 5% are shown according to the color scale. (E) Relative expression of mRNA in the naive to GC B-cell transition. mRNAs that were, on average, at least 2-fold differentially expressed at a false discovery rate of less than 1% are shown according to the color scale. (F) Relative expression of miRNA in the GC B-cell to plasma-cell transition. miRNAs that were, on average, at least 2-fold differentially expressed at a false discovery rate of less than 5% are shown according to the color scale. (G) Relative expression of mRNA in the GC B-cell to plasma-cell transition. mRNAs that were, on average, at least 2-fold differentially expressed at a false discovery rate of less than 1% are shown according to the color scale. (H) Relative expression of miRNA in the GC B cell to memory B–cell transition. miRNAs that were, on average, at least 2-fold differentially expressed at a false discovery rate of less than 5% are shown according to the color scale. (I) Relative expression of mRNA in the GC B-cell to memory B–cell transition. mRNAs that were, on average, at least 2-fold differentially expressed at a false discovery rate of less than 1% are shown according to the color scale. (J) Expression of key miRNA processing genes DGCR8, DICER1, EIF2C2, DROSHA, and XPO5 is unchanged among the B-cell subsets (P > .1 in all cases).
Mature B-cell subsets demonstrate distinct miRNA profiles. (A) Overall schema of mature B-cell differentiation. (B) Selection of the B-cell subsets with the use of flow cytometry. Cells were previously gated on CD19+ cells. Naive and memory B cells were distinguished from GC and plasma cells based on surface CD38 and IgD expression. (C) Distinction of naive and memory B cells based on IgD and CD27 expression with the use of flow cytometry. (D) Relative expression of miRNA in the naive to GC B-cell transition. miRNAs that were, on average, at least 2-fold differentially expressed at a false discovery rate of less than 5% are shown according to the color scale. (E) Relative expression of mRNA in the naive to GC B-cell transition. mRNAs that were, on average, at least 2-fold differentially expressed at a false discovery rate of less than 1% are shown according to the color scale. (F) Relative expression of miRNA in the GC B-cell to plasma-cell transition. miRNAs that were, on average, at least 2-fold differentially expressed at a false discovery rate of less than 5% are shown according to the color scale. (G) Relative expression of mRNA in the GC B-cell to plasma-cell transition. mRNAs that were, on average, at least 2-fold differentially expressed at a false discovery rate of less than 1% are shown according to the color scale. (H) Relative expression of miRNA in the GC B cell to memory B–cell transition. miRNAs that were, on average, at least 2-fold differentially expressed at a false discovery rate of less than 5% are shown according to the color scale. (I) Relative expression of mRNA in the GC B-cell to memory B–cell transition. mRNAs that were, on average, at least 2-fold differentially expressed at a false discovery rate of less than 1% are shown according to the color scale. (J) Expression of key miRNA processing genes DGCR8, DICER1, EIF2C2, DROSHA, and XPO5 is unchanged among the B-cell subsets (P > .1 in all cases).
Malignancies derived from mature B cells are common and constitute the majority of leukemias and lymphomas and reflect defined stages of normal B-cell differentiation. MicroRNAs (miRNA) are 18- to 22-nucleotide-long RNA molecules that regulate the expression of genes. There is an increasing recognition of the role of miRNAs in oncogenesis,6,7 lineage-selection,8,9 and immune cell function,10-12 including early B-cell differentiation.13 However, the full extent and function of miRNA expression during mature B-cell differentiation and in B-cell malignancies are not known. Researchers are beginning to address the role of miRNAs in hematologic malignancies.14-17 A complete delineation of miRNA and their target expression in normal B cells is essential to understanding the role of miRNAs in B-cell malignancies.
Through concomitant miRNA and mRNA profiling, we have identified a potential regulatory role for miRNAs at each stage in mature B-cell differentiation. We have experimentally identified a direct role for the miRNA-mediated regulation of oncogenes and key transcription factors in B-cell differentiation: LMO2 and PRDM1 (Blimp1). Our work establishes the landscape of normal miRNA expression in mature B cells and suggests a role in regulating normal B-cell differentiation. Furthermore, our work demonstrates that, in contrast to their described down-regulation in other malignancies, stage-specific miRNAs are retained in B-cell malignancies. The lineage of common B-cell malignancies can be predicted on the basis of miRNA profiles of normal B cells, pointing to a role for miRNAs in the maintenance of mature B-cell phenotypes in normal and malignant B cells.
Methods
Patient sample processing
B-cell populations were obtained from young patients undergoing routine tonsillectomy with the use of a protocol approved by the Clinical Center at the National Institutes of Health (Bethesda, MD) in accordance with the precepts established by the Declaration of Helsinki. The tonsils of these patients were disaggregated and separated by Ficoll. The mononuclear cell layer was harvested, washed in phosphate-buffered saline (PBS), and resuspended in ACK lysing buffer to remove small numbers of red blood cells. After a wash and resuspension with 10 mL PBS with 10% bovine serum albumin, cells were counted and 200 million were stained with fluorochrome-tagged monoclonal antibodies to CD19, IgD, CD38, and CD27. The specific monoclonal antibodies used were anti–CD19-PE-Cy5.5, anti–IgD-FITC, anti–CD27-PE, and anti–CD38-APC, all from BD Biosciences and BD Pharmingen (San Jose CA). Cells were sorted with the MoFlo Cell sorter (Dako Cytomation, Colorado Springs, CO) into naive B cells (CD19+IgD+CD27−CD38+), GCC B cells (CD19+IgD−CD38++), memory B cells (CD19+IgD−CD27+CD38dim), and plasma cells(CD19dimIgD−CD27++CD38+++). Three replicates of each B-cell subset were obtained from separate patients. The sample purity was verified by FACS and found to be greater than 90% in all cases.
Tumor specimens were obtained from patients who were studied with use of a protocol approved by the Duke University Medical Center Institutional Review Board. The pathologic diagnosis of the samples was verified before analysis. Samples from patients with diffuse large B-cell lymphoma were further subclassified as described previously.18 Chronic lymphocytic leukemia samples were processed and purified as described previously.19 Total RNA was extracted with the phenol-chloroform method to preserve miRNAs by the use of Ambion (Applied Biosystems/Ambion, Austin, TX) reagents.
MiRNA profiling using multiplexed real-time polymerase chain reaction (PCR)
MiRNA expression profiling was conducted by use of the Applied Biosystems 384-well multiplexed real-time PCR assay with 400 ng of total RNA. Eight reactions, each containing 50 ng of RNA and a multiplex looped primer pool with endogenous small nucleolar (sno)-RNA controls, were used to reverse-transcribe the miRNAs in parallel fashion. Each completed reaction was loaded onto the 384-well plate per the manufacturer's instructions, and real-time PCR was run on the ABI 7900HT Prism (Applied Biosystems). For each 384-well plate, we used the automatically determined cycle threshold (CT) using the SDS 2.2.1 software (Applied Biosystems). Consistent with manufacturer recommendations, we considered CT greater than 36 as undetected. A miRNA was considered to be present in a subpopulation if the CT was less than 36 in all 3 biologic replicates. The probes deemed to be present were normalized to the average expression of a sno-RNA control. The expression values were calculated as 2−ΔCT, then median-centered to 500 and log2-transformed.
Gene expression (mRNA) profiling with the use of microarrays
Gene expression profiling and normalization were performed with the use of methods identical to those we have described previously.20 Array elements with median signal intensities of less than 7 log2 units across the samples were removed from analysis to exclude poorly measured genes and genes not appreciably expressed in the samples. Genes that were on average 2-fold or greater differentially expressed in a binary comparison of B-cell subsets, and appreciably expressed in at least 1 of the 2 B-cell subsets being compared, were selected for further analysis as described to follow. The data are accessible through Gene Expression Omnibus Series accession number GSE12366 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE12366).
MiRNA profiling with the use of microarray
MiRNA expression profiling from human B-cell lymphomas was conducted with the use of up to 1 μg total RNA from sample and reference (normal lymph node), which were labeled with Cy3 or Cy5 fluorescent dyes, using the miRNA/LNA labeling kit (Exiqon, Vedbæk, Denmark). The fluorescently labeled samples were combined and hybridized to a miRNA microarray (version 10.0; Exiqon) in a nitrogen atmosphere. The microarray slides were scanned with GenePix 4100 scanner (Axon Instruments, Union City, CA). The quantified signals were normalized using the global Lowess algorithm, using Genespring (Agilent Technologies, Santa Clara, CA) software. The intensity values for multiple spots were averaged and the normalized values were log2-transformed. Missing values were replaced with the lowest value for analysis.
miRNA target prediction
Annotated genes on the U133plus 2.0 array were matched to the miRNA target list downloaded from TargetScan (www.targetscan.org). For the purpose of this study, a target gene was defined by the presence of a seed sequence match (nucleotides 2-8) and conservation of the seed sequence and 3′UTR in humans, dog, rat, mouse, and chicken. Additional conservation was examined in miRNA target genes selected for experimental validation. The distribution of the mRNA expression for these genes was plotted as a density plot by use of the Splus statistical software (Insightful Corporation/TIBCO, Palo Alto, CA). The difference in distribution between the B-cell subsets was calculated with a 2-sample, 1-sided Kolmogorov-Smirnov test to examine the hypothesis that being a miRNA target conferred repression in the appropriate population (consistent with the known biology of miRNA effects).
The 3′UTRs of LMO2, MYBL1, and PRDM1 were aligned with Blastz alignment of Human, Chimp, Mouse, Rat, Dog, Chicken, Frog (Xenopus), and Zebrafish and were displayed with the UCSC genome browser. The conservation of miR-223 seed sequence and the 3′UTRs of LMO2 and MYBL1, as well as that of the miR-30 family and miR-9 on PRDM1, were thus verified.
Western blot
RIPA Lysis buffer (1× PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 10 mmol/L phenylmethylsulfonyl fluoride, 1 μg/mL aprotinin, and 100 mmol/L sodium orthovanadate) was added to 750 000 cells and incubated on ice for 30 minutes. The mixture was spun down, and the supernatant was transferred to a new tube as the whole cell extract. A total of 20 μg cell lysate was separated on a 4% to 18% Tris-Bis NuPAGE gel (Invitrogen, Carlsbad, CA) and transferred using the iBlot transfer device (Invitrogen) program 3 for 7 minutes (LMO2 detection) or program 2 for 6 minutes (PRDM1). The blots were probed with a 1:200 mouse–anti-LMO2 (Santa Cruz Biotechnologies; SC-65736), 1:750 mouse-anti–Blimp-1 (Santa Cruz Biotechnologies; SC-66 015), or 1:5000 goat-anti–B-actin (Santa Cruz Biotechnologies; SC-47 778) for 1 hour at room temperature. The antibodies were detected by the use off 1:10 000 goat-anti–mouse horseradish peroxidase–conjugated antibodies (Santa Cruz Biotechnologies). Western Blotting Luminol Reagent (Santa Cruz Biotechnologies) was used to visualize the bands corresponding to each antibody.
Single miRNA/mRNA expression with the use of real-time PCR
With 10 ng RNA per reaction, miRNAs of interest were reverse-transcribed with ABI individual stem-loop primers designed to only detect mature miRNA and measured by Taqman real-time PCR normalized to the small nucleolar RNA, RNU48. To assess mRNA expression using RT-PCR, 1 μg RNA was reverse-transcribed with the ABI High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Gene expression was measured with exon-spanning Taqman probes and normalized to beta-2 microglobulin expression.
Cell culture
BJAB and H929 were cultured in RPMI (Gibco/Invitrogen) supplemented with 10% fetal bovine serum (FBS), and U266 was cultured in RPMI supplemented with 15% FBS. 293T cells were grown in Dulbecco modified Eagle medium (DMEM; Gibco/Invitrogen) with 10% FBS. All cell lines were grown in 37°C humidified cell culture incubators with CO2 maintained at 5%.
miRNA transfection
miRNAs of interest were overexpressed in cell lines of interest by transfecting the appropriate miRNA precursors (Ambion) at 100 nmol by use of Amaxa's nucleofector system. In particular, BJAB was transfected with Nucleofector solution T, program T-016, U266 with Nucleofector C, program X-005, and H929 with Nucleofector V, program T-001. A total of 1.5 million cells were used per transfection and mixed with appropriate miRNA precursors (Ambion) for a concentration of 100 nmol/L.
Statistical analysis
Identifying differentially expressed miRNA and mRNA.
miRNAs were considered to be differentially expressed if the mean measured expression changed at least 2-fold and a false discovery rate (q) was less than 5% when significance analysis of microarrays (SAM)21 with 1000 permutations was used.
Differentially expressed genes (mRNA) in naive versus GC, GC versus plasma cells, and GC versus memory cells comparisons were identified by the use of SAM.21 Genes that were 2-fold differentially expressed at a false discovery rate (q) less than 1% with 1000 permutations were identified as significantly differentially expressed.
Transcription factors and miRNA target genes.
Transcription factors were identified based on the gene ontology (gene ontology [GO] search term “transcription factor”; http://www.geneontology.org/) and matched to the probes of the Affymetrix U133plus 2.0 microarray. Of the total of 938 transcription factor genes thus identified, we selected 364 genes that were differentially expressed in at least one of the B-cell stage transitions. We evaluated the breakdown of the differentially expressed transcription factors among miRNA targets versus nontargets. The P values were computed with the χ2 test separately in each B-cell stage transition.
B-cell malignancy sample classification.
The top 50 most differentially expressed miRNAs (P < .01) in each pairwise lymphoma type comparison were chosen as the initial predictor. Singular value decomposition was applied to reduce the list to 20 most informative miRNAs in each pairwise comparison.22 A Bayesian logistic regression was performed in Matlab (The Mathworks, Natick, MA) with the 20-predictor miRNAs for each pairwise comparison. Each sample was tested with the use of the miRNA-based predictor in a leave-one-out fashion to determine the accuracy of each prediction. For a sample to be classified as a particular lymphoma (or normal) type, it had to be predicted as such in every pairwise comparison.
Normal B-cell stage classification of B-cell malignancies.
We constructed a Bayesian predictor to distinguish normal naive from GC B cells based on the 32 miRNAs depicted in Figure 1D. We then applied the predictor without optimization to the microarray data generated for GC-like DLBCL (GCB DLCBL), Burkitt lymphoma, and chronic lymphocytic leukemia to render a Bayesian prediction of lineage, that is, naive versus GC B cell.
Western blot quantitative analysis.
Western blot scans were quantified with National Institutes of Health ImageJ software (Bethesda, MD). For each experiment, the ratios of protein of interest (LMO2, PRDM1) to actin were determined and mean centered to 100 across the experiment. The average and standard deviation of these values across the 3 experiments were calculated and displayed relative to the scrambled control expression.
Luciferase indicator assay
Firefly luciferase reporter constructs were created in the pL/SV40/GL3 vector for the LMO2 3′UTR and the LMO2 3′UTR with the predicted miR-223 binding site mutated. Mature miRNA expression of a pL/CMV/eGFP vector coding for pri-miR-223 from the 3′UTR of EGFP of the vector was confirmed by the use of Taqman-real time PCR in transfected 293T cells. Activity of gl3 was normalized in dual luciferase assays to pL/SV40/RLuc, with which it was cotransfected. The PRDM1 3′UTR also was cloned into the pL/SV40/GL3 vector. miRNA expression vectors and their respective seed sequence mutants were created for miR-9-2, miR-30b, and miR-30d. Additional details regarding the assays including the primer sequences, concentrations, and vector descriptions are described fully in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Ratios of firefly luciferase (GL3) to internal control Renilla luciferase (RLuc) from 293T cells transfected with pL/CMV/eGFP/miR-223 were divided by those obtained from 293T cells transfected with the pL/CMV/eGFP vector control. The average and standard deviation were taken across 5 experiments for the pL/SV40/gl3 empty, LMO2, and LMO2 mutant vectors.
Firefly luciferase (GL3) activity readings of the PRDM1 3′UTR construct were divided by internal control Renilla luciferase (RLuc) activity readings. The average and standard deviation of these ratios across 3 experiments were calculated and scaled relative to the empty vector (pL/CMV/eGFP) transfection.
IgVH mutation status of chronic lymphocytic leukemia samples
IgVH mutation status was determined as described19 by the use of genomic DNA. In brief, genomic DNA was isolated from purified CLL cells and isolated with the GenElute Mammalian DNA extraction kit from Sigma-Aldrich (St Louis, MO) according to the manufacturers' instructions. DNA was amplified with the use of nested PCR primers. PCR products were electrophoresed, purified, and sequenced with an automated DNA sequencer (Applied Biosystems) with the BigDye Terminator kit (PerkinElmer, Waltham, MA). Forward and reverse sequences were aligned into a single resolved sequence with Sequencher 4.1 software (Gene Codes Corporation, Ann Arbor, MI) and then aligned with germ line sequences derived from DNA Plot on the V BASE directory website (http://vbase.mrc-cpe.cam.ac.uk/). The percentage sequence identity was calculated by dividing the number of mutations from FR1 to FR3 by the total number of nucleotides in this region. Samples were considered somatically mutated if they had more than 2% mutations in this region.
Results
Mature B-cell stages display characteristic patterns of MiRNA expression
Mature B-cell subsets can be defined by the expression of surface CD19, IgD, CD38, and CD2723 and were obtained by fluorescence-activated cell sorting of tonsils from young patients who were undergoing routine tonsillectomies (Figure 1B,C). To determine whether mature B-cell subsets had unique patterns of miRNA expression, we used a 384-well multiplexed RT-PCR assay (Applied Biosystems)24,25 that allowed measurement of all 365 miRNAs in miRBase 9.2. We detected a total of 113 unique miRNAs in the B-cell populations (Table S1). This detection frequency compares favorably to the identification of 71 unique miRNAs (45 miRNAs with more than one clone) through the examination of 3101 sequences cloned from unselected CD19+ mature B cells.26 We identified differentially expressed miRNAs in mature B-cell subsets by the use of a false discovery rate of less than 5%21 (Figure 1D,F,H). The complete list of assayed miRNAs found to be expressed in the B-cell populations is included in Tables S1 and S2.
The B-cell subsets were profiled for gene expression at the whole genome level, as described previously.20 At each stage, we identified differentially expressed genes as those genes with a mean 2-fold difference in expression and a false discovery rate of less than 1% (Figure 1E,G,I). Genes that we found to be differentially expressed in each stage-transition were consistent with those in previous studies that examined gene expression in B-cell subsets by the use of microarrays with fewer probes,27,28 an overlap that was found to be highly statistically significant (P < .001, χ2 test).
In the naïve → GC B-cell transition, we identified 32 miRNAs that were differentially expressed. Interestingly, all but 4 miRNAs were found to be expressed more highly in GC cells than in naive B cells (Figure 1D). The mRNA expression patterns confirmed the expression of several genes that are known to be differentially expressed in the transition including BCL6, MME, MYBL1, as well as LMO2 (Figure 1E). LMO2 was found to be expressed more highly in GC B cells compared with both naive B cells and memory B cells (Figure 1E,I). In the GC → plasma-cell transition, we found 33 miRNAs that were differentially expressed. Once again, we noted a striking asymmetry, with all but 2 miRNAs found to be expressed highly in GC cells but down-regulated in plasma cells (Figure 1F). We also confirmed that the plasma cell-specific genes, PRDM1 (Figure 1G), XBP1, and IRF4, were highly differentially expressed in our data. In the GC → memory B–cell transition, there was a preponderance of the 27 significant miRNAs expressed at greater levels in memory cells (Figure 1H). A total of 5 miRNAs were expressed highly in GC cells compared with all the other B-cell types, including 3 members of the miR-17∼92 cluster (miR-17-5p, miR-20b, miR-93), as well as miR-28 and miR-181b.
The expression pattern of all the miRNAs that were measurable in at least one of the B-cell subsets is summarized in Figure S1. Notably, there were no differences in the expression of genes involved in miRNA processing,29 including DICER1, DROSHA, XPO5 (exportin5), EIF2C2 (ago2), and DGCR8, among the B-cell subsets (Figure 1J).
Separately, we examined the expression of predicted target genes of differentially expressed miRNAs. We also found that predicted miRNA target genes of miRNAs expressed highly in GC cells were expressed at lower levels in GC cells compared with other stages (Figure S2). We also found that a greater number of transcription factors were predicted miRNA targets (Figure S3).
MiR-223 regulates LMO2 in the naive → GC and GC → memory cell transitions
We found miR-223 to be expressed at nearly 8-fold greater levels in both naive and memory cells compared with GC cells. This miRNA has a highly conserved sequence complementarity to the 3′UTR of 2 transcription factors that are expressed highly during GC cell differentiation: LMO2 (Figure 2A)30 and MYBL1 (Figure S4A).31
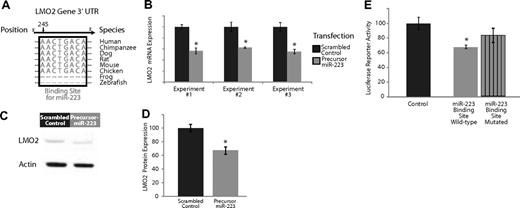
Experimental validation of the interaction of miR-223, which is expressed highly in naive and memory B cells compared with GC B cells, and targets the transcription factor LMO2. (A) Base-pairing of the 3′UTR LMO2 gene with nucleotides 1-8 of miR-223. This 8-mer is highly conserved across several species and serves as a potential binding site for miR-223. (B) Effects of overexpression of miR-223 in GC lymphoma-derived BJAB cells in 3 separate experiments. The dark gray bars depict expression of LMO2 24 hours after transfection with a scrambled control that does not possess complementarity to the human genome. The light gray bars depict the expression of LMO2 24 hours after transfection with a precursor for miR-223. The expression of LMO2 was consistently lower in the cells treated with the miR-223 precursor (P < .05 in all cases). (C) Relative LMO2 protein expression from a representative experiment (from 3 replicates) transfecting a scrambled control versus a precursor for miR-223 in BJAB cells. (D) Average expression of LMO2 relative to actin over 3 Western blots of BJAB transfected with a scrambled control versus a precursor for miR-223. LMO2 expression is lower in cells treated with miR-223 (P < .05). (E) Luciferase-expressing vectors were coupled to the 3′UTR of the LMO2 gene. The seed sequence mutant construct had consistently diminished miR-223 repression compared with the wild-type construct in 5 separate experiments (P < .05).
Experimental validation of the interaction of miR-223, which is expressed highly in naive and memory B cells compared with GC B cells, and targets the transcription factor LMO2. (A) Base-pairing of the 3′UTR LMO2 gene with nucleotides 1-8 of miR-223. This 8-mer is highly conserved across several species and serves as a potential binding site for miR-223. (B) Effects of overexpression of miR-223 in GC lymphoma-derived BJAB cells in 3 separate experiments. The dark gray bars depict expression of LMO2 24 hours after transfection with a scrambled control that does not possess complementarity to the human genome. The light gray bars depict the expression of LMO2 24 hours after transfection with a precursor for miR-223. The expression of LMO2 was consistently lower in the cells treated with the miR-223 precursor (P < .05 in all cases). (C) Relative LMO2 protein expression from a representative experiment (from 3 replicates) transfecting a scrambled control versus a precursor for miR-223 in BJAB cells. (D) Average expression of LMO2 relative to actin over 3 Western blots of BJAB transfected with a scrambled control versus a precursor for miR-223. LMO2 expression is lower in cells treated with miR-223 (P < .05). (E) Luciferase-expressing vectors were coupled to the 3′UTR of the LMO2 gene. The seed sequence mutant construct had consistently diminished miR-223 repression compared with the wild-type construct in 5 separate experiments (P < .05).
We evaluated the effects of miR-223 expression on its predicted target genes, LMO2, by transfecting precursors of miR-223 into a cell line derived from GC cell lymphoma cells (BJAB). Overexpression of miR-223 resulted in a consistent down-regulation of LMO2 at the transcript level (Figure 2B) compared with a transfection with a scrambled control with no sequence complementarity to the human genome, results that were statistically significant (P < .05 in each case, Student t test). There was no effect on the expression of a nontarget control, β-2 microglobulin, in these experiments (data not shown). Overexpression of miR-223 also resulted in a consistent down-regulation of LMO2 at the protein level (Figure 2C) compared with a transfection with a scrambled control with no sequence complementarity to the human genome. We quantified the results of 3 separate experiments examining LMO2 protein expression (Figure 2D) and found consistent down-regulation of LMO2 compared with cells transfected with scrambled controls, results that were statistically significant (P < .05, Student t test). The extent of down-regulation of LMO2 mRNA and protein by miR-223 was comparable, suggesting that miR-223 regulation of LMO2 occurs predominantly at the mRNA level. Similarly, overexpression of miR-223 resulted in a down-regulation of MYBL1 transcripts (Figure S4B).
As additional validation, we investigated whether the miR-223 had a direct effect on LMO2 by cloning the 3′UTR sequence of LMO2 3′ to the firefly luciferase ORF (Fluc).32 The resulting constructs and the unmodified vector were cotransfected into 293T cells along with a Renilla luciferase internal control and pL-CMV-eGFP constructs expressing either no miRNA or miR-223. Fluc expression from constructs bearing LMO2 3′UTR sequences were differentially down-regulated by miR-223 compared with those with mutated seed sequences (Figure 2E), results that were statistically significant (P < .05, Student t test). These observations provide evidence for an inhibitory role for miR-223 in regulating the transcription factors LMO2.
MiR-9 and the miR-30 family regulate PRDM1 (blimp-1) in the GC → plasma cell transition
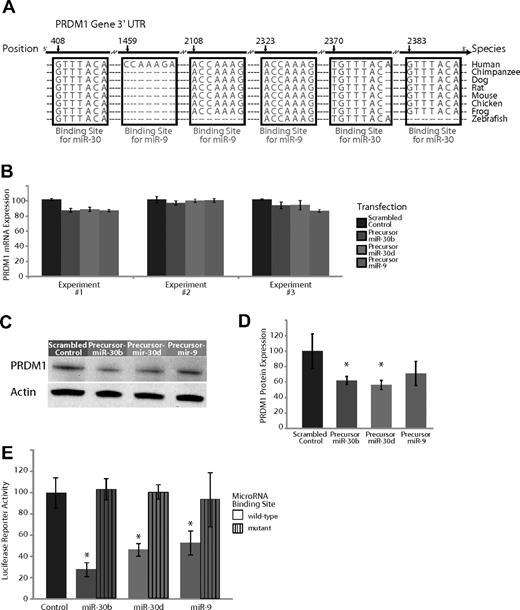
In the GC → plasma cell transition, we observed that several members of the miR-30 family were expressed at 2-fold or greater levels in GC cells (Figure 1F). The transcription factor PRDM1 is an essential regulator of plasma cell differentiation.33 The miR-30 family comprises 5 members (miR-30a, 30b, 30c, 30d, and 30e), of which 4 (all except 30e) were found to be expressed at greater levels in GC cells compared with plasma cells. Control transfection experiments documented excellent specificity of the RT-PCR probes for individual members of the miR-30 family with no discernible cross-hybridization (Figure S5).
The 3′UTR of PRDM1 contains 3 highly conserved binding sites complementary to the seed sequence of members of the miR-30 family, as well as 3 binding sites for the seed sequence of miR-9, 2 of which are highly conserved across multiple species (Figure 3A). To evaluate the effects of the miR-30 family and miR-9 on PRDM1 expression in plasma cells, we introduced precursors of miR-9, miR-30b, and miR-30d into the U266 multiple myeloma (plasma cell) cell line. Overexpression of miR-30 family members miR-30b and miR-30d, as well as miR-9, had no effect on PRDM1 at the mRNA level (Figure 3B). By contrast, there was a consistent down-regulation of PRDM1 at the protein level (Figure 3C,D), results that were statistically significant in each case (P < .05, Student t test), except for the transfections with the precursor to miR-9 (P = .08, Student t test). These exclusively posttranscriptional effects of miR-9 and miR-30 on PRDM1 expression are consistent with one mechanism of miRNA regulation and has been described previously7,32,34,35 in other systems. There was no effect on the expression of a nontarget control (actin). In addition, luciferase reporter activity of the PRDM1 3′UTR construct was decreased by overexpression of miR-9, miR-30b, and miR-30d, but not their respective seed sequence mutants (Figure 3E). The down-regulation of the luciferase reporter signal and its restoration in the mutant constructs was found to be statistically significant in each of the 3 miRNAs: miR-9, miR-30b, and miR-30d (P < .05, Student t test).
Experimental validation of the interaction of miR-9 and miR-30, which are expressed highly in GC B cells compared with plasma cells and target the transcription factor PRDM1. (A) Base-pairing of the 3′UTR of PRDM1 gene with the 5′ seed region of miR-9 and the miR-30 family. The miR-30 regions include 3 sites complementary to nucleotides 2-8 (UTR position 408), nucleotides 1-8 (UTR position 2370), and nucleotides 2-8 (UTR position 2383) on the miRNA, respectively. The miR-9 regions include 3 sites complementary to nucleotides 1-7 (UTR position 1459), nucleotides 2-8 (UTR position 2108), and nucleotides 2-8 (UTR position 2323) on the miRNA, respectively. These sites are highly conserved across several species, with the exception of one miR-9 site (UTR position 1459) that is present only in humans. (B) Effects of overexpression of miR-9 and 2 members of the miR-30 family, miR-30b and miR-30d, in plasma cell myeloma-derived U266 cells in 3 separate experiments. Expression of PRDM1 was measured 24 hours after transfection with a scrambled control with no complementarity to the human genome, a hairpin precursor for miR-30b, a hairpin precursor for miR-30d, or a hairpin precursor for miR-9. (C) Relative PRDM1 protein expression from a representative experiment (from 3 replicates) transfecting a scrambled control versus a precursor for miR-9, miR-30b, and miR-30d in U266 cells. (D) Average expression of PRDM1 relative to actin over 3 Western blots of U266 cells transfected with a scrambled control versus a precursor for miR-9, miR-30b, and miR-30d. P < .05 for miR-30b and miR-30d; P = .08 for miR-9. (E) A luciferase-expressing vector was coupled to the 3′UTR of PRDM1. Repression of luciferase activity was observed upon overexpression of miR-30b, miR-30d, and miR-9 (P < .05 in all 3 cases) but rescued to the activity level of the empty vector control (P > .5 in all 3 cases) when the seed sequence of the miRNAs was mutated. Displayed is the average of 3 separate experiments.
Experimental validation of the interaction of miR-9 and miR-30, which are expressed highly in GC B cells compared with plasma cells and target the transcription factor PRDM1. (A) Base-pairing of the 3′UTR of PRDM1 gene with the 5′ seed region of miR-9 and the miR-30 family. The miR-30 regions include 3 sites complementary to nucleotides 2-8 (UTR position 408), nucleotides 1-8 (UTR position 2370), and nucleotides 2-8 (UTR position 2383) on the miRNA, respectively. The miR-9 regions include 3 sites complementary to nucleotides 1-7 (UTR position 1459), nucleotides 2-8 (UTR position 2108), and nucleotides 2-8 (UTR position 2323) on the miRNA, respectively. These sites are highly conserved across several species, with the exception of one miR-9 site (UTR position 1459) that is present only in humans. (B) Effects of overexpression of miR-9 and 2 members of the miR-30 family, miR-30b and miR-30d, in plasma cell myeloma-derived U266 cells in 3 separate experiments. Expression of PRDM1 was measured 24 hours after transfection with a scrambled control with no complementarity to the human genome, a hairpin precursor for miR-30b, a hairpin precursor for miR-30d, or a hairpin precursor for miR-9. (C) Relative PRDM1 protein expression from a representative experiment (from 3 replicates) transfecting a scrambled control versus a precursor for miR-9, miR-30b, and miR-30d in U266 cells. (D) Average expression of PRDM1 relative to actin over 3 Western blots of U266 cells transfected with a scrambled control versus a precursor for miR-9, miR-30b, and miR-30d. P < .05 for miR-30b and miR-30d; P = .08 for miR-9. (E) A luciferase-expressing vector was coupled to the 3′UTR of PRDM1. Repression of luciferase activity was observed upon overexpression of miR-30b, miR-30d, and miR-9 (P < .05 in all 3 cases) but rescued to the activity level of the empty vector control (P > .5 in all 3 cases) when the seed sequence of the miRNAs was mutated. Displayed is the average of 3 separate experiments.
miRNAs and B-cell malignancies
To examine the expression of B-cell stage-specific miRNAs in B-cell malignancies, we undertook miRNA profiling of 75 tissue samples derived from normal lymph nodes (n = 5) as well as patients with B-cell malignancies including the molecular subsets of diffuse large B-cell lymphoma (DLBCL),36 GC B cell–like (GCB) DLBCL (n = 20), and activated B cell–like (ABC) DLBCL (n = 20), as well as cases of IgV mutated and unmutated chronic lymphocytic leukemia (n = 20) and Burkitt lymphoma (n = 10).
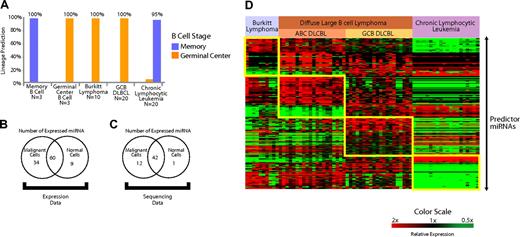
We constructed a Bayesian predictor from miRNAs that distinguished normal memory B cells from GC cells (Figure 1H). We tested the predictor in the B-cell malignancies derived from GC B cells (Burkitt lymphoma and GCB DLBCL) along with chronic lymphocytic leukemia, which is thought to arise from memory B cells.37 Using leave-one-out cross-validation, we found that the predictor constructed from miRNAs expressed in normal B cells was able to correctly identify the B cell–specific stage of the lymphoma type in more than 95% of the cases (Figure 4A).
Expression of miRNAs expressed in normal B cells is conserved in B-cell malignancies. (A) A predictor constructed of miRNAs differentially expressed in the normal naive B cells and GC B cells (miRNAs depicted in Figure 1D) was used to predict the normal counterpart B cell of both IgV mutated and unmutated chronic lymphocytic leukemia, GC B cell–derived DLBCL, and Burkitt lymphoma. The accuracy was greater than 95% in all cases. (B) Expression of miRNAs expressed in B cells that also were present and detectably measured on the microarrays (103/113) was examined in the B-cell malignancies (n = 70) and normal lymph nodes (n = 5) with use of the Student t test. miRNAs that were differentially expressed (P < .05) at greater levels in malignant cells, normal cells, as well as the miRNAs that were not differentially expressed are shown. (C) Cloning frequency of miRNAs was compared between unselected mature B cells (n = 3) compared with several B-cell malignancies (n = 42) from a previously published study26 (“sequencing data”) with use of the Student t test. miRNAs that were differentially expressed (P < .05) at greater levels in malignant and normal cells, as well as the miRNAs that were not differentially expressed, are shown. (D) Differentially expressed miRNAs that distinguish Burkitt lymphoma, activated B cell–like (ABC) diffuse large B-cell lymphoma (DLBCL), GC-like DLBCL (GCB DLBCL), and chronic lymphocytic leukemia. Predictor miRNAs from each pairwise comparison that distinguish each entity are shown in the boxes.
Expression of miRNAs expressed in normal B cells is conserved in B-cell malignancies. (A) A predictor constructed of miRNAs differentially expressed in the normal naive B cells and GC B cells (miRNAs depicted in Figure 1D) was used to predict the normal counterpart B cell of both IgV mutated and unmutated chronic lymphocytic leukemia, GC B cell–derived DLBCL, and Burkitt lymphoma. The accuracy was greater than 95% in all cases. (B) Expression of miRNAs expressed in B cells that also were present and detectably measured on the microarrays (103/113) was examined in the B-cell malignancies (n = 70) and normal lymph nodes (n = 5) with use of the Student t test. miRNAs that were differentially expressed (P < .05) at greater levels in malignant cells, normal cells, as well as the miRNAs that were not differentially expressed are shown. (C) Cloning frequency of miRNAs was compared between unselected mature B cells (n = 3) compared with several B-cell malignancies (n = 42) from a previously published study26 (“sequencing data”) with use of the Student t test. miRNAs that were differentially expressed (P < .05) at greater levels in malignant and normal cells, as well as the miRNAs that were not differentially expressed, are shown. (D) Differentially expressed miRNAs that distinguish Burkitt lymphoma, activated B cell–like (ABC) diffuse large B-cell lymphoma (DLBCL), GC-like DLBCL (GCB DLBCL), and chronic lymphocytic leukemia. Predictor miRNAs from each pairwise comparison that distinguish each entity are shown in the boxes.
An interesting aspect regarding the role of miRNAs in malignancies is their reported down-regulation in several malignancies compared with normal cells from the same lineage.38 To further examine this effect in B-cell malignancies, we examined their expression of 113 miRNAs that we had identified in normal B cells (Table S1). A total of 103 of 113 miRNAs were detected with the microarrays that we used to profile B-cell malignancies and normal lymph nodes. We applied a 2-sided Student t test to evaluate the relative expression of those 103 miRNAs in B-cell malignancies (n = 60) and normal lymph nodes (n = 5). A total of 34 miRNAs were differentially expressed (P < .05) at greater levels in malignant cells, and 9 miRNAs were expressed more highly in normal cells. Sixty miRNAs were not differentially expressed (Figure 4B).
As additional validation, we examined miRNA cloning frequencies26 for sequences cloned from normal and malignant B cells. MiRNAs for which a sequence was identified in at least 2 of the 3 available normal B-cell samples were used in the analysis. We applied a 2-sided Student t test (P < .05) to compare the differential cloning frequency of the miRNAs between normal B cells (n = 3) and a variety of mature B-cell lymphoma patient samples and cell lines (n = 42). In all, we found 56 miRNAs that were consistently expressed in normal B cells. We found 13 of those 56 miRNAs were differentially expressed (P < .05) between normal and malignant B cells, of which 12 miRNAs were expressed more highly in malignant cells and 1 miRNA was expressed more highly in normal cells (Figure 4C). To avoid effects from tumor-infiltrating nonmalignant cells, we repeated the analysis with 20 chronic lymphocytic leukemia samples in the malignant group. The results were virtually identical with as those obtained with the larger set of malignancies.
These results demonstrate that miRNAs are not down-regulated in B-cell malignancies compared with normal B cells and that normal B-cell stage–specific miRNAs are maintained in B-cell malignancies.
MiRNA profiling also revealed that each lymphoma type had a distinctive pattern of miRNA expression (Figure 4D). To evaluate the ability of miRNA profiles to distinguish different lymphoma types, we constructed Bayesian predictors from the most highly differentially expressed miRNAs (Tables S5Table S6. Predictor microRNAs that distinguish activated B-cell (ABC) DLBCL from other lymphoma types in each pair-wise comparison (PDF, 24.6 KB)–S7) for each pair-wise comparison. We tested the performance of the predictor using leave-one-out cross-validation and found it to be more than 90% accurate in the identification of each entity (Figure S6). The validity of the classifier will need to be assessed separately in an independent group of samples.
Discussion
Mature B-cell differentiation is important for the development of adaptive immunity. The process is also of interest because B-cell malignancies are common and they retain several features derived from their normal counterpart B-cell subsets. Unlike other maturation pathways in the hematopoietic and other lineages, successive stages of mature B cells do not simply signify progressive differentiation away from the stem cell stage. Rather, each stage represents a specialized state with specific functions.39 Thus, GC cells interact with CD4 T cells and dendritic cells and undergo somatic hypermutation and Ig-heavy chain class-switching. However, plasma cells secrete immunoglobulin, whereas memory cells are primed to proliferate and differentiate into plasma cells upon repeat contact with antigen. The specialized functions demand a finely tuned program of gene regulation. Our study provides initial evidence that miRNAs might play a role throughout the life cycle of mature B cells through the modulation of expression of key regulatory genes. By regulating the expression of multiple transcription factors,34,35,40 miRNAs could play a role in the tandem regulation of the large number of genes that are differentially expressed in B-cell stage transitions.
MiRNAs have been shown to exert their effects at the posttranscriptional level by destabilizing mRNA and at the posttranslational level by interfering with protein expression. Our data suggest that both modes may be active in the interaction of miRNAs and their targets in B cells. The complementarity of the miRNA seed sequence and the 3′UTR of genes has been shown experimentally to be an important determinant of miRNA target specificity.9 However, seed complementarity and other known factors are not sufficient for determining what miRNA–target interactions are biologically relevant. Such biologically relevant interactions are likely to represent a small minority among the computationally predicted targets and are likely to be context-dependent. These observations highlight the need for experimentally testing the interactions of miRNAs and their target genes in the appropriate biologic context.
MiR-223 has previously been described as being important in the commitment to myeloid lineage.8,41 We demonstrate that miR-223 also might play an important role in the B-lymphoid lineage by potentially targeting 2 separate transcription factors that are expressed highly in GC cells, LMO2 and MYBL1. Greater expression of miR-223 in the naive B-cell stage could inhibit the untimely expression of these transcription factors until the cell is ready to undergo the GC reaction.
We found that miR-9 and 4 separate members of the miR-30 family were overexpressed in the GC to plasma-cell transition. In our experiments, we tested the effects of overexpression of2 separate members of the miR-30 family and miR-9 on PRDM1 expression. We found that overexpression of each of these miRNAs had an average knockdown effect of approximately 40%. This effect is comparable with that achieved by RNA interference with shRNAs. The combined effect of 5 different miRNA species (miR-30a, miR-30b, miR-30c, miR-30d, and miR-9) is likely to be more potent than that of a single miRNA. The role of mutual repression of BCL6 and PRDM1 in the GC to plasma cell differentiation as been described previously.33 Our data suggest that miRNAs may bolster the effects of BCL6 in the inhibition of PRDM1. It would be interesting to evaluate whether BCL6 is involved in the regulation of these miRNAs.
Our data are consistent with a proposed role for miRNAs as a tunable rheostat that regulates gene expression.42 Activation of LMO2 has been associated with the development of leukemia in patients undergoing gene therapy.43 However, greater expression of PRDM1 alone is sufficient to induce plasma cell-differentiation.3 Inappropriate expression of such genes must be effectively turned off for a cell to maintain its state. This mode of regulation is reflected in the effects of miR-223, miR-9, and miR-30, which turn off the inappropriate expression of LMO2 and PRDM1 and might promote state maintenance and inhibition of lymphomagenesis. However, our data also identify several instances in which miRNAs are coexpressed with their predicted targets. It is possible that such interactions within the cell help to stabilize a defined expression level by dampening fluctuations. For example, in GC cells, we found that miR-181b was strongly coexpressed with its predicted target, BCL6. Such interactions could also be important in B-cell stage maintenance and curbing the oncogenic potential of genes involved in B-cell differentiation.44,45
Our data show that members of the miR-17∼92 family are consistently expressed in GC cells and may play a role in mature B-cell differentiation. Interestingly, the miR-17∼92 family has been implicated in early B-cell differentiation and mice lacking the loci that encode these miRNAs have arrested early B-cell development.13 The expression patterns of the miR-17∼92 family suggest that the regulatory motifs embedded in the interaction of this miRNA family and its targets might have an additional function in regulating mature B-cell differentiation.
A striking observation in this study is the high degree of asymmetry observed in relative expression of miRNAs in GC cells compared with naive and plasma cells. At least 2 hypotheses could account for these findings. First, miRNA expression may promote a highly regulated state that enables GC cells to interact with T cells and antigen presenting cells and to leave the GC cells poised for differentiation into memory or plasma cells. Second, miRNAs expressed highly in naive and plasma cells may be underrepresented in current miRNA libraries. Such libraries are often constructed from lymph nodes, which are typically enriched in GC cells. High-throughput sequencing of sorted populations of B cells could reveal novel miRNAs that are highly expressed in those populations. Interestingly, a larger number of miRNAs were highly expressed in memory cells compared with GC cells. This observation might stem from the fact that memory cells are known to be heterogeneous46 and standard methods used to select memory cells may capture a diverse group of memory subpopulations.
The consistent expression of several miRNAs in a diverse set of B-cell malignances also suggests a role for miRNAs in the maintenance of tumor phenotype. Assays for stage-specific B-cell markers such as BCL6, a marker for GC cells, are essential in the clinical diagnosis of B-cell malignancies. Our data suggest that stage-specific biology in B-cell malignancies is retained at the miRNA level. Recent work has demonstrated the utility of gene expression profiling in reliably distinguishing closely related lymphomas.47,48 However, clinical translation of gene expression profiling has proved to be difficult because of the need for freshly frozen tissue. Because intact miRNAs can be isolated from tissues preserved using standard methods,49,50 diagnostic methods based upon miRNA profiles could be fairly easy to translate to clinical use.
Interestingly, in contrast to a previous study,38 we did not note a decrease in the expression of the total number or overall expression levels of miRNAs in B-cell malignancies compared with normal lymph nodes. Although B-cell malignancies maintain the expression of several stage-specific miRNAs, their miRNA expression patterns are clearly deranged compared with normal lymph nodes. The consequences of altered miRNA expression in B-cell tumors would be important to explore in future studies.
In conclusion, our study demonstrates that mature B-cell subsets have distinct patterns of miRNA expression, suggesting a role for miRNAs in B-cell differentiation. We provide experimental evidence that transcription factors such as LMO2 and PRDM1 are direct targets of differentially expressed miRNAs. B-cell malignancies demonstrate a distinct pattern of miRNA expression that could be useful in distinguishing morphologically identical subtypes of these tumors. The conserved expression of stage-specific miRNAs in normal and malignant B cells suggests a role for miRNAs in the maintenance of the mature B-cell phenotype.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
S.S.D. was supported by the Doris Duke Charitable Foundation (New York, NY), National Institutes of Health grant K12, and a Duke Cancer Center Development Grant. We thank Dr Joseph Nevins for a critical review of the manuscript, and Laszlo Jakoi and Chaitanya Acharya for technical assistance. We also thank Wayne Terrell and Dr Raj Dash for assistance with pathology specimens, and Tri-Tin Le (Applied Biosystems), Chris Miller (Applied Biosystems), and John Alexandrou (Exiqon) for advice and technical support.
National Institutes of Health
Authorship
Contribution: J.Z. performed research, conducted data analysis, and wrote the paper; D.D.J. conducted data analysis and wrote the paper; C.J. performed research, conducted data analysis, and wrote the paper; R.F., E.G., G.H., P.L.L., A.S.L., D.A.R., D.R.F., and J.B.W. performed research; P.E.L. designed the study and wrote the paper; and S.S.D. designed the study, performed research, conducted data analysis, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sandeep S. Dave, MD, 101 Science Dr, Duke University, Box 3382, Durham, NC 27710; e-mail: sandeep.dave@duke.edu.
References
Author notes
*J.Z. and D.D.J. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal