Abstract
Pretreatment cytogenetics is a known predictor of outcome in hematologic malignancies. However, its usefulness in adult acute lymphoblastic leukemia (ALL) is generally limited to the presence of the Philadelphia (Ph) chromosome because of the low incidence of other recurrent abnormalities. We present centrally reviewed cytogenetic data from 1522 adult patients enrolled on the Medical Research Council (MRC) UKALLXII/Eastern Cooperative Oncology Group (ECOG) 2993 trial. The incidence and clinical associations for more than 20 specific chromosomal abnormalities are presented. Patients with a Ph chromosome, t(4;11)(q21;q23), t(8;14)(q24.1;q32), complex karyotype (5 or more chromosomal abnormalities), or low hypodiploidy/near triploidy (Ho-Tr) all had inferior rates of event-free and overall survival when compared with other patients. In contrast, patients with high hyperdiploidy or a del(9p) had a significantly improved outcome. Multivariate analysis demonstrated that the prognostic relevance of t(8;14), complex karyotype, and Ho-Tr was independent of sex, age, white cell count, and T-cell status among Ph-negative patients. The observation that Ho-Tr and, for the first time, karyotype complexity confer an increased risk of treatment failure demonstrates that cytogenetic subgroups other than the Ph chromosome can and should be used to risk stratify adults with ALL in future trials.
Introduction
Recurrent chromosomal abnormalities in the malignant cells of patients with acute leukemia are hallmarks of the disease.1 Specific aberrations, which are frequently indicative of consistent underlying molecular lesions, can assist or even establish the diagnosis and determine optimal therapy. In childhood acute lymphoblastic leukemia (ALL) numerous good and high-risk cytogenetic subgroups have been identified which are regularly used to stratify patients to particular therapies.2 However, in adult ALL the role of cytogenetics in patient management has largely been centered on the presence of the Philadelphia (Ph) chromosome which usually arises from t(9;22)(q34;q11.2) and results in BCR-ABL fusion.3 Although the overall incidence of Ph+ ALL in adults is approximately 25%, it is correlated with age and rises to greater than 50% among patients older than the age of 55 years.4 Even though other recurrent chromosomal abnormalities have been described in adult ALL, their frequency has been low and their prognostic relevance unclear. Indeed, some aberrations have been reported variously as good and poor risk by different study groups.5-10
One of the principal hurdles in developing a more sophisticated cytogenetic profile of adult ALL and assessing the utility of cytogenetics in predicting outcome is the rarity of this disease (less than 1 case per 100 000 person-years).11 This situation is exacerbated by the fact that apart from the Ph chromosome each of the other recurrent abnormalities accounts for less than 10% of the total. These 2 factors necessitate the acquisition of large cohorts of patients which have undergone prospective cytogenetic analysis. The majority of previously published studies have either concentrated solely on the presence of the Ph chromosome or comprised cohorts too small to accurately assess the outcome of rare cytogenetic subgroups. In this report we present cytogenetic results from a large multicenter international treatment trial of adult ALL: Medical Research Council (MRC) UKALLXII/Eastern Cooperative Oncology Group (ECOG) 2993.12-14 A total of 1522 patients has been registered on this trial, and pretreatment cytogenetic analysis was attempted in 90% of cases. The frequency, clinical features, and prognostic relevance of more than 20 cytogenetic subgroups are reported.
Patients, materials, and methods
Patients
The MRC UKALLXII/ECOG E2993 trial started recruiting patients diagnosed with ALL in January 1993.12-14 Patients between 15 and 55 (MRC) or 65 (ECOG) years of age were eligible for this study irrespective of their prognostic factors at presentation, including those with Ph+ or mature-B ALL. Clinical details, including age, white cell count (WCC), immunophenotype, and survival were collected by the Clinical Trial Service Unit (CTSU), University of Oxford, United Kingdom.
Full details of the protocol have been previously published.12-14 Briefly, patients received 2 phases of standard induction therapy. Patients with an HLA-matched family donor were assigned to receive an allogeneic bone marrow transplant. Ph+ patients could also receive an allogeneic transplant from a matched unrelated donor. All other patients were randomly assigned to standard consolidation/maintenance chemotherapy or to receive a single autologous transplant. Prior to the assigned or randomized therapy, all patients received intensification with 3 courses of high-dose methotrexate. The study was approved by the institutional review board of each treatment center and informed consent was given. The current study focuses on 1522 patients registered before the revision to incorporate imatinib for Ph+ patients (MRC, May 2003; ECOG, May 2004).
Cytogenetic and molecular genetic analysis
Pretreatment bone marrow (BM) or peripheral blood (PB) samples taken at diagnosis were cultured and analyzed by standard cytogenetic methods at local laboratories. Slides and/or karyograms were centrally reviewed either by the Leukaemia Research Cytogenetics Group (LRCG)15 or the ECOG Cytogenetics Subcommittee. All karyotypes were described according to the International System for Human Cytogenetic Nomenclature.16 Where possible, diagnostic samples were also tested for the presence of the BCR-ABL fusion gene by reverse-transcription polymerase chain reaction (RT-PCR) or fluorescence in situ hybridization (FISH). RT-PCR was performed using Invitrogen (Carlsbad, CA) reagents and protocols, following a TRIzol-based RNA extraction, using previously published primer sets (MRC17 ; ECOG18 ). FISH was performed using the LSI BCR-ABL Dual-Color Single-Fusion or Dual-Color Dual-Fusion probe (Abbot Diagnostics/Vysis, Des Plaines, IL).
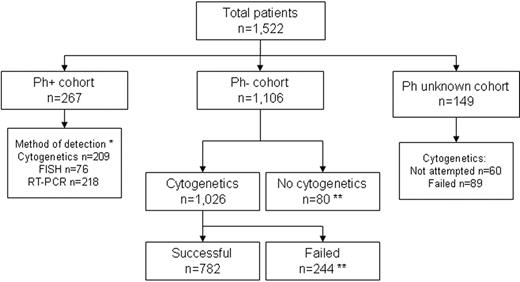
Patients were classified according to the presence of the following chromosomal abnormalities: (1) t(9;22)(q34;q11.2)/BCR-ABL fusion (Ph+); (2) t(4;11)(q21;q23); (3) other MLL/11q23 translocations; (4) t(1;19)(q21;p13.3); (5) t(8;14)(q24.1;q32)/t(8;22)(q24.1;q11.2)/t(2;8)(p12;q24.1); (6) t(10;14)(q24;q11.2); (7) other T-cell receptor translocations (TCRs)—that is, those involving 14q11.2, 7p14∼15, 7q34∼36; (8) 14q32 translocations [t14q32]; (9) 6q deletions [del(6q)]; (10) 7p deletions [del(7p)]; (11) monosomy 7 (−7); (12) trisomy 8 (+8); (13) gain of chromosome X (+X); (14) 9p deletions [del(9p)]; (15) 11q abnormalities [abn(11q)]; (16) 12p deletions [del(12p)]; (17) monosomy 13/13q deletions [del(13q)]; (18) 17p deletions [del(17p)]; (19) complex karyotype (5 or more chromosomal abnormalities) excluding those patients with an established translocation; (20) low hypodiploidy (30-39 chromosomes)/near triploidy (60-78 chromosomes) (Ho-Tr)19,20 ; (21) high hyperdiploidy (51-65 chromosomes) (HeH), karyotypes with 60 to 65 chromosomes were examined individually and classified as HeH or Ho-Tr according to whether the pattern of chromosomal gain was closest to the classical description of HeH21 or Ho-Tr19 ; (22) tetraploidy (80 or more chromosomes) (Tt); (23) other abnormality (any abnormal karyotype without any of the abnormalities listed earlier); and (24) normal karyotype (20 or more normal metaphases observed from a bone marrow sample in the absence of a clonal abnormality). Patients were classified into all relevant subgroups. Cytogenetic analysis was considered unacceptable if fewer than 20 normal metaphases were analyzed in the absence of an abnormal clone. In addition, 3 cohorts of patients were defined according to Ph status (Figure 1). The first comprised Ph+ patients, identified by cytogenetics, FISH, or RT-PCR. The second comprised patients where the presence of the Ph chromosome had been excluded on the basis of a successful cytogenetic result or a negative BCR-ABL result by FISH or RT-PCR. Finally, the third cohort comprised patients with an indeterminable Ph status (ie, no cytogenetic, FISH, or RT-PCR result).
Philadelphia chromosome status and outcome of cytogenetic analysis among 1522 patients registered on MRC UKALLXII/ECOG 2993. *Some cases were tested by more than one technique; **BCR-ABL negative by RT-PCR, FISH, or both.
Philadelphia chromosome status and outcome of cytogenetic analysis among 1522 patients registered on MRC UKALLXII/ECOG 2993. *Some cases were tested by more than one technique; **BCR-ABL negative by RT-PCR, FISH, or both.
Statistical analysis
The relationships between cytogenetic subgroups and sex and T-cell status were analyzed using the chi-square test, whereas the t test was used to compare continuous variables such as age and WCC [log(WCC + 1)]. The main survival analyses of event-free survival (EFS) and overall survival (OS) were defined as time from diagnosis to the first adverse event, whether relapse or death (including patients who failed to achieve a complete remission and those who died in remission) or death. Patients who did not have an event or died within the follow-up period were censored at the date of last contact or October 31, 2005, if earlier. The median follow-up time for survivors was 7 years (MRC) or 5 years (ECOG). For the whole cohort, the 5-year EFS and OS were 34% (95% confidence interval [CI], 31%-36%) and 38% (95% CI, 35%-40%), respectively. There was no difference in EFS or OS between MRC and ECOG patients. Ph+ and Ph− patients were considered separately in all analyses, whereas those in the Ph unknown cohort were excluded. Kaplan-Meier life tables and curves were constructed by means of the log-rank method.22 All EFS and OS rates are quoted at 5 years. The observed-to-expected (O/E) ratios presented are from unadjusted log-rank tests comparing 2 groups. Multivariate analysis was performed using the Cox proportional hazards model.23 Cytogenetic variables, T-cell status, and sex were included in the model as Boolean categorical variables, whereas age and WCC [log(WCC + 1)] were treated as continuous variables. Models were fitted using stepwise forward selection with variables added to the model if P was less than .01 but removed if P was greater than .05. All statistical analysis was performed using Intercooled Stata v8.2 for Windows (Stata, College Station, TX).
Results
Conventional cytogenetic analysis
Cytogenetic analysis was attempted in 1366 (90%) of 1522 patients: 501 (90%) of 557 on ECOG and 865 (90%) of 965 on MRC. This rate did not vary by year of diagnosis or patient age. Cytogenetic analysis was successful in 1003 cases (73%) but was considered unacceptable in 363 cases (27%). Among those patients with successful cytogenetics an abnormal clone was detected in 796 (79%). There was no trend toward a higher rate of unacceptable cytogenetics in the early part of the trial compared with the later years. Among MRC patients the rate of unacceptable cytogenetics was significantly higher in PB samples (27 of 85 [32%]) compared with BM samples (156 of 772 [20%]) (P < .02).
Patients with Philadelphia chromosome/t(9;22)(q34;q11.2)/BCR-ABL
A total of 267 Ph+ patients (19%) were detected among 1373 that were tested by conventional cytogenetics, FISH, RT-PCR, or a combination of these methods (Figure 1). The incidence of Ph+ was significantly lower among MRC patients (142 of 872 [16%]) compared with ECOG patients (125 of 501 [25%]) (P < .001). Overall, the incidence of Ph+ increased with patient age, 15 to 19 years (12 of 267 [4%]), 20 to 29 years (53 of 375 [14%]), 30 to 39 years (68 of 288 [24%]), 40 to 49 years (88 of 269 [33%]), and 50 years and older (46 of 174 [26%]).
Among 209 Ph+ patients with abnormal cytogenetics an additional aberration was observed in 141 patients (67%) (Table 1). The most frequent additional anomaly was gain of a Ph chromosome [+der(22)t(9;22)] in 49 cases (23%). Monosomy 7, +8, +X, del(9p), and HeH occurred in 10% to 15% of cases (Table 1). The minor BCR breakpoint was more prevalent, occurring in 101 (70%) of 143 patients, than the major breakpoint, which was present in 43 (30%) of 143 patients. A single patient showed the presence of both breakpoints.
Cytogenetic distribution of patients with adult acute lymphoblastic leukemia on MRC UKALLXII/ECOG 2993
| Cytogenetic subgroup* . | t(9;22) . | t(4;11) . | t(11q23) . | t(1;19) . | t(8;14) . | t(10;14) . | TCR . | t(14q32) . | del(6q) . | del(7p) . | −7 . | +8 . | +X . | del(9p) . | abn(11q) . | del(12p) . | del(13q) . | del(17p) . | Complex . | Ho-Tr . | HeH . | Tt . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| t(9;22)(q24;q11.2)† | 209 | |||||||||||||||||||||
| t(4;11)(q21;q23) | 0 | 54 | ||||||||||||||||||||
| Other MLL/11q23 translocations | 0 | 0 | 15 | |||||||||||||||||||
| t(1;19)(q21;p13.3) | 0 | 0 | 0 | 24 | ||||||||||||||||||
| t(8;14)(q24.1;q32) | 0 | 0 | 0 | 0 | 16 | |||||||||||||||||
| t(10;14)(q24;q11.2) | 0 | 0 | 0 | 0 | 0 | 16 | ||||||||||||||||
| Other TCR translocations | 0 | 0 | 0 | 0 | 0 | 0 | 18 | |||||||||||||||
| 14q32 translocations | 4 | 0 | 0 | 0 | 3 | 0 | 2 | 49 | ||||||||||||||
| del(6q) | 6 | 0 | 1 | 4 | 0 | 0 | 3 | 8 | 61 | |||||||||||||
| del(7p) | 7 | 3 | 1 | 2 | 0 | 0 | 0 | 2 | 0 | 30 | ||||||||||||
| −7‡ | 31 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 2 | 62 | |||||||||||
| +8‡ | 24 | 1 | 1 | 2 | 0 | 1 | 1 | 5 | 5 | 3 | 6 | 85 | ||||||||||
| +X‡ | 23 | 10 | 1 | 0 | 2 | 0 | 0 | 8 | 8 | 3 | 3 | 35 | 129 | |||||||||
| del(9p) | 24 | 0 | 0 | 4 | 0 | 5 | 1 | 5 | 4 | 11 | 7 | 7 | 0 | 95 | ||||||||
| Abnormality of 11q | 7 | 0 | 1 | 0 | 1 | 0 | 1 | 3 | 4 | 1 | 3 | 1 | 4 | 4 | 36 | |||||||
| del(12p) | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 3 | 4 | 2 | 1 | 0 | 1 | 1 | 1 | 30 | ||||||
| del(13q)/−13 | 5 | 0 | 2 | 2 | 2 | 0 | 1 | 5 | 4 | 1 | 10 | 4 | 6 | 7 | 2 | 3 | 45 | |||||
| del(17p) | 3 | 1 | 0 | 0 | 2 | 0 | 0 | 3 | 4 | 0 | 10 | 9 | 11 | 3 | 3 | 2 | 16 | 43 | ||||
| Complex karyotype | NA | NA | NA | NA | NA | NA | NA | NA | 10 | 1 | 5 | 3 | 4 | 8 | 7 | 7 | 5 | 6 | 41 | |||
| Low hypodiploidy/near triploidy | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 9 | 18 | 16 | 3 | 0 | 0 | 12 | 12 | 0 | 31 | ||
| High hyperdiploidy | 29 | 1 | 1 | 0 | 2 | 0 | 0 | 6 | 5 | 2 | 3 | 31 | 77 | 0 | 1 | 1 | 3 | 7 | 0 | 0 | 106 | |
| Tetraploidy | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 2 | 2 | 0 | 1 | 3 | 2 | 2 | 3 | 0 | 0 | 0 | 17 |
| Cytogenetic subgroup* . | t(9;22) . | t(4;11) . | t(11q23) . | t(1;19) . | t(8;14) . | t(10;14) . | TCR . | t(14q32) . | del(6q) . | del(7p) . | −7 . | +8 . | +X . | del(9p) . | abn(11q) . | del(12p) . | del(13q) . | del(17p) . | Complex . | Ho-Tr . | HeH . | Tt . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| t(9;22)(q24;q11.2)† | 209 | |||||||||||||||||||||
| t(4;11)(q21;q23) | 0 | 54 | ||||||||||||||||||||
| Other MLL/11q23 translocations | 0 | 0 | 15 | |||||||||||||||||||
| t(1;19)(q21;p13.3) | 0 | 0 | 0 | 24 | ||||||||||||||||||
| t(8;14)(q24.1;q32) | 0 | 0 | 0 | 0 | 16 | |||||||||||||||||
| t(10;14)(q24;q11.2) | 0 | 0 | 0 | 0 | 0 | 16 | ||||||||||||||||
| Other TCR translocations | 0 | 0 | 0 | 0 | 0 | 0 | 18 | |||||||||||||||
| 14q32 translocations | 4 | 0 | 0 | 0 | 3 | 0 | 2 | 49 | ||||||||||||||
| del(6q) | 6 | 0 | 1 | 4 | 0 | 0 | 3 | 8 | 61 | |||||||||||||
| del(7p) | 7 | 3 | 1 | 2 | 0 | 0 | 0 | 2 | 0 | 30 | ||||||||||||
| −7‡ | 31 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 2 | 62 | |||||||||||
| +8‡ | 24 | 1 | 1 | 2 | 0 | 1 | 1 | 5 | 5 | 3 | 6 | 85 | ||||||||||
| +X‡ | 23 | 10 | 1 | 0 | 2 | 0 | 0 | 8 | 8 | 3 | 3 | 35 | 129 | |||||||||
| del(9p) | 24 | 0 | 0 | 4 | 0 | 5 | 1 | 5 | 4 | 11 | 7 | 7 | 0 | 95 | ||||||||
| Abnormality of 11q | 7 | 0 | 1 | 0 | 1 | 0 | 1 | 3 | 4 | 1 | 3 | 1 | 4 | 4 | 36 | |||||||
| del(12p) | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 3 | 4 | 2 | 1 | 0 | 1 | 1 | 1 | 30 | ||||||
| del(13q)/−13 | 5 | 0 | 2 | 2 | 2 | 0 | 1 | 5 | 4 | 1 | 10 | 4 | 6 | 7 | 2 | 3 | 45 | |||||
| del(17p) | 3 | 1 | 0 | 0 | 2 | 0 | 0 | 3 | 4 | 0 | 10 | 9 | 11 | 3 | 3 | 2 | 16 | 43 | ||||
| Complex karyotype | NA | NA | NA | NA | NA | NA | NA | NA | 10 | 1 | 5 | 3 | 4 | 8 | 7 | 7 | 5 | 6 | 41 | |||
| Low hypodiploidy/near triploidy | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 9 | 18 | 16 | 3 | 0 | 0 | 12 | 12 | 0 | 31 | ||
| High hyperdiploidy | 29 | 1 | 1 | 0 | 2 | 0 | 0 | 6 | 5 | 2 | 3 | 31 | 77 | 0 | 1 | 1 | 3 | 7 | 0 | 0 | 106 | |
| Tetraploidy | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 2 | 2 | 0 | 1 | 3 | 2 | 2 | 3 | 0 | 0 | 0 | 17 |
NA indicates not applicable.
See “Patients, materials, and methods” for a full definition of the individual cytogenetic subgroups.
Only cases with a cytogenetically visible t(9;22)(q34;q11.2) are included.
Gain or loss of whole chromosomes from diploid or tetraploid.
Compared with Ph− patients, Ph+ patients were significantly older (mean age, 38 years versus 31 years, P < .01), had a higher WCC (mean, 58 versus 49 × 109/L, P < .01), and a lower incidence of T-cell ALL (< 1% versus 22%, P < .01). Ph+ patients had significantly inferior 5-year EFS and OS compared with Ph− patients: EFS, 16% (95% CI, 12%-21%) versus 36% (95% CI, 33%-39%); OS, 22% (95% CI, 17%-27%) versus 41% (95% CI, 38%-44%) (both P < .001, adjusting for age, sex, and WCC). There was no survival difference by BCR breakpoint status or the presence of an extra Ph, −7, +8, or del(9p). Ph+ patients with a HeH karyotype had a higher EFS (31% [95% CI, 16%-48%] versus 15% [95% CI, 10%-21%], P = .04) and OS at 5 years (37% [95% CI, 20%-54%] versus 19% [95% CI, 13%-25%], P = .09). However, the effect was not independent of age or WCC and disappeared when patients who received a transplant were removed from the analysis.
Patients with t(4;11)(q21;q23) and other MLL/11q23 translocations
A total of 69 Ph− patients (9%) had a translocation involving the MLL gene located at 11q23. The majority (n = 54) had t(4;11) (Table 1), whereas the remainder had other translocations, including 6 with t(11;19)(q23;p13.3). All patients with an 11q23 translocation had a significantly higher WCC, but only those with a t(4;11) showed a female predominance or were older (Table 2). Although none of the patients with t(4;11) had T-ALL, 40% of patients with other 11q23 translocations were classified as T-ALL. Patients with t(4;11) had significantly inferior EFS and OS compared with other Ph− patients (Table 3; Figure 2). Six patients with t(4;11) (11%) failed to achieve a CR. Each of the 20 patients with t(4;11) who relapsed had isolated BM relapses and each one subsequently died. The majority (17 of 20) of these patients relapsed within 1 year of diagnosis with a median time to relapse of 6.5 months. A further 16 patients with t(4;11) died without relapsing; again mostly (12 of 16) within 1 year of diagnosis. Among the 12 patients with t(4;11) in continuing complete remission (CCR) 7 have, however, survived more than 5 years. Patients with other MLL/11q23 translocations did not have a significantly inferior outcome compared with other Ph− patients (Table 3).
Overall survival by cytogenetic subgroup of patients registered on MRC UKALLXII/ECOG 2993.
Overall survival by cytogenetic subgroup of patients registered on MRC UKALLXII/ECOG 2993.
Clinical features by cytogenetic subgroup of patients with Philadelphia-negative adult acute lymphoblastic leukemia on MRC UKALLXII/ECOG 2993
| Cytogenetic subgroup* . | No. of cases (%) . | Male cases, %† . | Mean age, y‡ . | Mean WCC, × 109/L‡ . | T-cell cases, %† . |
|---|---|---|---|---|---|
| Total§ | 782 (100) | 62 | 31 | 53 | 22 |
| t(4;11)(q21;q23) | 54 (7) | 33‖ | 38‖ | 234‖ | 0¶ |
| Other MLL/11q23 translocations | 15 (2) | 67 | 34 | 128‖ | 40 |
| t(1;19)(q21;p13.3) | 24 (3) | 58 | 31 | 46 | 0‖ |
| t(8;14)(q24.1;q32) | 16 (2) | 69 | 39‖ | 33 | 0‖ |
| t(10;14)(q24;q11.2) | 16 (2) | 63 | 35 | 65 | 100¶ |
| Other TCR translocations | 18 (2) | 89¶ | 27 | 97‖ | 100¶ |
| 14q32 translocations | 45 (6) | 58 | 33 | 50 | 12 |
| del(6q) | 55 (7) | 64 | 28‖ | 63‖ | 35‖ |
| del(7p) | 23 (3) | 70 | 30 | 49 | 17 |
| −7# | 19 (2) | 68 | 41‖ | 12‖ | 22 |
| +8# | 23 (3) | 57 | 30 | 44 | 25 |
| +X# | 34 (4) | 56 | 35¶ | 70¶ | 3‖ |
| del(9p) | 71 (9) | 65 | 29¶ | 37 | 16 |
| abnormality of 11q | 29 (4) | 72 | 33 | 70 | 34 |
| del(12p) | 29 (4) | 62 | 30 | 38 | 19 |
| del(13q)/−13 | 40 (5) | 63 | 30 | 46 | 19 |
| del(17p) | 40 (5) | 58 | 31 | 33 | 10 |
| Complex karyotype | 41 (5) | 73 | 33 | 40 | 25 |
| Low hypodiploidy/near triploidy | 31 (4) | 52 | 34 | 17‖ | 3¶ |
| High hyperdiploidy | 77 (10) | 60 | 27‖ | 15‖ | 4¶ |
| Tetraploidy | 15 (2) | 67 | 33 | 11¶ | 20 |
| Other abnormality** | 102 (13) | 72¶ | 30 | 45 | 24 |
| Normal karyotype | 195 (25) | 62 | 31 | 34‖ | 23 |
| Cytogenetic subgroup* . | No. of cases (%) . | Male cases, %† . | Mean age, y‡ . | Mean WCC, × 109/L‡ . | T-cell cases, %† . |
|---|---|---|---|---|---|
| Total§ | 782 (100) | 62 | 31 | 53 | 22 |
| t(4;11)(q21;q23) | 54 (7) | 33‖ | 38‖ | 234‖ | 0¶ |
| Other MLL/11q23 translocations | 15 (2) | 67 | 34 | 128‖ | 40 |
| t(1;19)(q21;p13.3) | 24 (3) | 58 | 31 | 46 | 0‖ |
| t(8;14)(q24.1;q32) | 16 (2) | 69 | 39‖ | 33 | 0‖ |
| t(10;14)(q24;q11.2) | 16 (2) | 63 | 35 | 65 | 100¶ |
| Other TCR translocations | 18 (2) | 89¶ | 27 | 97‖ | 100¶ |
| 14q32 translocations | 45 (6) | 58 | 33 | 50 | 12 |
| del(6q) | 55 (7) | 64 | 28‖ | 63‖ | 35‖ |
| del(7p) | 23 (3) | 70 | 30 | 49 | 17 |
| −7# | 19 (2) | 68 | 41‖ | 12‖ | 22 |
| +8# | 23 (3) | 57 | 30 | 44 | 25 |
| +X# | 34 (4) | 56 | 35¶ | 70¶ | 3‖ |
| del(9p) | 71 (9) | 65 | 29¶ | 37 | 16 |
| abnormality of 11q | 29 (4) | 72 | 33 | 70 | 34 |
| del(12p) | 29 (4) | 62 | 30 | 38 | 19 |
| del(13q)/−13 | 40 (5) | 63 | 30 | 46 | 19 |
| del(17p) | 40 (5) | 58 | 31 | 33 | 10 |
| Complex karyotype | 41 (5) | 73 | 33 | 40 | 25 |
| Low hypodiploidy/near triploidy | 31 (4) | 52 | 34 | 17‖ | 3¶ |
| High hyperdiploidy | 77 (10) | 60 | 27‖ | 15‖ | 4¶ |
| Tetraploidy | 15 (2) | 67 | 33 | 11¶ | 20 |
| Other abnormality** | 102 (13) | 72¶ | 30 | 45 | 24 |
| Normal karyotype | 195 (25) | 62 | 31 | 34‖ | 23 |
See “Patients, materials, and methods” for a full definition of the individual cytogenetic subgroups.
The percentage of male and T-cell cases were compared with all other Philadelphia-negative cases with a cytogenetic result using a chi-square test.
The age (y) and log(WCC + 1) of patients was compared with all other Philadelphia-negative cases with a cytogenetic result using a t test.
Philadelphia-negative cases with a successful cytogenetic result.
P < .05.
P < .01.
These groups exclude cases with low hypodiploid/near triploidy, high hyperdiploidy, and tetraploidy.
Abnormal karyotypes excluding those with any of the above abnormalities.
Event-free and overall survival by cytogenetic subgroup of patients with Philadelphia-negative adult acute lymphoblastic leukemia on MRC UKALLXII/ECOG 2993
| Cytogenetic subgroup* . | Event-free survival . | Overall survival . | ||||
|---|---|---|---|---|---|---|
| 5 y, no. (95% CI) . | O/E† . | P† . | 5 y, no. (95% CI) . | O/E† . | P† . | |
| Total‡ | 38 (34-41) | 1.02 | .365 | 42 (38-45) | 1.02 | .458 |
| t(4;11)(q21;q23) | 24 (13-36) | 1.70 | < .001 | 24 (13-36) | 1.86 | < .001 |
| Other MLL/11q23 translocations | 29 (9-52) | 1.28 | .432 | 33 (12-56) | 1.26 | .462 |
| t(1;19)(q21;p13.3) | 29 (13-48) | 1.31 | .254 | 32 (14-51) | 1.26 | .342 |
| t(8;14)(q24.1;q32) | 13 (2-33) | 3.15 | < .001 | 13 (2-33) | 3.22 | < .001 |
| t(10;14)(q24;q11.2) | 34 (13-58) | 0.91 | .758 | 41 (17-64) | 0.86 | .652 |
| Other TCR translocations | 33 (14-55) | 1.24 | .458 | 33 (14-55) | 1.39 | .252 |
| 14q32 translocations | 33 (20-47) | 1.21 | .277 | 35 (22-49) | 1.18 | .366 |
| del(6q) | 29 (18-41) | 1.31 | .072 | 36 (23-48) | 1.26 | .145 |
| del(7p) | 22 (8-40) | 1.27 | .306 | 26 (11-45) | 1.41 | .137 |
| −7§ | 36 (16-57) | 1.15 | .617 | 36 (16-57) | 1.32 | .328 |
| +8§ | 22 (8-40) | 1.45 | .108 | 22 (8-40) | 1.50 | .078 |
| +X§ | 24 (11-40) | 1.22 | .300 | 27 (13-44) | 1.38 | .096 |
| del(9p) | 49 (37-60) | 0.73 | .043 | 58 (46-69) | 0.70 | .032 |
| Abnormality of 11q | 40 (22-57) | 0.99 | .964 | 48 (29-65) | 0.96 | .858 |
| del(12p) | 34 (18-51) | 1.05 | .844 | 41 (24-58) | 0.98 | .933 |
| del(13q)/−13 | 32 (19-47) | 1.09 | .650 | 41 (26-56) | 1.07 | .731 |
| del(17p) | 32 (18-46) | 1.08 | .681 | 36 (21-51) | 1.12 | .547 |
| Complex karyotype | 21 (10-35) | 1.57 | .009 | 28 (15-43) | 1.48 | .027 |
| Low hypodiploidy/near triploidy | 18 (7-34) | 1.78 | .003 | 22 (9-38) | 1.86 | .001 |
| High hyperdiploidy | 50 (38-60) | 0.67 | .007 | 53 (41-64) | 0.69 | .015 |
| Tetraploidy | 46 (20-68) | 0.71 | .335 | 65 (35-84) | 0.58 | .170 |
| Other abnormality‖ | 38 (28-48) | 0.96 | .727 | 39 (29-49) | 0.96 | .731 |
| Normal karyotype | 43 (36-50) | 0.86 | .064 | 48 (40-55) | 0.83 | .025 |
| Cytogenetic subgroup* . | Event-free survival . | Overall survival . | ||||
|---|---|---|---|---|---|---|
| 5 y, no. (95% CI) . | O/E† . | P† . | 5 y, no. (95% CI) . | O/E† . | P† . | |
| Total‡ | 38 (34-41) | 1.02 | .365 | 42 (38-45) | 1.02 | .458 |
| t(4;11)(q21;q23) | 24 (13-36) | 1.70 | < .001 | 24 (13-36) | 1.86 | < .001 |
| Other MLL/11q23 translocations | 29 (9-52) | 1.28 | .432 | 33 (12-56) | 1.26 | .462 |
| t(1;19)(q21;p13.3) | 29 (13-48) | 1.31 | .254 | 32 (14-51) | 1.26 | .342 |
| t(8;14)(q24.1;q32) | 13 (2-33) | 3.15 | < .001 | 13 (2-33) | 3.22 | < .001 |
| t(10;14)(q24;q11.2) | 34 (13-58) | 0.91 | .758 | 41 (17-64) | 0.86 | .652 |
| Other TCR translocations | 33 (14-55) | 1.24 | .458 | 33 (14-55) | 1.39 | .252 |
| 14q32 translocations | 33 (20-47) | 1.21 | .277 | 35 (22-49) | 1.18 | .366 |
| del(6q) | 29 (18-41) | 1.31 | .072 | 36 (23-48) | 1.26 | .145 |
| del(7p) | 22 (8-40) | 1.27 | .306 | 26 (11-45) | 1.41 | .137 |
| −7§ | 36 (16-57) | 1.15 | .617 | 36 (16-57) | 1.32 | .328 |
| +8§ | 22 (8-40) | 1.45 | .108 | 22 (8-40) | 1.50 | .078 |
| +X§ | 24 (11-40) | 1.22 | .300 | 27 (13-44) | 1.38 | .096 |
| del(9p) | 49 (37-60) | 0.73 | .043 | 58 (46-69) | 0.70 | .032 |
| Abnormality of 11q | 40 (22-57) | 0.99 | .964 | 48 (29-65) | 0.96 | .858 |
| del(12p) | 34 (18-51) | 1.05 | .844 | 41 (24-58) | 0.98 | .933 |
| del(13q)/−13 | 32 (19-47) | 1.09 | .650 | 41 (26-56) | 1.07 | .731 |
| del(17p) | 32 (18-46) | 1.08 | .681 | 36 (21-51) | 1.12 | .547 |
| Complex karyotype | 21 (10-35) | 1.57 | .009 | 28 (15-43) | 1.48 | .027 |
| Low hypodiploidy/near triploidy | 18 (7-34) | 1.78 | .003 | 22 (9-38) | 1.86 | .001 |
| High hyperdiploidy | 50 (38-60) | 0.67 | .007 | 53 (41-64) | 0.69 | .015 |
| Tetraploidy | 46 (20-68) | 0.71 | .335 | 65 (35-84) | 0.58 | .170 |
| Other abnormality‖ | 38 (28-48) | 0.96 | .727 | 39 (29-49) | 0.96 | .731 |
| Normal karyotype | 43 (36-50) | 0.86 | .064 | 48 (40-55) | 0.83 | .025 |
O/E indicates observed-to-expected events ratio from the log-rank test.
See “Patients, materials, and methods” for a full definition of the individual cytogenetic subgroups.
Observed-to-expected ratios and P value derived from the log-rank test comparing the outcome of each cytogenetic group to all other Philadelphia-negative patients with a successful cytogenetic result, except for the total group which was compared with patients with a failed cytogenetic result or without cytogenetics.
Philadelphia-negative cases with a successful cytogenetic result.
These groups exclude cases with low hypodiploid/near triploidy, high hyperdiploidy, and tetraploidy.
Abnormal karyotypes, excluding those with any of the above abnormalities.
Patients with t(8;14)(q24.1;q32)
Only 16 patients with t(8;14) or variant of this translocation were registered on this trial, accounting for 2% of the Ph− cohort. Although the majority (10 of 16 [63%]) had the classic t(8;14), 5 had t(8;22)(q24.1;q11) and 1 had t(2;8)(p12;q24.1). Eight of these patients had a mature-B immunophenotype, whereas the remaining patients were classified as either common (n = 5) or pre-B (n = 3). The outcome of this small subgroup was extremely poor compared with other Ph− patients (Table 3; Figure 2). A total of 14 (88%) have died, 10 following a relapse. All but one of the deaths occurred in the first year after diagnosis.
Patients with low hypodiploidy/near triploidy
The 2 ploidy subgroups, low hypodiploidy and near triploidy, were combined into a single subgroup (Ho-Tr) because patients with these karyotypes have been shown to represent a single distinct subtype of adult ALL.19 No patients with a near-haploid clone (< 30 chromosomes) were detected in this cohort. Within the Ph− cohort, 31 patients (4%) had a Ho-Tr karyotype and none harbored any established translocation (Table 1). Cytogenetic analysis revealed the presence of both subclones in 6 patients, whereas in the remaining 25 patients only the low-hypodiploid (n = 6) or near-triploid (n = 19) subclone was detected. Patients in this subgroup had a significantly lower WCC compared with other Ph− patients and were less likely to have T-ALL (Table 2). Patients with Ho-Tr had significantly inferior EFS and OS compared with other Ph− patients (Table 3). Five patients (16%) failed to achieve remission. Among patients that did achieve a CR, 13 (50%) of 26 relapsed and a further 7 (27%) died in remission within 1 year of diagnosis. The majority of relapses involved an isolated BM relapse (12 of 13). The median time to relapse was 9 months and was followed by death in every case. The 5-year OS for Ho-Tr patients is 22% and only 6 (19%) are still in CCR (Table 3). There was no difference in outcome between those that presented with a low-hypodiploid subclone and those that presented solely with a near-triploid subclone.
Patients with a complex karyotype
In the Ph− cohort, a total of 41 patients (5%), without an established translocation, had a complex karyotype with 5 or more chromosomal abnormalities (Table 1). Although patients in this subgroup were not associated with any particular sex, age, WCC, or T-cell status (Table 2), they had a significantly inferior EFS and OS (Table 3). The majority (32 of 41 [78%]) of these patients had an adverse event. Four patients (10%) failed to achieve a CR, and among those that did, 19 (51%) relapsed. Most of these patients (17 of 19) had an isolated BM relapse with the other 2 having an isolated CNS or combined relapse. The majority of relapses (16 of 19) occurred within 2 years of diagnosis and was invariably (17 of 19) followed by death. A further 9 patients with complex karyotypes who achieved CR died in remission (8 in the first year).
Patients with high hyperdiploidy
HeH was the most prevalent specific chromosomal abnormality in the Ph− cohort occurring in 77 patients (10%), occasionally in conjunction with established translocations (Table 1). HeH patients were significantly younger and had a lower WCC and incidence of T-cell disease compared with other Ph− patients (Table 2). They had significantly improved EFS and OS compared with other patients in this cohort (Table 3). Virtually all HeH patients who relapsed subsequently died (19 of 21) but their relapse rate at 5 years was significantly lower than other Ph− patients (34% [95% CI, 24%-48%] versus 50% [95% CI, 45%-54%], P = .007). The median time to relapse among HeH patients was 1.4 years compared with less than 1 year for non-HeH patients.
Patients with deleted 9p
Deletions of 9p were observed in 71 Ph− patients (9%), were the second most prevalent abnormality, and were frequently a secondary aberration (Table 1). Patients with del(9p) were marginally younger than other Ph− patients and had an improved EFS (Tables 2-3). They were significantly less likely to die (OS at 5 years, 58% [95% CI, 45%-69%] versus 40% [95% CI, 36%-44%], P = .03) compared with other Ph− patients.
Patients with other chromosomal abnormalities
In addition to the chromosomal abnormalities already discussed we examined the demographic, clinical, and survival profiles of Ph− patients with 16 other specific cytogenetic abnormalities (Tables 1,Table 2-3). Although several subgroups showed distinct sex, age, WCC, or T-cell status profiles, none of these cytogenetic subgroups showed any significant association with disease outcome.
Multivariate analysis
We used a Cox proportional hazards model to assess the prognostic relevance of cytogenetic variables within the Ph− cohort in the context of other established survival indicators: sex, age, WCC, and T-cell status (Table 4). Age, WCC, and T-cell status were strong predictors of outcome for both EFS and OS, but sex was only relevant with respect to OS. Females were more likely to have died compared with males (64% versus 56%, P = .022); however, this is, in part, explained because females in this cohort were significantly older than their male counterparts (33 years versus 30 years; P < .001). The adverse effect of t(8;14), Ho-Tr, and complex karyotype was shown to be independent of age, sex, WCC, and T-cell status (Table 4). This was true of both EFS and OS. Patients with t(8;14) had more than a 2-fold increased risk of having an adverse event, whereas those with a Ho-Tr or complex karyotype were approximately 80% and approximately 70% more likely to relapse or die, respectively. Both good-risk cytogenetic variables, HeH and del(9p), failed to retain their significance in multivariate analysis, suggesting that factors such as age and WCC are more important.
Final Cox regression models for the risk of relapse or death and the risk of death among patients with adult acute lymphoblastic leukemia on MRC UKALLXII/ECOG 2993
| Variable* . | Hazard ratio (95% CI) . | SE . | P . |
|---|---|---|---|
| Risk of relapse or death | |||
| Age | 1.02 (1.01-1.03) | 0.004 | < .001 |
| WCC | 1.20 (1.12-1.28) | 0.04 | < .001 |
| T-cell status | 0.71 (0.56-0.89) | 0.09 | .004 |
| t(8;14)(q24.1;q32) | 2.55 (1.48-4.38) | 0.70 | .001 |
| Low hypodiploidy/near triploidy | 1.90 (1.26-2.86) | 0.40 | .002 |
| Complex karyotype | 1.75 (1.22-2.52) | 0.32 | .002 |
| Risk of death | |||
| Age | 1.03 (1.02-1.03) | 0.004 | < .001 |
| WCC | 1.20 (1.12-1.28) | 0.04 | < .001 |
| T-cell status | 0.75 (0.59-0.96) | 0.09 | .020 |
| Male sex | 0.81 (0.67-0.98) | 0.08 | .027 |
| t(8;14)(q24.1;q32) | 2.73 (1.58-4.72) | 0.76 | < .001 |
| Low hypodiploidy/near triploidy | 1.90 (1.26-2.86) | 0.40 | .002 |
| Complex karyotype | 1.70 (1.17-2.47) | 0.32 | .005 |
| Variable* . | Hazard ratio (95% CI) . | SE . | P . |
|---|---|---|---|
| Risk of relapse or death | |||
| Age | 1.02 (1.01-1.03) | 0.004 | < .001 |
| WCC | 1.20 (1.12-1.28) | 0.04 | < .001 |
| T-cell status | 0.71 (0.56-0.89) | 0.09 | .004 |
| t(8;14)(q24.1;q32) | 2.55 (1.48-4.38) | 0.70 | .001 |
| Low hypodiploidy/near triploidy | 1.90 (1.26-2.86) | 0.40 | .002 |
| Complex karyotype | 1.75 (1.22-2.52) | 0.32 | .002 |
| Risk of death | |||
| Age | 1.03 (1.02-1.03) | 0.004 | < .001 |
| WCC | 1.20 (1.12-1.28) | 0.04 | < .001 |
| T-cell status | 0.75 (0.59-0.96) | 0.09 | .020 |
| Male sex | 0.81 (0.67-0.98) | 0.08 | .027 |
| t(8;14)(q24.1;q32) | 2.73 (1.58-4.72) | 0.76 | < .001 |
| Low hypodiploidy/near triploidy | 1.90 (1.26-2.86) | 0.40 | .002 |
| Complex karyotype | 1.70 (1.17-2.47) | 0.32 | .005 |
All variables were entered to the model as binary categorical variables except age and WCC [log(WCC + 1)], which were used as continuous variables.
Although patients with t(4;11) had a very poor outcome, they were older and had a higher WCC (Table 2); hence, t(4;11) did not remain statistically significant within an overall multivariate model. Stratified log-rank tests showed that among patients with a low WCC (< 100 × 109/L) those with t(4;11) had a worse OS at 5 years compared with other Ph− patients (13% [95% CI, 2%-33%] versus 44% [95% CI, 40%-48%], respectively; O/E = 2.09; P = .004). Similarly, among younger patients (< 35 years) those with t(4;11) had an inferior OS at 5 years (35% [95% CI, 16%-55%] versus 49% [95% CI, 45%-54%], respectively; O/E = 1.70; P = .04).
Ph− patients in this study were either biologically selected or randomly assigned to receive a BM transplant as part of their treatment, and cytogenetics was not used to assign therapy. To account for treatment heterogeneity, we rebuilt the final EFS and OS models, excluding patients who received a transplant in first CR (n = 289) from the EFS model and all patients who received a transplant (n = 359) from the OS model. These exclusions made no difference to the prognostic relevance of the 3 significant cytogenetic variables: t(8;14), Ho-Tr, and complex karyotype.
Discussion
The results of this study provide compelling evidence for the importance of cytogenetics in predicting prognosis in adult ALL. Ph+ and Ph− patients were analyzed separately with respect to survival because the presence of the Ph chromosome was used to direct therapy. This approach meant that the prognostic relevance of cytogenetic subgroups could be ascertained accurately without their effect being masked by the strong and established poor outcome associated with Ph+ ALL. Univariate analysis showed that within the Ph− cohort patients with t(4;11), t(8;14), complex karyotype, and Ho-Tr had an inferior outcome, whereas those with HeH and del(9p) fared better. Moreover, the 3 cytogenetic variables, t(8;14), complex karyotype, and Ho-Tr, were shown to be independent prognostic factors. Because this was a randomized trial, patients were either biologically selected or randomly assigned to receive a BM transplant.12 Therefore, the multivariate analysis (Table 4) is valid despite treatment heterogeneity. The large number of patients available for analysis enabled us to exclude from these models patients who received a transplant. The observation that the exclusion of these patients did not alter the prognostic relevance of 3 cytogenetic subgroups confirms that transplantation-related mortality was not biasing our results. Too few patients in the prognostically relevant cytogenetic subgroups underwent a transplantation for us to evaluate whether an allogenic bone marrow transplantation (BMT) would alter the outcome of these high-risk patients. The rarity of these cytogenetic subgroups means that a meta-analysis is needed to clarify the effect of transplantation.
Patients with t(4;11) in this study, like previous studies,5-7,10 had a shorter survival, but the effect was not independent of age and WCC. However, both age and WCC are strongly correlated with this abnormality. Although based on relatively small numbers of patients, additional analyses showed that patients with t(4;11) with a low WCC still had a poor outcome, whereas younger patients with t(4;11) probably fared worse than their age-matched counterparts. This analysis highlights the difficulty of confirming the independent prognostic importance of relatively small cytogenetics subgroups which are strongly correlated with other risk factors such as WCC and age. Our evidence coupled with results from previous studies5-7,10 indicates that all patients with t(4;11) have an inferior outcome. This conclusion is consistent with the view that cytogenetics, which is a fundamental biological marker of leukemogenesis, is likely to emerge as a more important indicator of poor prognosis than are surrogate markers such as WCC.
A total of 16 patients with t(8;14) or a variant were treated on the early part of this trial. Interestingly, only half of these patients had a mature-B immunophenotype, and among patients with common/pre-B ALL the involvement of the MYC gene had been confirmed in 2 patients (data not shown). This is not the first time that the t(8;14) has been reported outside the context of mature-B ALL.24 The poor outcome of this subgroup reflects the fact that patients with t(8;14)/mature-B ALL are now usually treated on lymphoma-like protocols.25,26 There were too few survivors from this cohort to ascertain whether t(8;14) with or without mature-B ALL had a differential survival.
Charrin et al19 reported that patients with low hypodiploidy and near triploidy represent 2 sides of a distinct biological subgroup. Therefore, we incorporated this novel grouping into our classification system and showed that, like the LALA94 patients, our patients with Ho-Tr had a significantly reduced EFS and OS. However, we were also able to demonstrate that this adverse effect was independent of age and WCC. These observations are consistent with previous studies which have examined the prognosis of these ploidy subgroups separately.5,6 These patients had no other adverse features, such as high WCC or increasing age; hence, cytogenetics represents the only method of identifying them.
This is the first study to classify patients with ALL according to karyotype complexity. The definition of karyotype complexity in this study used 5 or more chromosomal abnormalities; this is the same criteria used by the MRC for patients with acute myeloid leukemia (AML).27 However, we excluded patients with an established translocation because these patients belonged to a distinct cytogenetic subgroup. This strategy is consistent with the most recent and comprehensive definition of a complex karyotype in AML.28 We report for the first time that patients with ALL and a complex karyotype have a poor prognosis, both in terms of EFS and OS, which is independent of age and WCC. Like patients with Ho-Tr, those with a complex karyotype did not have distinctive age or WCC profiles; hence, cytogenetics remains the sole detection method.
Only 2 cytogenetic subgroups, HeH and del(9p), were associated with an improved EFS or OS, although the del(9p) association was marginal. However, these 2 subgroups did occur in approximately 10% of Ph− patients, albeit occasionally in conjunction with other abnormalities. Both these subgroups were associated with good-risk features (low WCC or younger age), and neither subgroup retained its significance in multivariate analysis. HeH is common in childhood ALL and is associated with a very good outcome.21 Therefore, it is not surprising that adults with HeH should be younger than the rest of the cohort and have a better survival. In contrast, del(9p) in childhood ALL is associated with a poor outcome; however, it is more prevalent among patients with T-ALL who in turn have a poor outcome.29,30 Good-risk cytogenetic abnormalities have rarely been reported in adult ALL, but, where these 2 subgroups have been studied before, the trend has been toward an improved outcome.5-7,10
The incidence of Ph+ ALL was higher among ECOG patients compared with MRC patients. This is explained, in part, by the facts that ECOG patients were significantly older than their MRC counterparts (mean age, 36 versus 31 years, P < .01) and that its incidence increased with age. The proportion of Ph+ patients in previous studies ranges from 11% to 29%.5-10 In keeping with our data, those studies with a low incidence of Ph+ ALL also showed a lower median age and vice versa. As expected, Ph+ patients had significantly inferior EFS and OS compared with Ph− patients. This effect was independent of age and WCC, both of which are significantly higher among Ph+ patients. In contrast to the CALGB study31 we did not find heterogeneity of outcome among Ph+ patients according to the presence of −7 or an extra Ph. There are differences between the 2 cohorts and in the methods of analysis. In the CALGB study the association of −7 with a poor outcome was restricted to a reduced CR rate and limited to patients with −7 as the sole additional abnormality, which meant the result was based on just 9 patients. In addition, the median age of the CALGB cohort was 46 years compared with 39 years in ours. The results from our study are strengthened by being based on a larger cohort all treated on a single protocol.
Over the past decade there have been only 4 large trial-based cytogenetic studies of adult ALL,5-7,10 although 2 other studies have incorporated limited cytogenetic data.8,9 The paucity of studies is due to the rarity of the disease and the assumption that cytogenetic indicators of prognosis are limited to the Ph+ and possibly t(4;11). The prognostic relevance of other cytogenetic subgroups has been uncertain, with the same abnormality being reported as both good and poor risk in different studies.5-7,10 The scale of this trial coupled with prospective central karyotype review contributed to the creation of the largest and most comprehensive cytogenetic dataset of adult ALL. The absolute number of cases with cytogenetics exceeds that accrued by similar studies, whereas the proportion of cases that met the cytogenetic entry criteria compares favorably.5-7,10 Despite this, 27% cases did not meet our entry requirements, and, although this figure is comparable with other studies,5-7,10 it is too high and greater than seen in childhood ALL. Interestingly, the proportion of cases meeting the entry criteria did not increase over the length of the study, in contrast to the improving results over the some period in childhood ALL. Data from MRC patients strongly indicated that BM samples are preferable to blood samples. The abnormality rate within this cohort was nearly 80%, which again compares favorably with previous studies which have reported rates of 66% to 85%.5-7,10 Routine FISH screening of future cases with probes to genes known to be involved in ALL should help to increase the usefulness of cytogenetics in adult ALL as similar strategies have done in childhood ALL.32
The cytogenetic classification system presented identified a wide spectrum of specific structural and numerical chromosomal abnormalities in this disease. We did not restrict the classification to the expected common abnormalities included in most studies but also grouped karyotypes according to the presence of 14q32 translocations, del(7p), del(13q)/−13, Ho-Tr, and complex karyotype. The classification used in this study and the large number of patients accrued allowed the prognostic relevance of numerous other abnormalities to be examined with a higher degree of accuracy than previously possible.5-7,10,33
We did not find a significant association between t(1;l9), 11q23 translocations other than t(4;11), +8, −7, del(6q), and prognosis as previously reported.5-7,10,33 Although we approached our analysis differently by considering Ph+ and Ph− patients separately, this should not have affected our findings. The majority of aberrations listed earlier were negatively associated with outcome; hence. the removal of poor-risk patients from the analysis should have served to accentuate these differences rather than diminish them. Both the GFCH5 and MRC UKALLXA6 studies found that the prognosis of patients with 11q23 translocations other than t(4;11) was also poor. In this study these patients did not have a significantly inferior survival. The prognostic relevance of t(1;19) is ambiguous with the GFCH and GIMEMA studies5,10 reporting it as a poor-risk abnormality, whereas the MRC UKALLXA study6 found it to be associated with a favorable outcome. However, these studies were based on just 11, 7, and 10 patients, respectively. In our cohort, the outcome of 28 patients with t(1;19) was not significantly different from other Ph− patients.
Monosomy 7 has been reported to be associated with shorter survival whether it occurs with or without a Ph chromosome.7 However, in our analysis Ph− patients with −7 fared equally as well as other Ph− patients. Similarly, patients with +8 have been reported to have an inferior outcome by CALGB,7 and, although our patients with +8 did have a slightly lower EFS and OS, the difference was not statistically significant. Mancini et al33 reported that patients with a del(6q) were associated with a T-cell phenotype and inferior prognosis. Although we confirmed the association between del(6q) and T-cell ALL, we did not observe a similar decrease in survival.
In conclusion, this large cohort of adult ALL with high-quality cytogenetic data demonstrates the value of cytogenetics for identifying patients at greatest and least risk of treatment failure. Future randomized clinical trials of adult ALL can and should use cytogenetic data to stratify patients into appropriate risk groups so that they may receive the most effective therapy. Additional cytogenetic and molecular genetic studies of adult ALL are urgently required to further characterize this disease, thereby increasing the number of patients that can benefit from alternative treatment strategies.
Authorship
Contribution: A.V.M. designed and performed the study, analyzed the data, and wrote the manuscript. G.W.D., C.J.H., and L.M.S.-W. directed and designed the overall cytogenetic study and reviewed the cytogenetic data. M.M., G.H.V., A.M.C., and R.R.H. reviewed the cytogenetic data. S.M.R. and G.A.N.B. were the trial statisticians and collected the clinical and survival data. L.F. and E.P. directed the molecular analyses. A.H.G., J.M.R., M.R.L., M.S.T., P.H.W., and A.K.F. designed the treatment protocol and coordinated the clinical trial. All authors critically reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A full list of the members of the Medical Research Council (MRC) UKALLXII/Eastern Cooperative Oncology Group (ECOG) 2993 trial appears as Document S1, available on the Blood website; see the Supplemental Materials link at the top of the online article.
Correspondence: Anthony V. Moorman, Leukaemia Research Cytogenetics Group, Cancer Sciences Division, University of Southampton, MP 822, Duthie Building, Southampton General Hospital, Southampton, SO16 6YD, United Kingdom; e-mail: avm@soton.ac.uk.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
A.V.M., C.J.H., M.M., and L.M.S.-W. thank the laboratories of the UK Cancer Cytogenetics Group (UKCCG) for providing cytogenetic data and the past and present staff members of the Leukaemia Research Cytogenetics Group for collecting and reviewing the cytogenetic data. G.W.D., G.H.V., A.M.C., and R.R.H. thank the ECOG cytogenetic laboratories for providing cytogenetic data. We thank the doctors who participated in the MRC UKALLXII/ECOG 2993 trial.
This work was supported by Leukaemia Research and the Medical Research Council.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal