Abstract
Cytogenetic aberrations are important prognostic factors in acute myeloid leukemia (AML). Of adults with de novo AML, 45% lack cytogenetic abnormalities, and identification of predictive molecular markers might improve therapy. We studied the prognostic impact of BAALC (Brain And Acute Leukemia, Cytoplasmic), a novel gene involved in leukemia, in 86 de novo AML patients with normal cytogenetics who were uniformly treated on Cancer and Leukemia Group B 9621. BAALC expression was determined by comparative real-time reverse transcriptase–polymerase chain reaction in pretreatment blood samples, and patients were dichotomized at BAALC's median expression into low and high expressers. Low expressers had higher white counts (P = .03) and more frequent French-American-British M5 morphology (P = .007). Compared to low expressers, high BAALC expressers showed significantly inferior overall survival (OS; median, 1.7 vs 5.8 years, P = .02), event-free survival (EFS; median, 0.8 vs 4.9 years, P = .03), and disease-free survival (DFS; median, 1.4 vs 7.3 years, P = .03). Multivariable analysis confirmed high BAALC expression as an independent risk factor. For high BAALC expressers the hazard ratio of an event for OS, EFS, and DFS was respectively 2.7, 2.6, and 2.2. We conclude that high BAALC expression predicts an adverse prognosis and may define an important risk factor in AML with normal cytogenetics.
Introduction
Clonal cytogenetic abnormalities are one of the most important factors predicting clinical outcome in acute myeloid leukemia (AML) and are used to guide risk-adapted treatment strategies.1-3 However, approximately 45% of adults younger than 60 with de novo AML have normal cytogenetics and therefore lack informative chromosome markers.4,5 With current therapies 42% to 43% of these patients are long-term survivors, and 40% to 47% of complete responders remain in continuous remission at 5 years.3,6,7 To date, several pretreatment clinical or laboratory prognostic factors have been identified in AML patients with intermediate risk cytogenetics,8-13 but very few markers have been consistently shown to be predictive in patients with normal cytogenetics.14-17 Consequently, in patients with a normal karyotype new molecular markers identifying those who are at risk to fail standard therapeutic approaches are warranted to optimize treatment strategies.
In a search for novel genes involved in leukemia, we identified BAALC (Brain And Acute Leukemia, Cytoplasmic), a gene located on chromosome 8q22.3.18 DNA sequence and expression pattern were highly conserved among mammals, whereas no orthologs were found in lower organisms. The protein sequence showed no homology to any known proteins or functional domains. Expression of BAALC was found mainly in neuroectoderm-derived tissues and hematopoietic precursor cells. In hematopoietic cells, BAALC expression was restricted to the compartment of progenitor cells, whereas no expression was detected in mature bone marrow or circulating white blood cells.19 In leukemias, we found high BAALC expression in a subset of patients with AML, acute lymphoblastic leukemia (ALL), and chronic myelogenous leukemia (CML) in blast crisis, whereas no BAALC expression could be detected in patients with chronic-phase CML or chronic lymphocytic leukemia (CLL).18
High BAALC expression levels were first identified in a study of AML patients with trisomy 8 as a sole abnormality.18 Trisomy 8 has been associated with poor prognosis in AML,2,5 and we therefore hypothesized that BAALC expression might assist in prognosis of AML patients lacking cytogenetic aberrations. To do this, we examined a well-defined cohort of adult patients younger than 60 years of age, diagnosed with de novo AML, and treated on a single dose-intensive protocol within the Cancer and Leukemia Group B (CALGB 9621). Since we have previously shown that a FLT3 internal tandem duplication (ITD) in the absence of a FLT3 wild-type (WT) allele identifies AML patients with a normal karyotype with a very poor prognosis on CALGB 9621,14 in the current study we focused on 86 AML patients with normal cytogenetics who lacked this adverse prognostic factor. We used quantitative real-time reverse transcriptase–polymerase chain reaction (RT-PCR) to analyze messenger RNA levels of BAALC in pretreatment blood samples. Our data show that high BAALC expression is an independent adverse prognostic factor among intensively treated younger adults with cytogenetically normal de novo AML.
Patients and methods
Patients
This study included 86 patients enrolled on CALGB treatment trial 9621 who had a primary diagnosis of AML, a centrally confirmed pretreatment normal bone marrow karyotype, a centrally determined pretreatment bone marrow FLT3 genotype containing a wild-type allele, and available pretreatment blood stored in the CALGB Leukemia Tissue Bank on CALGB protocol 9665.20-22 This represents 74% of all patients enrolled on CALGB 9621 who met the eligibility criteria. There were no significant differences in outcome (P ≥ .54 for all endpoints) or clinical features for the remaining 31 patients not included in this study, except that patients not included had a higher median percentage of bone marrow blasts (67% versus 58%, P = .03). Pathologic diagnoses were reviewed centrally and classified according to the French-American-British (FAB) schema. As required by CALGB 9621, all patients were between ages 16 and 60, and patients with a prior history of myelodysplasia, other antecedent hematologic malignancies, prior nonsteroidal cytotoxic chemotherapy or radiation therapy, pre-existing liver disease, or uncontrolled infection were excluded. Written informed consent was obtained from all patients. Of the 86 AML patients, 26 were included in the first published report on BAALC.18 Approval was obtained from the Ohio State University institutional review board for these studies according to the Declaration of Helsinki.
Cytogenetic and FLT3 studies
All patients included in this analysis were enrolled on CALGB 8461, a prospective cytogenetics protocol. Cytogenetic analyses of bone marrow were performed in institutional CALGB cytogenetics laboratories, and karyotypes were reviewed centrally. Specimens were obtained at diagnosis from all patients and processed using unstimulated short-term (24-, 48-, and 72-hour) cultures with or without a direct method. G-banding was usually done, although Q-banding also was acceptable. The criteria used to describe a cytogenetic clone and description of karyotype followed the recommendations of the International System for Human Cytogenetic Nomenclature (ISCN; 1995).23 A minimum of 20 bone marrow metaphases was required to be examined for a patient to be classified as having normal cytogenetics.5
The FLT3 genotype was determined using previously described techniques.14 Briefly, PCR and RT-PCR were carried out using primers that detect the length mutations discovered for the FLT3 gene.24 Long-range DNA PCRs were performed in the FLT3 gene extending from exon 10 to the 3′-end of exon 12. Two genotypes including the WT allele were distinguished, one with the ITD, designated FLT3ITD/WT, and one without, designated FLT3WT/WT.
Treatment
Patients received induction chemotherapy consisting of cytarabine, daunorubicin, and etoposide (ADE) with or without the multidrug resistance protein modulator, PSC-833.20 For patients achieving complete remission (CR), this was followed by autologous peripheral blood stem cell transplantation (PBSCT) using high-dose etoposide and cytarabine for “in vivo purging” and stem cell mobilization followed by a myeloablative regimen of busulfan and etoposide.21 Patients unable to receive PBSCT were consolidated with a regimen consisting of the “in vivo purging” portion of the PBSCT sequence followed by 2 cycles of high-dose cytarabine. Following consolidation, patients received a 90-day low-dose subcutaneous interleukin-2 (IL-2, Proleukin, Chiron, Emeryville, CA) regimen interrupted with intermediate dose pulsing of subcutaneous IL-2 every 2 weeks.
BAALC determination
Mononuclear cells from pretreatment blood samples were enriched by Ficoll-Hypaque gradient and frozen in liquid nitrogen. Total RNA was extracted from thawed samples using Trizol reagent (Invitrogen, Carlsbad, CA) following the manufacturer's directions. Complementary DNA (cDNA) was synthesized using 0.5 μg RNA, avian myeloblastosis virus reverse transcriptase (Roche, Indianapolis, IN), and gene-specific primers for glucose-phosphate isomerase (GPI) and BAALC at 50°C for 60 minutes in the presence of RNase inhibitor (RNasin; Roche).
Comparative real-time RT-PCR assays were performed for each sample in triplicate in a final reaction volume of 25 μL. GPI and BAALC were coamplified in the same tube using 2 μL cDNA, 1 × universal master mix (Applied Biosystems, Foster City, CA), 250 nM human GPI probe (VIC-labeled) with 600 nM each of the GPI forward and reverse primers, and 250 nM of BAALC probe (6-FAM-labeled) with 900 nM each of the BAALC forward and reverse primers. Final concentrations of primers and probes were chosen based on optimization experiments. Primers for GPI and BAALC were intron spanning. Probes were labeled with quencher TAMRA at the 3′-end. Amplification was carried out at 50°C for 2 minutes, 95°C for 10 minutes, followed by 40 PCR cycles at 95°C for 15 seconds, and 60°C for 1 minute. All reactions were done in MicroAmp optical 96-well plates using an ABI Prism 7700 sequence detection system (Applied Biosystems). Sequences of primers and probes are available upon request. The comparative cycle threshold (CT) method was used to determine the relative expression levels of BAALC (Applied Biosystems). The threshold cycles for BAALC and GPI were determined, and the cycle number difference (ΔCT = GPI-BAALC) was calculated for each replicate. Relative BAALC expression values were calculated using the mean of ΔCT from the 3 replicates, that is, μ(ΔCT) = (ΣΔCT)/3, and expressed as 2μ(ΔCT).
Positive and negative controls were included in all assays. To assess reproducibility of the assay, samples from 28 AML patients with high and low BAALC expression values were split and evaluated on different days. Data generated showed high reproducibility of the results (Pearson correlation coefficient, r = 0.98).
In addition to real-time RT-PCR, comparative semiquantitative RT-PCR was carried out in a final volume of 50 μL containing 1 μL cDNA, 2.5 U AmpliTaq Gold, 100 nM of each dNTP (all Roche), and 20 pmol of each primer. Conditions were 95°C for 10 minutes, 32 cycles of 95°C for 15 seconds, 58°C for 15 seconds, and 72°C for 1 minute with a final step of 72°C for 5 minutes. Levels of BAALC amplification products were estimated relative to the expression levels of the housekeeping gene GPI on nondenaturing polyacrylamide gel electrophoresis after staining with silver nitrate.18,25
Statistical methods
To evaluate the impact of BAALC expression values on clinical outcome without seeking an optimal cut point, AML samples were dichotomized at the median value and divided into 2 expression groups: a low BAALC group with BAALC values less than 0.166 (n = 43; median expression, 0.050; range, 0.000 03-0.163) and a high BAALC group consisting of patients with BAALC values of more than 0.166 (n = 43; median expression, 0.547; range, 0.170-4.532). BAALC values were calculated as described in “BAALC determination.”
Baseline clinical features were compared for patients expressing low BAALC and those expressing high BAALC. Categorical variables were compared using Fisher exact test and continuous variables using the Wilcoxon rank sum test. The 2-sided level of significance was set at .05. Complete remission required an absolute neutrophil count of at least 1500/μL, a platelet count of at least an α value of 100 × 109/L, no leukemic blasts in the blood, bone marrow cellularity greater than 20% with maturation of all cell lines, no Auer rods, fewer than 5% bone marrow blast cells, and no extramedullary leukemia, with persistence for at least one month.26 Relapse was defined as the reappearance of circulating blast cells not attributable to “overshoot” following recovery from myelosuppressive therapy, or greater than 5% blasts in the marrow not attributable to another cause, or development of extramedullary leukemia.
Overall survival (OS) was measured from the protocol on-study date until the date of death regardless of cause, censoring for patients alive at last follow-up. Event-free survival (EFS) was defined for those achieving CR as the time from on-study until relapse or death regardless of cause, censoring for those alive at last follow-up. If a patient did not achieve CR but expired within 2 months of the on-study date, then EFS was defined as the time from on-study until death, regardless of cause. Otherwise, EFS was set at 2 months. Disease-free survival (DFS) was defined only for those patients achieving a CR. It was measured from the CR date until date of relapse or death, regardless of cause, censoring for patients alive at last follow-up. Overall, event-free and disease-free survival all were analyzed using the Kaplan-Meier method, and the log-rank test was used to compare differences between survival curves.27,28
To further investigate the effect of increasing expression of BAALC, a Cox proportional hazards model was constructed, adjusting for potential confounding covariates, using backward elimination.29 Fractional polynomials were used to identify the most appropriate scaling for the continuous covariates.30 Univariate models integrating an artificial time-dependent covariate, expressed as the product of the fixed-time covariate and the log of time, were fit to check the proportional hazards assumption in conjunction with assessing the scaled Schoenfeld residuals.31 The log-transformed BAALC failed to satisfy the proportional hazards assumption, so BAALC was dichotomized at its median value, where the assumption of proportional hazards was met, to represent high BAALC expression versus low BAALC expression (see first paragraph in this section). All covariates, including the BAALC indicator, whose univariate models reflected a P value less than .20 from the a likelihood ratio test were included in a full model. Variables with least significance from the Wald statistic were eliminated one at a time until the only variables in the model were significant at a P value of .05. Any variables that were initially excluded from the model were added back into the model to confirm that they were neither statistically significant nor an important confounder, defined by a change in the estimated coefficients of at least 20%. Lastly, interactions were considered important if the associated P value was less than .05. Statistical analyses were performed by the CALGB Statistical Center.
Results
BAALC expression in adult de novo AML with normal karyotype
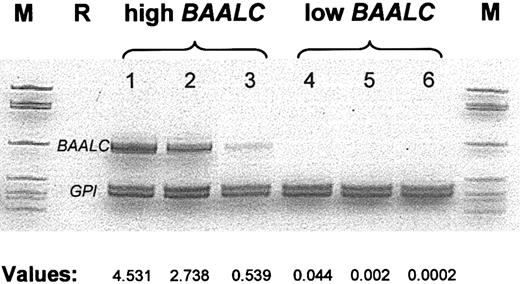
Pretreatment blood samples of 86 patients were evaluated for BAALC expression by comparative real-time RT-PCR. BAALC values were calculated as described in “BAALC determination.” BAALC expression levels represented a continuum ranging from 0.00003 to 4.532. High BAALC expression was defined as the upper 50% of BAALC values and low BAALC expression as the lower 50% of BAALC values. In addition to the real-time RT-PCR assay, conventional semiquantitative RT-PCR was performed and confirmed the results obtained by the real-time RT-PCR (Figure 1).
BAALC mRNA expression in pretreatment AML samples as determined by comparative RT-PCR and real-time RT-PCR.The gel illustrates the comparative RT-PCR. The values shown below (expressed as 2μ(ΔCT); see “Patients and methods”) are from real-time RT-PCR experiments on samples from the same individuals. AML samples 1 to 3 represent cases with high level of BAALC transcripts. Samples 4 to 6 represent AML cases from the low BAALC group. M indicates size marker; R, reagent control.
BAALC mRNA expression in pretreatment AML samples as determined by comparative RT-PCR and real-time RT-PCR.The gel illustrates the comparative RT-PCR. The values shown below (expressed as 2μ(ΔCT); see “Patients and methods”) are from real-time RT-PCR experiments on samples from the same individuals. AML samples 1 to 3 represent cases with high level of BAALC transcripts. Samples 4 to 6 represent AML cases from the low BAALC group. M indicates size marker; R, reagent control.
Clinical and laboratory characteristics at presentation and treatment
There were no significant differences between high BAALC- and low BAALC-expressing patients with respect to age, sex, hemoglobin, platelet count, percentage of blasts in bone marrow or blood, race, FLT3 genotype, or other presenting physical findings such as gum hypertrophy, lymphadenopathy, splenomegaly, and hepatomegaly (Table 1). Low BAALC expression was associated with a significantly higher white blood count (WBC) (median, 31.6 × 109/L versus 13.8 × 109/L, P = .03), and M5 FAB morphology (24% versus 3%, P = .007).
Presenting characteristics by BAALC expression group
. | Low BAALC n = 43 . | High BAALC n = 43 . | P . |
|---|---|---|---|
| Age, y, median (range) | 48 (20-59) | 45 (18-59) | .21 |
| Sex, n (% males) | 24 (56) | 22 (51) | .83 |
| Hemoglobin level, g/dL, median (range) | 8.95 (4.7-12.8) | 8.7 (4.6-12.9) | .84 |
| Platelet count, ×109/L, median (range) | 50 (5-311) | 61 (7-378) | .71 |
| WBC, ×109/L, median (range) | 31.6 (1.4-295.0) | 13.8 (0.8-96.5) | .03 |
| Percent blood blasts, median (range) | 53 (0-97) | 46 (0-95) | .33 |
| Percent bone marrow blasts, median (range) | 58 (28-90) | 55 (26-90) | .46 |
| Race, n (%) | .10 | ||
| White | 35 (81) | 40 (93) | |
| Hispanic | 6 (14) | 1 (2) | |
| African American | 1 (2) | 1 (2) | |
| Oriental | 1 (2) | 0 (0) | |
| Other | 0 (0) | 1 (2) | |
| FAB classification, n (%) | .03 | ||
| M0 | 1 (2) | 2 (5) | |
| M1 | 7 (17) | 11 (28) | |
| M2 | 12 (29) | 16 (40) | |
| M4 | 12 (29) | 9 (23) | |
| M5 | 10 (24) | 1 (3) | |
| M6 | 0 (0) | 1 (3) | |
| Unknown | 1 | 3 | |
| Gum hypertrophy, n (%) | 10 (23) | 6 (14) | .41 |
| Lymphadenopathy, n (%) | 3 (7) | 5 (12) | .71 |
| Skin infiltrates, n (%) | 8 (19) | 3 (7) | .20 |
| Hepatomegaly, n (%) | 2 (5) | 2 (5) | .99 |
| Splenomegaly, n (%) | 1 (2) | 4 (9) | .36 |
| FLT3 genotype, n (%) | |||
| FLT3WT/WT | 35 (81) | 35 (81) | .99 |
| FLT3ITD/WT | 8 (19) | 8 (19) |
. | Low BAALC n = 43 . | High BAALC n = 43 . | P . |
|---|---|---|---|
| Age, y, median (range) | 48 (20-59) | 45 (18-59) | .21 |
| Sex, n (% males) | 24 (56) | 22 (51) | .83 |
| Hemoglobin level, g/dL, median (range) | 8.95 (4.7-12.8) | 8.7 (4.6-12.9) | .84 |
| Platelet count, ×109/L, median (range) | 50 (5-311) | 61 (7-378) | .71 |
| WBC, ×109/L, median (range) | 31.6 (1.4-295.0) | 13.8 (0.8-96.5) | .03 |
| Percent blood blasts, median (range) | 53 (0-97) | 46 (0-95) | .33 |
| Percent bone marrow blasts, median (range) | 58 (28-90) | 55 (26-90) | .46 |
| Race, n (%) | .10 | ||
| White | 35 (81) | 40 (93) | |
| Hispanic | 6 (14) | 1 (2) | |
| African American | 1 (2) | 1 (2) | |
| Oriental | 1 (2) | 0 (0) | |
| Other | 0 (0) | 1 (2) | |
| FAB classification, n (%) | .03 | ||
| M0 | 1 (2) | 2 (5) | |
| M1 | 7 (17) | 11 (28) | |
| M2 | 12 (29) | 16 (40) | |
| M4 | 12 (29) | 9 (23) | |
| M5 | 10 (24) | 1 (3) | |
| M6 | 0 (0) | 1 (3) | |
| Unknown | 1 | 3 | |
| Gum hypertrophy, n (%) | 10 (23) | 6 (14) | .41 |
| Lymphadenopathy, n (%) | 3 (7) | 5 (12) | .71 |
| Skin infiltrates, n (%) | 8 (19) | 3 (7) | .20 |
| Hepatomegaly, n (%) | 2 (5) | 2 (5) | .99 |
| Splenomegaly, n (%) | 1 (2) | 4 (9) | .36 |
| FLT3 genotype, n (%) | |||
| FLT3WT/WT | 35 (81) | 35 (81) | .99 |
| FLT3ITD/WT | 8 (19) | 8 (19) |
No significant differences were seen between high BAALC- and low BAALC-expressing patients in the type of induction, consolidation, or maintenance therapy received (Table 2). Fifty-three percent of patients in both groups were randomized to induction chemotherapy with PSC-833. Sixty-six percent of low BAALC expressers and 76% of high BAALC expressers received autologous PBSCT in first CR. Sixteen high and 17 low BAALC expressers received low-dose subcutaneous IL-2 maintenance therapy.
Treatment by BAALC expression group
. | Low BAALC n = 43 . | High BAALC n = 43 . | P . |
|---|---|---|---|
| Induction chemotherapy, n (%) | |||
| ADE* | 20 (47) | 20 (47) | |
| ADE* plus PSC-833 | 23 (53) | 23 (53) | .99 |
| Consolidation therapy | |||
| Auto PBSCT,† n (%) | 23 (66) | 25 (76) | .43 |
| Maintenance therapy | |||
| Subcutaneous IL-2,† n (%) | 16 (46) | 17 (52) | .81 |
| Salvage therapy | |||
| Primary refractory or relapsed patients, n | 18 | 27 | |
| Autologous PBSCT, n (%) | 1 (6) | 1 (4) | |
| Allogeneic SCT, n (%) | 3 (17) | 5 (19) | |
| No SCT outside of first CR, n (%) | 14 (78) | 21 (78) | .99 |
. | Low BAALC n = 43 . | High BAALC n = 43 . | P . |
|---|---|---|---|
| Induction chemotherapy, n (%) | |||
| ADE* | 20 (47) | 20 (47) | |
| ADE* plus PSC-833 | 23 (53) | 23 (53) | .99 |
| Consolidation therapy | |||
| Auto PBSCT,† n (%) | 23 (66) | 25 (76) | .43 |
| Maintenance therapy | |||
| Subcutaneous IL-2,† n (%) | 16 (46) | 17 (52) | .81 |
| Salvage therapy | |||
| Primary refractory or relapsed patients, n | 18 | 27 | |
| Autologous PBSCT, n (%) | 1 (6) | 1 (4) | |
| Allogeneic SCT, n (%) | 3 (17) | 5 (19) | |
| No SCT outside of first CR, n (%) | 14 (78) | 21 (78) | .99 |
SCT indicates stem cell transplantation.
ADE comprises cytarabine, daunorubicin, and etoposide.20
Percent among patients in CR.
Correlation between BAALC expression and clinical outcome
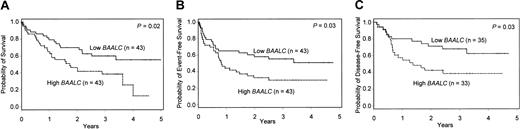
There was no significant difference in response to induction therapy between the 2 cohorts: 35 (81%) of low BAALC and 33 (77%) of high BAALC expressers achieved CR (P = .79, Table 3). AML patients with high BAALC expression tended to relapse more frequently than those with low BAALC expression (52% versus 29%, P = .08). High BAALC expression predicted significantly shorter OS (median, 1.7 versus 5.8 years, P = .02; Table 3). The probability of survival at 3 years was 39% in the high BAALC group versus 60% in the low BAALC group (Table 3; Figure 2A). Event-free survival also was significantly shorter in patients with high BAALC expression compared to those with low BAALC expression (median, 0.8 versus 4.9 years, P = .03; Figure 2B), as was DFS (median, 1.4 versus 7.3 years, P = .03; Figure 2C).
Impact of BAALC expression on clinical outcome
. | Low BAALC n = 43 . | High BAALC n = 43 . | P . |
|---|---|---|---|
| Complete remission, n (%) | 35 (81) | 33 (77) | .79 |
| Relapse rate, n (%) | 10 (29) | 17 (52) | .08 |
| Death in CR, n (%) | 2 (6) | 3 (9) | .67 |
| Overall survival | |||
| Median, y | 5.8* | 1.7 | |
| Percent alive at 3 y (95% CI) | 60 (45-75) | 39 (24-54) | .02 |
| Event-free survival | |||
| Median, y | 4.9* | 0.8 | |
| Percent event-free at 3 y (95% CI) | 55 (40-70) | 30 (16-43) | .03 |
| Disease-free survival | |||
| Median, y | 7.3* | 1.4 | |
| Percent disease-free at 3 y (95% CI) | 68 (52-84) | 39 (22-56) | .03 |
. | Low BAALC n = 43 . | High BAALC n = 43 . | P . |
|---|---|---|---|
| Complete remission, n (%) | 35 (81) | 33 (77) | .79 |
| Relapse rate, n (%) | 10 (29) | 17 (52) | .08 |
| Death in CR, n (%) | 2 (6) | 3 (9) | .67 |
| Overall survival | |||
| Median, y | 5.8* | 1.7 | |
| Percent alive at 3 y (95% CI) | 60 (45-75) | 39 (24-54) | .02 |
| Event-free survival | |||
| Median, y | 4.9* | 0.8 | |
| Percent event-free at 3 y (95% CI) | 55 (40-70) | 30 (16-43) | .03 |
| Disease-free survival | |||
| Median, y | 7.3* | 1.4 | |
| Percent disease-free at 3 y (95% CI) | 68 (52-84) | 39 (22-56) | .03 |
CI indicates confidence interval.
Nonparametric estimate of the median is not available. Tabled value represents the median, assuming the survival estimate after the last observation follows an exponential curve. The median follow-up for patients alive was 3.3 years (range, 1.6-5.0 years, n = 40).
Kaplan-Meier analysis of OS, EFS, and DFS for de novo AML patients with normal cytogenetics. AML patients with high BAALC expression show significantly inferior OS (A), EFS (B), and DFS (C) compared to low BAALC patients.
Kaplan-Meier analysis of OS, EFS, and DFS for de novo AML patients with normal cytogenetics. AML patients with high BAALC expression show significantly inferior OS (A), EFS (B), and DFS (C) compared to low BAALC patients.
We also looked at fitting a model based upon the 4 quartiles of BAALC expression; this resulted in 4 fairly small groups. If the first quartile was used as the reference group, the second quartile was similar in outcome, whereas the third quartile had a worse outcome, and the fourth quartile had an even worse outcome. Thus, we preferred to use the median, grouping the second and first quartiles and then grouping the third and fourth quartiles. However, there was strong evidence, using the test for trend,32 that a worse outcome was observed in at least one quartile of patients when compared to patients in the preceding quartile (OS and DFS, P < .01; EFS, P = .01), indicating that the higher the BAALC expression, the higher the likelihood of an event.
Multivariable analysis of BAALC expression for OS, EFS, and DFS
A multivariable analysis was conducted to determine if high BAALC expression was a significant independent prognostic factor for OS, EFS, and DFS once the model was adjusted for other characteristics. Variables considered for model inclusion were log-transformed WBC, platelets, hemoglobin, percentage of blasts in bone marrow and blood, age, FAB (M4/M5 versus others), sex, race (white versus other), treatment (ADE plus autologous PBSCT versus ADE/PSC-833 plus autologous PBSCT), FLT3 genotype, and BAALC expression. The variables included in the final models were BAALC (OS, EFS, DFS), log-transformed WBC (OS, EFS), and age (EFS). Controlling for all other covariates in the models, older patients (EFS) and those with a higher WBC (OS, EFS) had a worse prognosis. After adjusting for other covariates, BAALC expression remained a significant predictor for OS and EFS.
Patients with high BAALC expression were 2.7 (95% CI: 1.4-5.0) times more likely to die than patients with low BAALC expression. The hazard ratio of an event for the high BAALC group in EFS was 2.6 (95% CI: 1.4-4.7). BAALC expression was the only significant predictor for DFS. The hazard ratio for high BAALC patients to fail treatment after achieving CR, in the form of relapse or death, was 2.2 (95% CI: 1.1-4.5; Table 4).
Multivariable analysis of BAALC expression for overall survival, event-free survival, and disease-free survival
. | Overall survival . | . | Event-free survival . | . | Disease-free survival . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| Variable . | Hazard ratio (95% Cl) . | P . | Hazard ratio (95% Cl) . | P . | Hazard ratio (95% Cl) . | P . | |||
| High BAALC | 2.7 (1.4-5.0) | .002 | 2.6 (1.4-4.7) | .002 | 2.2 (1.1-4.5) | .03 | |||
. | Overall survival . | . | Event-free survival . | . | Disease-free survival . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| Variable . | Hazard ratio (95% Cl) . | P . | Hazard ratio (95% Cl) . | P . | Hazard ratio (95% Cl) . | P . | |||
| High BAALC | 2.7 (1.4-5.0) | .002 | 2.6 (1.4-4.7) | .002 | 2.2 (1.1-4.5) | .03 | |||
Hazard ratios and P values are given for high BAALC expression after controlling for WBC (OS, EFS) and age (EFS).
Discussion
In this well-defined cohort of uniformly treated AML patients with a normal karyotype, we have identified high BAALC expression in pretreatment blood samples as an independent adverse prognostic factor for OS, EFS, and DFS. Within this group of relatively young de novo AML patients who lack other important risk factors, such as adverse chromosomal aberrations and the ITD of FLT3 in the absence of a FLT3 WT allele, we were able to identify a new subset of patients with a relatively unfavorable outcome. Even in the setting of intensive chemotherapy and autologous transplantation, at 3 years only 39% of patients with high BAALC expression were alive and only 39% of those achieving CR remained in continuous CR. In contrast, 60% of low BAALC expressers were alive and 68% of those achieving CR remained continuously disease-free at 3 years. Indeed, high BAALC expression helped identify patients otherwise classified as “standard risk” who were more than twice as likely to fail to achieve long-term disease-free survival despite dose-intensive chemotherapy and autologous peripheral stem cell transplantation.
Chromosomal abnormalities provide a powerful tool to stratify AML patients into different prognostic risk groups. Patients lacking cytogenetic aberrations, accounting for approximately 45% of newly diagnosed de novo AML cases, are contained in an intermediate risk group.1-3 In reality this is a heterogeneous cohort of patients with either favorable, intermediate, or relatively poor clinical outcome.5 Little is known about underlying molecular mechanisms contributing to the clinical heterogeneity of AML with a normal karyotype. For these patients the identification of novel molecular markers is necessary to overcome the limitations of current risk assessment and to design new risk-adapted treatment strategies. Additional stratification could prevent overtreatment and undertreatment by discriminating patients who would benefit from more aggressive procedures including allogeneic stem cell transplantation. Furthermore, molecular markers may elucidate underlying mechanisms involved in leukemogenesis and may also serve as potential targets for new therapies.
Molecular studies have provided additional insights into prognostically relevant markers in cytogenetically normal AML patients.14-17 Most important have been mutations of the FLT3 gene.14,15 For the purpose of this study we excluded patients without a wild-type FLT3 allele, as we have already documented very poor outcome on this same CALGB treatment protocol 9621 for this small group of patients.14 We included patients with the more favorable FLT3WT/WT and FLT3ITD/WT genotypes and found that the 2 groups were equally frequent in the low and high BAALC expressers and did not differ in clinical outcome. High BAALC expression remained a significant adverse prognostic factor for FLT3ITD/WT and FLT3WT/WT patients, compared to those with low BAALC expression (OS, P = .002; hazard ratio [HR] = 2.7; DFS, P = .04, HR = 2.2), even when the 8 additional patients harboring a FLT3ITD/– genotype (4 showed high BAALC expression and 4 showed low BAALC expression) with normal cytogenetics and treated on CALGB 9621 were included in the multivariable analyses for OS and DFS. However, no difference in OS and DFS between the FLT3ITD/– patients with high BAALC expression compared to those with low BAALC expression was observed. High BAALC was an independent adverse prognostic factor for EFS regardless of the FLT3 status (P = .008, HR = 2.2). Thus, identification of high BAALC expression within AML patients harboring the more favorable FLT3 genotypes further identified AML patients with normal cytogenetics who are at higher risk to fail chemotherapy and autologous transplantation.
Future studies may facilitate the design of new molecular-based risk stratification for AML patients with normal cytogenetics. In this respect, determination of the BAALC status may be useful to predict long-term disease-free survival and to guide postremission therapy. Low levels of BAALC expression identified patients with a favorable outcome treated with chemotherapy and autologous stem cell transplantation. In contrast, high BAALC expression predicted a relatively poor outcome in this study of intensively treated patients. For these high-risk patients, more aggressive procedures including early allogeneic transplantation or alternative investigative therapies might be beneficial.
Several lines of evidence suggest that increased expression of BAALC identifies a distinct subset among the leukemic phenotypes. Whereas normal blood and bone marrow show very low levels of BAALC expression, high levels of BAALC transcript can be detected in hematopoietic progenitor cells as well as in leukemic blasts in some AML patients. Furthermore, BAALC expression can be detected in patients with ALL and CML in blast crisis, but BAALC transcripts are absent in chronic phase CML and in CLL.18 Given the fact that BAALC expression in normal bone marrow is restricted to the compartment of progenitor cells and that it shows high expression in a subset of leukemic blasts, BAALC may be seen as a stage-specific marker regulated during hematopoiesis and aberrantly expressed in leukemogenesis. The function of the BAALC protein in hematopoiesis and leukemogenesis contributing to a more aggressive behavior of AML has yet to be identified.
In summary, in AML patients with normal cytogenetics we have found that high-level expression of the novel gene BAALC identified patients less likely to achieve prolonged survival. Obviously, additional prospective studies, including ones using standard less-intensive treatment regimens, are needed to confirm and expand our results before BAALC expression can be used routinely as a potential marker for risk stratification in adult de novo AML. Furthermore, investigations are indicated to understand how the BAALC protein contributes to a more aggressive leukemic phenotype.
Appendix
The following Cancer and Leukemia Group B institutions, principal investigators, and cytogeneticists participated in this study:
Wake Forest University School of Medicine, Winston-Salem, NC: David D. Hurd, Mark J. Pettenati, and Wendy L. Flejter (grant no. CA03927); North Shore–Long Island Jewish Health System, Manhasset, NY: Daniel R. Budman and Prasad R. K. Koduru (grant no. CA35279); Duke University Medical Center, Durham, NC: Jeffrey Crawford and Mazin B. Qumsiyeh (grant no. CA47577); The Ohio State University Medical Center, Columbus, OH: Clara D. Bloomfield and Karl S. Theil (grant no. CA77658); Roswell Park Cancer Institute, Buffalo, NY: Ellis G. Levine and AnneMarie W. Block (grant no. CA02599); University of Massachusetts Medical Center, Worcester, MA: Mary Ellen Taplin and Vikram Jaswaney (grant no. CA37135); Vermont Cancer Center, Burlington, VT: Hyman B. Muss and Elizabeth F. Allen (grant no. CA77406); University of Iowa Hospitals, Iowa City, IA: Gerald H. Clamon and Shivanand R. Patil (grant no. CA47642); Washington University School of Medicine, St Louis, MO: Nancy L. Bartlett and Michael S. Watson (grant no. CA77440); Mount Sinai School of Medicine, New York, NY: Lewis R. Silverman and Vesna Najfeld (grant no. CA04457); University of Chicago Medical Center, Chicago, IL: Gini Fleming, Michelle M. LeBeau, and Diane Roulston (grant no. CA41287); Christiana Care Health Services, Inc., Newark, DE: Stephen S. Grubbs, Jeanne M. Meck, and Digamber S. Borgaonkar (grant no. CA45418); Dana-Farber Partners Cancer Care, Boston, MA: George P. Canellos, Paola Dal Cin, Cynthia C. Morton, Leonard L. Atkins, and Ramana Tantravahi (grant no. CA32291); Eastern Maine Medical Center CCOP, Bangor, ME: Philip L. Brooks and Laurent J. Beauregard (grant no. CA35406); Ft. Wayne Medical Oncology/Hematology, Ft. Wayne, IN: Sreenivasa Nattam and Patricia I. Bader; University of Illinois at Chicago: Thomas E. Lad and Maureen M. McCorquodale (grant no. CA74811); University of Missouri/Ellis Fischel Cancer Center, Columbia, MO: Michael C. Perry and Tim Huang (grant no. CA12046); University of North Carolina at Chapel Hill: Thomas C. Shea and Kathleen W. Rao (grant no. CA47559); University of Puerto Rico School of Medicine, San Juan, PR: Enrique Velez-Garcia, Paola Dal Cin, Cynthia C. Morton, and Leonard L. Atkins; Weill Medical College of Cornell University, New York, NY: Scott Wadler and Prasad R. K. Koduru (grant no. CA07968); University of California at San Diego: Stephen L. Seagren and Marie L. Dell'Aquila (grant no. CA11789); Southern Nevada Cancer Research Foundation, Las Vegas, NV: John Ellerton and Renée Bernstein (grant no. CA35421); University of Tennessee Memphis Cancer Center: Harvey B. Niell and Sugandhi A. Tharapel (grant no. CA47555); Walter Reed Army Medical Center, Washington, DC: Joseph J. Drabeck and Karl S. Theil (grant no. CA26806).
Prepublished online as Blood First Edition Paper, May 15, 2003; DOI 10.1182/blood-2003-02-0359.
Supported by National Cancer Institute grants CA16058, CA101140, CA31946, CA77658, CA03927, CA41287, and CA35279, and the Coleman Leukemia Research Fund. C. D. Baldus was supported by a grant from the Deutsche Krebshilfe. Additional grant support for participating CALGB institutions is listed in the “Appendix.”
C. D. Baldus and S.M.T. contributed equally to this work.
Participating institutions and principal investigators are listed in the “Appendix.”
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to John Byrd for critical reading of the manuscript.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal