Abstract
Overexpression of BAALC is an adverse prognostic factor in adults with cytogenetically normal acute myeloid leukemia and T-cell acute lymphoblastic leukemia (ALL). Here, we analyzed the prognostic significance of BAALC in B-precursor ALL. BAALC MRNA expression was determined in 368 primary adult B-precursor ALL patients enrolled on the 06/99 and 07/03 GMALL trials. Patients were grouped into tertiles according to BAALC expression (T1-T3). Higher BAALC expression (T3 vs T2 vs T1) was associated with higher age (P < .001), a higher white blood cell count (P = .008), CD34 (P = .001), BCR-ABL (P < .001), and MLL-AF4 (P < .001). Higher BAALC expression predicted primary therapy resistance in the overall cohort (P = .002) and in the BCR-ABL− and MLL-AF4− subgroup (P = .01). In BCR-ABL− and MLL-AF4− patients, higher BAALC expression was associated with a shorter overall survival (OS; 5-year OS: T3, 38%; T2, 52%; T1, 70%; P = .004) and independently predicted OS in multivariate models (P = .03). Gene-expression profiling revealed an up-regulation of stem cell markers and genes involved in chemoresistance (TSPAN7 and LYN) in the high BAALC group. Thus, high BAALC expression is associated with an immature, chemoresistant leukemic phenotype and identifies patients with inferior OS. Determination of BAALC might contribute to risk assessment of molecularly undefined adult B-precursor ALL.
Introduction
Outcome of adult B-precursor acute lymphoblastic leukemia (ALL) has considerably improved because of identification of clinical and genetic risk factors stratifying patients to different treatment groups. With the current risk-adapted treatment protocols, complete remission (CR) rates range between 74% and 93%; however, long-term survival remains limited to only 27% to 54% of adult patients.1 Commonly accepted risk factors in B-precursor ALL include white blood cell (WBC) count, the immunophenotype of pro-B ALL, response to induction therapy, level of minimal residual disease, age, and cytogenetic as well as molecular genetic aberrations.1,2 Thus far, high-risk cytogenetics in B-precursor ALL mainly compose the Philadelphia chromosome with the balanced translocation t(9;22)(q34;q11) and BCR-ABL fusion protein in approximately 20% to 36%,1,3 and the translocation t(4;11)(q21;q23) with the MLL-AF4 fusion protein in approximately 6% to 9%1,4 of cases. Patients lacking clinical and molecular risk factors are considered standard risk (SR); however, the definition of risk groups varies between different study protocols. Outcome for SR patients is still unsatisfactory with a long-term survival of only 40% to 60%,1,4,5 indicating the clinical and biologic heterogeneity of these patients. Therefore, the identification of novel predictive molecular markers in B-precursor ALL might be particularly useful for patients yet unclassified by (molecular) risk factors and may improve treatment stratification of this subgroup.
The Brain And Acute Leukemia, Cytoplasmic (BAALC) gene is a marker of early hematopoietic progenitor cells and is aberrantly expressed in a subset of acute leukemias.6,7 The BAALC gene is located on chromosome 8q22.3. The protein sequence shows no homology to any other known proteins or functional domains. Its role in hematopoiesis and leukemogenesis is unknown. BAALC has been associated with an immature leukemic subtype in adult cytogenetically normal (CN) acute myeloid leukemia (AML) and acute T-lymphoblastic leukemia (T-ALL).6,8,9 The prognostic significance of BAALC was first shown for CN-AML, with a poor CR rate, overall survival (OS), and disease-free survival for patients with high BAALC expression.8,10 In adult T-ALL, high BAALC expression was associated with inferior overall and relapse-free survival (RFS).9 Because of the prognostic relevance of BAALC in different leukemic subgroups, we hypothesized that BAALC might also show an aberrant expression pattern and prognostic significance in adult B-lineage ALL and may help to risk-stratify molecularly unclassified patients with B-precursor ALL.
To assess this, we analyzed BAALC mRNA expression in 368 adult patients with B-precursor ALL enrolled on multicenter treatment protocols of the German Multicenter ALL (GMALL) study group in the context of established risk factors. Moreover, gene-expression profiling (GEP) was performed in 51 adult B-precursor ALL specimens to gain insights into biologic pathways of BAALC and mechanisms of chemoresistance.
Methods
Patients
We analyzed 368 patients with newly diagnosed adult B-precursor ALL enrolled between 1999 and 2006 on the 06/99 (n = 199) and 07/03 (n = 169) GMALL multicenter trials.11 Specimens were selected from consecutive patients who had sufficient material available. Patient characteristics and outcome were comparable with those from the total study population.12,13 The 06/99 and 07/03 study protocols included intensive chemotherapy, radiotherapy, and autologous or allogeneic stem cell transplantation (SCT) according to risk-adapted classification, using the following risk factors: WBC more than 30 × 109/L, late CR (> 3 weeks), pro-B ALL, t(9;22)/BCR-ABL, and t(4;11)/MLL-AF4. Risk groups were assigned as follows: SR (no risk factor), high risk (≥ 1 risk factor), and very high risk (presence of t(9;22)/BCR-ABL). High-risk and very-high-risk patients with a sibling or matched unrelated donor were scheduled for allogeneic SCT in first CR (CR1). The short form of the study protocol in the European Leukemia Trial Registry contains details.14
The studies were approved by the ethics board of the Johann Wolfgang Goethe-Universität Frankfurt/Main, Germany, and informed consent was given according to the Declaration of Helsinki.
Immunophenotyping
Immunophenotypic analyses were performed by flow cytometry on fresh pretreatment bone marrow (BM) and peripheral blood (PB) samples at the GMALL central reference laboratory at the Charité Berlin, Germany, as previously described.15 A cell-surface antigen was considered positive when at least 20% of cells showed fluorescence intensity greater than the negative control.
Molecular analyses
RNA isolation and synthesis of complementary DNA
Total RNA isolation from mononuclear cells was carried out from diagnostic BM samples using RNeasy Mini Kit (QIAGEN) following the manufacturer's instruction. Synthesis of complementary DNA was performed using 500 ng of total RNA, avian myeloblastosis virus-reverse transcriptase, RNase inhibitor, and oligo dT20-primer at 50°C for 60 minutes (Roche Diagnostics).
Real-time reverse-transcribed polymerase chain reaction assays
BAALC expression was analyzed by multiplex quantitative real-time reverse transcribed polymerase chain reaction (PCR) on the Rotorgene RG-3000 real time cycler (Corbett Research) with coamplification of the housekeeping gene Glucose-Phosphate Isomerase (GPI) as previously reported.18 Relative BAALC expression was calculated with the mean of the cycle number difference of the 2 replicates (ΔCT = GPI-BAALC), expressed as 2μ(ΔCT). For each sample, polymerase chain reaction assays were performed in duplicate. Positive and negative controls were included in each assay. RNA from the cell line KG1a was included in each run for calibration between runs. If amplification of GPI did not reach the threshold within 30 cycles, samples were excluded from this study.
Oligonucleotide microarray studies
Microarray-based GEP was performed on an additional set of 51 BM samples from adult patients with newly diagnosed t(9;22)/BCR-ABL− and t(4;11)/MLL-AF4− B-precursor ALL, which had been included in the Microarray Innovations in LEukemia multicenter study (MILE).19 Oligonucleotide microarray hybridization (HG-U133 Plus 2.0; Affymetrix) was performed as described previously.20
GeneSpring software, Version 4.2 (Silicon Genetics), was used for data analyses. All microarrays fulfilled the MIAME standards.21 BAALC expression was represented by 2 probe sets (218899_s_at, 222780_s_at), and samples were grouped into tertiles (t1-t3) according to the median expression level of the 2 probe sets. To obtain gene-expression signatures that sufficiently discriminated between high and low BAALC expressers, samples in the upper BAALC expression tertile were compared with samples in the lower tertile (t3 vs t1) and named high and low BAALC, respectively. Genes were considered to be differentially expressed if their expression showed an at least 3-fold change and probe sets were called present by the Affymetrix data analysis in at least 75% of samples. Statistical significance of changes was calculated by the nonparametric t test (P ≤ .05). The microarray data can be accessed at the Gene Expression Omnibus public database under accession number GSE13204 (http://www.ncbi.nlm.nih.gov/geo/).
Statistical analyses
BAALC mRNA expression levels determined by real-time reverse-transcribed polymerase chain reaction were grouped into tertiles (T1-T3) based on the trend observed in clinical outcome (Figure 1). BAALC expression ranged between 0 and 91 2μ(ΔCT) (median T1, 0.2; median T2, 2.0; median T3, 9.0). T1, T2, and T3 are capitalized to distinguish the data from microarray-based expression (t1, t2, and t3).
Kaplan-Meier analyses of OS according to BAALC expression. Patients undergoing SCT in CR1 were censored at the time of transplantation. (A) Overall cohort of B-precursor ALL. (B) BCR-ABL− and MLL-AF4− ALL.
Kaplan-Meier analyses of OS according to BAALC expression. Patients undergoing SCT in CR1 were censored at the time of transplantation. (A) Overall cohort of B-precursor ALL. (B) BCR-ABL− and MLL-AF4− ALL.
Remission status was assessed after completion of induction chemotherapy. CR was defined as follows: granulocyte count of at least 1.5 × 109/L, platelet count of at least 100 × 109/L, no PB blasts, BM cellularity of at least 20% with maturation of all cell lines and less than 5% blasts, and no extramedullary leukemia. Primary therapy failure was defined as persistence of PB blasts or at least 25% blasts in BM after induction therapy. Relapse was defined as reappearance of PB blasts, more than 5% blasts in BM, or appearance of extramedullary manifestations after CR was achieved.
OS was determined from the first day of therapy until death or date of last follow-up. RFS was measured from the date of first CR until the date of relapse. Patients without reported relapse were censored by the end of the follow-up. Patients with withdrawal or death in CR were censored at the respective dates. Survival curves were calculated using the Kaplan-Meier method, with the log-rank comparing differences between survival curves. The median follow-up for living patients was 44 months. For 1 patient, no clinical data were available. Patients who received SCT in CR1 were censored at the time of transplantation for survival analyses.
Comparisons of molecular and clinical features across groups were made using the χ2 or a 2-sided Fisher exact test for categorical data and the nonparametric Mann-Whitney U test for continuous variables. A P value less than or equal to .05 (2-sided) was considered significant. Multivariate analyses were performed according to the Cox proportional hazards model for OS and with logistic regression for response to induction therapy, both with stepwise forward selection, including the following variables in the full model: BAALC expression (T1 vs T2 vs T3), WBC (> 30 × 109/L vs ≤ 30 × 109/L), age (10-year increase), CD20 (positive vs negative), BCR-ABL (presence vs absence), MLL-AF4 (presence vs absence), and immunophenotype (pre-B vs common/pro-B). When analyzing SR patients, WBC count was treated as a categorical variable with 1 × 109/L increase. SPSS software package (Version 17.0 for Windows; SPSS) was used for all calculations.
Results
BAALC expression and correlation to clinical and molecular features
BAALC mRNA expression was analyzed in pretreatment BM samples of 368 patients with newly diagnosed B-precursor ALL. Patients were divided into 3 BAALC expression groups as described in “Statistical analyses” (T1-T3). Patients with higher BAALC expression (T3 vs T2 vs T1) were significantly older (P < .001) and had higher WBC (P = .008) at initial diagnosis compared with patients with lower BAALC expression (Table 1). Higher BAALC expression was associated with the presence of BCR-ABL (P < .001) and MLL-AF4 (P < .001). In addition, higher BAALC expression correlated with a more immature CD34+ leukemic phenotype (P = .001). There was no significant association between BAALC expression level and CD20 positivity, coexpression of myeloid markers, or immunophenotypic subgroups of B-precursor ALL (Table 1).
Clinical and molecular characteristics at diagnosis according to BAALC expression in B-precursor ALL
| Characteristic . | BAALC T1 (n = 123) . | BAALC T2 (n = 123) . | BAALC T3 (n = 122) . | P . |
|---|---|---|---|---|
| Median age, y (range) | 32 (15-64) | 36 (15-65) | 42 (18-64) | < .001 |
| Male sex, percentage | 54 | 57 | 49 | .51 |
| WBC, ×109/L (n = 342) | ||||
| Median (range) | 10 (0.4-317) | 15 (0.8-472) | 20 (0.5-594) | .008 |
| More than 30 × 109/L, no. (%) | 34 (29) | 43 (37) | 46 (43) | .09 |
| CNS involvement, no. /total (%) (n = 272) | 4/96 (4) | 6/96 (6) | 3/80 (4) | .77 |
| Molecular genetics | ||||
| BCR-ABL, no. (%) | < .001 | |||
| Positive | 29 (24) | 41 (33) | 70 (57) | |
| Negative | 94 (76) | 82 (67) | 52 (43) | |
| MLL-AF4, no. (%) | < .001 | |||
| Positive | 0 (0) | 14 (11) | 11 (9) | |
| Negative | 123 (100) | 109 (89) | 111 (91) | |
| CD34 positivity, no. (%) | 79 (64) | 103 (84) | 95 (78) | .001 |
| CD20 positivity, no. (%) | 43 (35) | 49 (40) | 36 (30) | .23 |
| Myeloid markers, no. (%)* | 50 (41) | 49 (40) | 58 (48) | .42 |
| Immunophenotypic subtype, no. (%) | .10† | |||
| Common ALL | 77 (63) | 81 (66) | 89 (73) | |
| Pre-B ALL | 37 (30) | 25 (20) | 23 (19) | |
| Pro-B ALL | 9 (7) | 17 (14) | 10 (8) |
| Characteristic . | BAALC T1 (n = 123) . | BAALC T2 (n = 123) . | BAALC T3 (n = 122) . | P . |
|---|---|---|---|---|
| Median age, y (range) | 32 (15-64) | 36 (15-65) | 42 (18-64) | < .001 |
| Male sex, percentage | 54 | 57 | 49 | .51 |
| WBC, ×109/L (n = 342) | ||||
| Median (range) | 10 (0.4-317) | 15 (0.8-472) | 20 (0.5-594) | .008 |
| More than 30 × 109/L, no. (%) | 34 (29) | 43 (37) | 46 (43) | .09 |
| CNS involvement, no. /total (%) (n = 272) | 4/96 (4) | 6/96 (6) | 3/80 (4) | .77 |
| Molecular genetics | ||||
| BCR-ABL, no. (%) | < .001 | |||
| Positive | 29 (24) | 41 (33) | 70 (57) | |
| Negative | 94 (76) | 82 (67) | 52 (43) | |
| MLL-AF4, no. (%) | < .001 | |||
| Positive | 0 (0) | 14 (11) | 11 (9) | |
| Negative | 123 (100) | 109 (89) | 111 (91) | |
| CD34 positivity, no. (%) | 79 (64) | 103 (84) | 95 (78) | .001 |
| CD20 positivity, no. (%) | 43 (35) | 49 (40) | 36 (30) | .23 |
| Myeloid markers, no. (%)* | 50 (41) | 49 (40) | 58 (48) | .42 |
| Immunophenotypic subtype, no. (%) | .10† | |||
| Common ALL | 77 (63) | 81 (66) | 89 (73) | |
| Pre-B ALL | 37 (30) | 25 (20) | 23 (19) | |
| Pro-B ALL | 9 (7) | 17 (14) | 10 (8) |
CNS indicates central nervous system.
Coexpression of CD13, CD33, CD65s, or CD15.
Overall P value for the frequency of the 3 immunophenotypes across the BAALC tertiles.
Within the subgroup of BCR-ABL− and MLL-AF4− B-precursor ALL, higher BAALC expression significantly correlated with CD34 positivity (P = .03). No other association was seen between BAALC expression and clinical features in this subgroup (data not shown).
Outcome in B-precursor ALL patients with respect to BAALC expression (overall cohort)
Patients with higher BAALC expression showed a significantly lower CR rate (BAALC T3, 79%; BAALC T2, 86%; BAALC T1, 92%; P = .03) and a higher incidence of primary resistant disease (BAALC T3, 15%; BAALC T2, 8%; BAALC T1, 3%; P = .004). In multivariate analysis, BAALC remained the only predictive factor for primary therapy resistance (P = .002; odds ratio = 2.4; 95% confidence interval [CI], 1.4-4.3).
OS was significantly shorter for patients with higher BAALC expression compared with lower BAALC expression (5-year OS, 95% CI: BAALC T3, 29%, 18%-40%; BAALC T2, 38%, 25%-50%; BAALC T1, 65%, 54%-75%; P < .001; Figure 1A). However, BAALC expression was not independently predictive for OS in multivariate analysis. Significant adverse factors for OS were WBC and age (data not shown). No influence on the relapse rate (P = .94) or RFS (P = .52) was observed with respect to BAALC expression. There were no significant differences regarding deaths in induction therapy (P = .96) or deaths in CR (P = .28) between the 3 BAALC groups.
BAALC expression and outcome in BCR-ABL− and MLL-AF4− B-precursor ALL
We next analyzed the prognostic impact of BAALC expression in the molecular low-risk subgroup of BCR-ABL− and MLL-AF4− B-precursor ALL (n = 202). In this group, patients with higher BAALC expression had a lower CR rate (BAALC T3, 81%; BAALC T2, 91%; BAALC T1, 94%; P = .09; Table 2). BCR-ABL− and MLL-AF4− patients with higher BAALC expression had a significantly higher frequency of refractory disease compared to patients with lower BAALC expression (BAALC T3, 15%; BAALC T2, 6%; BAALC T1, 1%; P = .009; Table 2). On multivariate analysis, higher BAALC expression was independently predictive for primary resistant disease (P = .01) as well as the immunophenotype (Table 3).
Clinical outcome according to BAALC expression in BCR-ABL− and MLL-AF4− patients
| Clinical outcome . | BAALC T1 (n = 94) . | BAALC T2 (n = 67) . | BAALC T3 (n = 41) . | P . |
|---|---|---|---|---|
| Complete remission, no./total (%) (n = 193) | 86/92 (94) | 59/65 (91) | 29/36 (81) | .09‡ |
| Refractory disease, no./total (%) (n = 184) | 1/87 (1) | 4/63 (6) | 5/34 (15) | .009 |
| Overall survival at 5 y, percentage (95% CI) (n = 202)* | 70 (59-81) | 52 (36-67) | 38 (22-54) | .004 |
| Relapse rate, no./total (%) (n = 114)† | 26/53 (49) | 16/36 (44) | 10/25 (40) | .76 |
| Clinical outcome . | BAALC T1 (n = 94) . | BAALC T2 (n = 67) . | BAALC T3 (n = 41) . | P . |
|---|---|---|---|---|
| Complete remission, no./total (%) (n = 193) | 86/92 (94) | 59/65 (91) | 29/36 (81) | .09‡ |
| Refractory disease, no./total (%) (n = 184) | 1/87 (1) | 4/63 (6) | 5/34 (15) | .009 |
| Overall survival at 5 y, percentage (95% CI) (n = 202)* | 70 (59-81) | 52 (36-67) | 38 (22-54) | .004 |
| Relapse rate, no./total (%) (n = 114)† | 26/53 (49) | 16/36 (44) | 10/25 (40) | .76 |
Patients who received SCT in CR1 were censored at the time of transplantation.
Patients who received SCT in CR1 were excluded.
P value across the 3 BAALC tertiles. Statistical significance was reached comparing BAALC T1 vs T3 (P = .047).
Multivariate analysis of BAALC expression for clinical outcome in BCR-ABL− and MLL-AF4− patients
| Variable . | HR . | OR . | 95% CI . | P . |
|---|---|---|---|---|
| Refractory disease* | ||||
| BAALC expression, T1 vs T2 vs T3 | 4.2 | 1.4-12.3 | .01 | |
| Immunophenotype | .01‡ | |||
| Common vs pre-B | 1.0 | 0.1-9.1 | .98 | |
| Pro-B vs pre-B | 14 | 1.1-176 | .04 | |
| Overall survival*† | ||||
| BAALC expression, T1 vs T2 vs T3 | 1.4 | 1.0-2.0 | .03 | |
| WBC more than 30 × 109/L | 3.9 | 2.2-6.9 | < .001 | |
| Age, 10-year increase | 1.5 | 1.3-1.8 | < .001 |
| Variable . | HR . | OR . | 95% CI . | P . |
|---|---|---|---|---|
| Refractory disease* | ||||
| BAALC expression, T1 vs T2 vs T3 | 4.2 | 1.4-12.3 | .01 | |
| Immunophenotype | .01‡ | |||
| Common vs pre-B | 1.0 | 0.1-9.1 | .98 | |
| Pro-B vs pre-B | 14 | 1.1-176 | .04 | |
| Overall survival*† | ||||
| BAALC expression, T1 vs T2 vs T3 | 1.4 | 1.0-2.0 | .03 | |
| WBC more than 30 × 109/L | 3.9 | 2.2-6.9 | < .001 | |
| Age, 10-year increase | 1.5 | 1.3-1.8 | < .001 |
HR indicates hazard ratio; and OR, odds ratio.
Variables considered for model inclusion are described in "Statistical analyses." The final model is shown.
Patients who received SCT in CR1 were censored at the time of transplantation.
Overall P value for the variable immunophenotype.
Higher BAALC expression was significantly associated with an inferior OS for BCR-ABL− and MLL-AF4− patients (5-year OS: BAALC T3, 38%; BAALC T2, 52%; BAALC T1, 70%; P = .004; Table 2; Figure 1B). In the Cox regression analysis, BAALC expression was of independent prognostic significance for OS (P = .03); the other factors in the final model predicting OS were WBC and age (Table 3). BAALC expression did not have an impact on relapse rate (P = .76; Table 2) or RFS (P = .94) in BCR-ABL− and MLL-AF4− patients.
Moreover, within the SR group as defined by GMALL (BCR-ABL− and MLL-AF4− patients with CR after first induction therapy and WBC ≤ 30 × 109/L at initial diagnosis), higher BAALC expression remained predictive for a shorter OS (P = .04; hazard ratio = 1.5; 95% CI, 1.0-2.3) as well as age.
We further examined the clinical outcome of BCR-ABL− and MLL-AF4− patients undergoing allogeneic SCT in CR1 with respect to BAALC expression. Because of the small number of patients and the stronger discrimination of T1 versus T2 compared with T2 versus T3 in OS (Figure 1), BAALC T2 and T3 were defined as high BAALC and BAALC T1 was defined as low BAALC for this analysis. BCR-ABL− and MLL-AF4− patients were assigned to allogeneic SCT if they fulfilled one of the following high-risk criteria: late therapy response, the immunophenotype of pro-B ALL, or high WBC count (“Patients”). In this subgroup, BAALC was no longer an adverse factor regarding OS, as patients with high BAALC expression tended to have an improved survival with a 5-year OS of 62% (data not shown). No selection bias was found between the BAALC groups in terms of age, WBC count, sex, indication for SCT, or other clinical factors. Thus, allogeneic SCT might overcome the adverse impact of BAALC and thereby leveling the differences in survival curves between the BAALC groups.
GEP of BCR-ABL− and MLL-AF4− B-precursor ALL
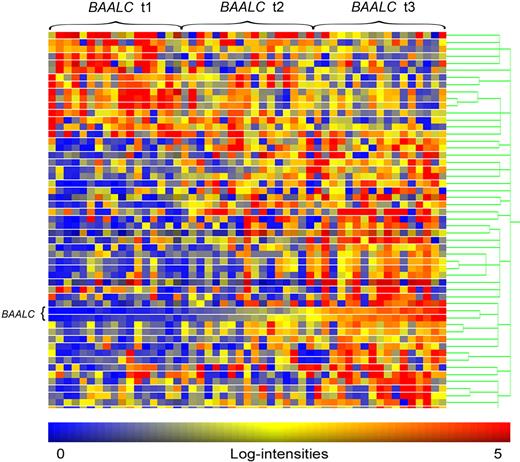
A microarray-based GEP of 51 B-precursor ALL pretreatment BM samples was performed to gain insights into biologic mechanisms of BAALC and its potential role in B-lymphoblastic leukemia and chemoresistance. Because BAALC was of particular prognostic relevance in the BCR-ABL− and MLL-AF4− subgroup and both aberrations, BCR-ABL and MLL-AF4, have previously been shown to drive specific gene-expression patterns,22,23 we restricted our GEP analysis to BCR-ABL− and MLL-AF4− samples. Samples were grouped into high and low BAALC (t3 vs t1) based on the microarray expression data as described in “Oligonucleotide microarray studies.” Expression level of BAALC was 22-fold increased in t3 samples compared with t1. Between the high and low BAALC expression groups, 54 differentially expressed probe sets corresponding to 43 genes and hypothetical proteins were identified (Figure 2); 32 were up-regulated and 11 were down-regulated (Table 4).
Heat map of differentially expressed probe sets between high and low BAALC expression groups in BCR-ABL− and MLL-AF4− ALL. Columns represent samples and rows represent genes ordered by hierarchical cluster analysis. Colors indicate relative expression of each gene with respect to the median expression level: red indicates above median value; yellow, equal; blue, below median value. Heat map of differentially expressed probe sets between high and low BAALC expression groups in BCR-ABL− and MLL-AF4− ALL.
Heat map of differentially expressed probe sets between high and low BAALC expression groups in BCR-ABL− and MLL-AF4− ALL. Columns represent samples and rows represent genes ordered by hierarchical cluster analysis. Colors indicate relative expression of each gene with respect to the median expression level: red indicates above median value; yellow, equal; blue, below median value. Heat map of differentially expressed probe sets between high and low BAALC expression groups in BCR-ABL− and MLL-AF4− ALL.
Differentially expressed genes in BCR-ABL− and MLL-AF4− patients between BAALC expression groups (t3 vs t1)
| Gene symbol . | Description . | Fold change . |
|---|---|---|
| Genes overexpressed in the high BAALC group (t3 vs t1) | ||
| BAALC* | Brain and acute leukemia, cytoplasmic | 22.8 |
| ARHGEF17* | Rho guanine nucleotide exchange factor (GEF) 17 | 10.2 |
| KCNK12 | Potassium channel, subfamily K, member 12 | 8.2 |
| IGJ | Immunoglobulin J polypeptide, linker protein for immunoglobulin-α and -μ polypeptides | 7.7 |
| LOC285628 | Hypothetical protein LOC285628 | 6.3 |
| STARD13 | Start domain containing 13 | 6.0 |
| LOC26010 | Viral DNA polymerase-transactivated protein 6 | 5.6 |
| SEMA6A | Sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A | 5.6 |
| MMP28 | Matrix metallopeptidase 28 | 5.2 |
| SDC2 | Syndecan 2 | 5.0 |
| STYK1 | Serine/threonine/tyrosine kinase 1 | 5.0 |
| KLF9* | Kruppel-like factor 9 | 4.8 |
| CD34 | CD34 antigen | 4.1 |
| CD99* | CD99 antigen | 3.9 |
| CYTL1 | Cytokine-like 1 | 3.9 |
| LOC144481 | Hypothetical protein LOC144481 | 3.8 |
| NRN | Neuritin 1 | 3.8 |
| SESN1* | Sestrin 1 | 3.7 |
| EMP1* | Epithelial membrane protein 1 | 3.7 |
| NUDT4 | NUDIX (nucleoside diphosphate-linked moiety X)-type motif 4 | 3.5 |
| TBXA2R | Thromboxane A2 receptor | 3.5 |
| TMEM156 | Transmembrane protein 156 | 3.5 |
| PPM1F* | Protein phosphatase (PP2C domain containing) | 3.3 |
| TSPAN7 | Tetraspanin 7 | 3.3 |
| MDFIC | MYOD family inhibitor domain containing | 3.3 |
| LYN | V-YES-1 Yamaguchi sarcoma viral oncogene homolog 1 | 3.1 |
| CYGB | Cytoglobin | 3.1 |
| SERPINB9 | Serpin peptidase inhibitor, clade B (ovalbumin), member 9 | 3.1 |
| CLEC14A | C-type lectin domain family 14, member A | 3.1 |
| FHL1 | Four-and-a-half LIM domains 1 | 3.1 |
| MADD | MAP-kinase activating death domain | 3.1 |
| FZD6 | Frizzled homolog 6 (Drosophila) | 3.0 |
| Genes underexpressed in the high BAALC group (t3 vs t1) | ||
| BCL11B* | B-cell CLL/lymphoma 11B (zinc finger protein) | 5.1 |
| CAMK2D | Calcium/calmodulin-dependent protein kinase (CAM kinase) IIδ | 4.6 |
| CDKN1C | Cyclin-dependent kinase inhibitor 1C | 3.8 |
| FLJ34870 | Hypothetical protein FLJ34870 | 3.6 |
| MGC10744* | Transmembrane protein 107 | 3.4 |
| COL27A1 | Collagen, type 27, α 1 | 3.3 |
| GATA3 | GATA binding protein 3 | 3.2 |
| AEBP1 | AE binding protein 1 | 3.1 |
| GLT1D1 | Glycosyltransferase 1 domain containing 1 | 3.1 |
| PYCARD | PYD and CARD domain containing | 3.0 |
| RGS2 | Regulator of G-protein signaling 2, 24 kDa | 3.0 |
| Gene symbol . | Description . | Fold change . |
|---|---|---|
| Genes overexpressed in the high BAALC group (t3 vs t1) | ||
| BAALC* | Brain and acute leukemia, cytoplasmic | 22.8 |
| ARHGEF17* | Rho guanine nucleotide exchange factor (GEF) 17 | 10.2 |
| KCNK12 | Potassium channel, subfamily K, member 12 | 8.2 |
| IGJ | Immunoglobulin J polypeptide, linker protein for immunoglobulin-α and -μ polypeptides | 7.7 |
| LOC285628 | Hypothetical protein LOC285628 | 6.3 |
| STARD13 | Start domain containing 13 | 6.0 |
| LOC26010 | Viral DNA polymerase-transactivated protein 6 | 5.6 |
| SEMA6A | Sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A | 5.6 |
| MMP28 | Matrix metallopeptidase 28 | 5.2 |
| SDC2 | Syndecan 2 | 5.0 |
| STYK1 | Serine/threonine/tyrosine kinase 1 | 5.0 |
| KLF9* | Kruppel-like factor 9 | 4.8 |
| CD34 | CD34 antigen | 4.1 |
| CD99* | CD99 antigen | 3.9 |
| CYTL1 | Cytokine-like 1 | 3.9 |
| LOC144481 | Hypothetical protein LOC144481 | 3.8 |
| NRN | Neuritin 1 | 3.8 |
| SESN1* | Sestrin 1 | 3.7 |
| EMP1* | Epithelial membrane protein 1 | 3.7 |
| NUDT4 | NUDIX (nucleoside diphosphate-linked moiety X)-type motif 4 | 3.5 |
| TBXA2R | Thromboxane A2 receptor | 3.5 |
| TMEM156 | Transmembrane protein 156 | 3.5 |
| PPM1F* | Protein phosphatase (PP2C domain containing) | 3.3 |
| TSPAN7 | Tetraspanin 7 | 3.3 |
| MDFIC | MYOD family inhibitor domain containing | 3.3 |
| LYN | V-YES-1 Yamaguchi sarcoma viral oncogene homolog 1 | 3.1 |
| CYGB | Cytoglobin | 3.1 |
| SERPINB9 | Serpin peptidase inhibitor, clade B (ovalbumin), member 9 | 3.1 |
| CLEC14A | C-type lectin domain family 14, member A | 3.1 |
| FHL1 | Four-and-a-half LIM domains 1 | 3.1 |
| MADD | MAP-kinase activating death domain | 3.1 |
| FZD6 | Frizzled homolog 6 (Drosophila) | 3.0 |
| Genes underexpressed in the high BAALC group (t3 vs t1) | ||
| BCL11B* | B-cell CLL/lymphoma 11B (zinc finger protein) | 5.1 |
| CAMK2D | Calcium/calmodulin-dependent protein kinase (CAM kinase) IIδ | 4.6 |
| CDKN1C | Cyclin-dependent kinase inhibitor 1C | 3.8 |
| FLJ34870 | Hypothetical protein FLJ34870 | 3.6 |
| MGC10744* | Transmembrane protein 107 | 3.4 |
| COL27A1 | Collagen, type 27, α 1 | 3.3 |
| GATA3 | GATA binding protein 3 | 3.2 |
| AEBP1 | AE binding protein 1 | 3.1 |
| GLT1D1 | Glycosyltransferase 1 domain containing 1 | 3.1 |
| PYCARD | PYD and CARD domain containing | 3.0 |
| RGS2 | Regulator of G-protein signaling 2, 24 kDa | 3.0 |
Gene was represented by more than one probe set. The fold change corresponds to the mean value of all probe sets.
Differentially up-regulated genes in patients with high BAALC expression included markers of hematopoietic stem cells (CD34 and CD99)24 and genes that have previously been associated with drug resistance in chronic myeloid leukemia and AML (LYN and TSPAN7).25-27 Interestingly, SERPINB9 was up-regulated in the high BAALC group (BCR-ABL− patients), a gene overexpressed in adult BCR-ABL ALL22 and differentially expressed in pediatric ALL.28 Within the up-regulated genes, we further found the frizzled receptor family member FZD6 that has already been associated with high BAALC expression in CN-AML.10 Among the down-regulated genes in the high BAALC group, we identified genes that have been related to a favorable outcome in adult ALL and AML (CDKN1C/p57Kip2 and RGS2).29,30 Thus, our BAALC-associated GEP signature supports the clinical observation of a chemoresistant disease with inferior survival defined by high BAALC expression.
Discussion
The identification of novel predictive molecular markers in B-precursor ALL is particularly important for patients lacking high-risk markers (eg, BCR-ABL, MLL-AF4) and might in the future improve the outcome of this clinically and biologically heterogeneous subgroup. In this study, we have evaluated the prognostic significance of BAALC expression in B-precursor ALL, focusing on BCR-ABL− and MLL-AF4− patients.
As we have shown here, high BAALC expression independently predicted primary therapy resistance in adult B-precursor ALL patients. The risk of primary therapy failure in the overall cohort was 5 times higher for patients in the upper BAALC tertile (T3) compared with the lower tertile (T1). In BCR-ABL− and MLL-AF4− patients, the incidence of resistant disease was even 15-fold higher in BAALC T3 versus T1. In addition, higher BAALC expression was independently predictive for OS in BCR-ABL− and MLL-AF4− patients with a 5-year OS of only 38% in the BAALC T3 group.
In previous studies, high BAALC expression has already been predictive for an unfavorable therapy response and inferior long-term survival in adult CN-AML and T-ALL.8-10,18 Our results in B-precursor ALL further highlight the adverse prognostic significance of aberrant BAALC expression in acute leukemias independent of the lineage. Moreover, BAALC is associated with an immature CD34+ leukemic subtype throughout the leukemic lineages. In B-precursor ALL, higher BAALC expression was further strongly correlated with known risk factors, such as high WBC count, BCR-ABL, and MLL-AF4. Thus, BAALC characterizes a more aggressive, immature, highly proliferative, and chemoresistant leukemic phenotype. BAALC as marker for normal hematopoietic progenitor cells may thereby reflect a marker of putative leukemic stem cells conferring drug resistance. Although the functional role of BAALC remains unknown, these findings support the possible involvement of BAALC in leukemogenesis.
These clinical observations were further supported by the GEP signature associated with high BAALC expression. In the high BAALC group, we observed up-regulation of genes associated with hematopoietic stem and progenitor cells (CD34, CD99, and FZD6)24,31 and genes involved in therapy resistance in myeloid leukemias (LYN and TSPAN7).25-27 This is similar to findings of Langer et al,10 who reported an up-regulation of CD34, FZD6, and the multidrug resistance gene ABCB1 in CN-AML with high BAALC expression. Overexpression of the tyrosine kinase LYN appears to be important for proliferation and survival of leukemic blasts.32 LYN signaling was also shown to play a crucial role in AML with FLT3 internal tandem duplication (ITD).33,34 In addition, RGS2, one of the differentially down-regulated genes in the high BAALC group, was demonstrated to be repressed by FLT3-ITD in AML.30
The cyclin-dependent kinase inhibitor CDKN1C/p57Kip2, a cell-cycle regulatory gene and putative tumor suppressor,35 was down-regulated in patients with high BAALC expression. CDKN1C/p57Kip2 is aberrantly methylated in adult ALL, and inactivation of CDKN1C/p57Kip2, p73, and p15 has been related to a worse outcome in BCR-ABL− patients.36 Thus, the GEP signature associated with high BAALC expression designates a highly proliferative subgroup of B-precursor ALL. This is further strengthened by similarities with molecular pathways and expression profiles found in other highly proliferative and resistant leukemic subtypes with inferior prognosis, such as BCR-ABL ALL and FLT3-ITD AML. In addition, the overlap of BAALC-associated signatures in B-precursor ALL and CN-AML underscores the lineage-independent impact of BAALC in acute leukemias.
For patients with primary therapy failure, new strategies during induction therapy are needed to overcome chemoresistance and to improve long-term outcome. General approaches to increase initial treatment response in B-lineage ALL include dose intensification of cytostatic agents37,38 and the addition of monoclonal antibodies.39-41 Intensification of myelosuppressive therapy has to be carefully considered as the rate of early deaths is increased and dose intensity of subsequent treatments may be compromised. In this regard, determination of BAALC expression may help to better select patients at diagnosis with a poor response to induction therapy for which dose intensification or the use of new agents without multidrug resistance may be advantageous.
We demonstrate that higher BAALC expression independently predicts a shorter OS in BCR-ABL− and MLL-AF4− patients as well as in the SR group. Our findings indicate that determination of BAALC expression may contribute to a more detailed risk stratification of these patients. We focused on BCR-ABL− and MLL-AF4− patients for survival analyses because BCR-ABL and MLL-AF4, as confirmed unfavorable molecular markers, already direct specific treatment strategies.1,42-44 We did not consider other high-risk cytogenetics, such as complex karyotypes,44 because these aberrations are less frequent in B-precursor ALL and are not yet consistently integrated in ALL risk stratification throughout different study groups.
Interestingly, in BCR-ABL− and MLL-AF4− high-risk patients undergoing allogeneic SCT, high BAALC expressers showed a tendency toward an improved outcome compared with the low BAALC group (data not shown). Similar findings have been made in CN-AML, where the beneficial effect of allogeneic SCT was most evident in patients with high BAALC expression.18 The adverse impact of BAALC might be overcome by intensive treatment strategies, such as allogeneic SCT. Further prospective clinical trials and experimental studies are necessary to address the impact of BAALC expression on outcome of patients undergoing allogeneic SCT.
Identification of molecular markers in acute leukemias is important to elucidate molecular pathways of leukemogenesis and to direct the development of targeted therapies. The integration of prognostically relevant molecular markers into the risk assignment of ALL may improve clinical outcome by attributing to a precise risk-adapted treatment strategy. Recent approaches to identify new risk groups in B-precursor ALL included the determination of CTGF expression,45 analyses of methylation status,46 the integration of rare cytogenetic subgroups,44,47 and genome-wide profiling of genetic alterations.48,49 Deletions or mutations of the IKAROS gene have been associated with poor outcome in BCR-ABL− high-risk childhood B-precursor ALL.49 However, prospective studies are needed to confirm the prognostic significance of these new markers in the context of established risk factors.
This is the first study that investigated BAALC expression with respect to clinical features, treatment outcome, and GEP in adult B-precursor ALL. Our data suggest that BAALC may provide a new marker that identifies patients with an immature, chemoresistant leukemic phenotype associated with an unfavorable therapy response and inferior survival. Determination of BAALC expression might in the future facilitate treatment stratification for molecularly undefined subgroups of adult B-precursor ALL.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the members of the GMALL study group for their support and the participating centers for enrolling patients and providing the clinical data for the study as well as Liliana Mochmann for critical reading of the manuscript.
This work was supported by the Deutsche Krebshilfe (Max Eder Nachwuchsförderung) and the Deutsche Forschungsgesellschaft (C.D.B.). The GMALL studies were supported by the Deutsche Krebshilfe.
A complete list of the members of the GMALL study group appears in a supplemental Appendix (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Authorship
Contribution: A.K. performed research, analyzed data, and wrote the manuscript; N.G. supervised the GMALL study center and reviewed the manuscript; A.S. performed statistical analyses; T.B. provided diagnostic molecular genetic data; M.N. and S.H. contributed to study analysis; T.H. provided samples and expression data; D.H. supervised the GMALL study group; W.-K.H. contributed to the study analysis and reviewed the manuscript; E.T. supervised the central immunophenotyping and molecular genetics laboratory and reviewed the manuscript; and C.D.B. designed the research, contributed to study analysis, and reviewed the manuscript.
Conflict-of-interest disclosure: The Microarray Innovations in LEukemia (MILE) study was in part supported by Roche Molecular Systems Inc. T.H. is part owner of the MLL Münchner Leukämie Labor GmbH. The remaining authors declare no competing financial interests.
Correspondence: Claudia D. Baldus, Charité Universitätsmedizin Berlin, Campus Benjamin Franklin, Medizinische Klinik III, Hindenburgdamm 30, 12203 Berlin, Germany; e-mail: claudia.baldus@charite.de.